Abstract
The bcl-2 proto-oncogene is overexpressed in a variety of human cancers and plays an important role in programmed cell death. Recent reports implied that the 3′-untranslated region (3′UTR) functions effectively in the regulation of gene expression. Here, we attempt to assay the ability of triplex forming oligonucleotides (TFOs) to inhibit expression of a target gene in vivo and to examine the potential of the 3′UTR of the bcl-2 proto-oncogene in the regulation of bcl-2 gene expression. To do this, we have developed a novel cellular system that involves transfection of a Doxycyclin inducible expression plasmid containing the bcl-2 ORF and the 3′UTR together with a TFO targeted to the 3′UTR of the bcl-2 proto-oncogene. Phosphorothioate-modified TFO targeted to the 3′UTR of the bcl-2 gene significantly downregulated the expression of the bcl-2 gene in HeLa cells as demonstrated by western blotting. Our results indicate that blocking the functions of the 3′UTR using the TFO can downregulate the expression of the targeted gene, and suggest that triplex strategy is a promising approach for oligonucleotide-based gene therapy. In addition, triplex-based sequence targeting may provide a useful tool for studying the regulation of gene expression.
INTRODUCTION
Triplex forming oligonucleotides (TFOs) have attracted a great deal of attention because of their ability to specifically bind double-stranded DNA and their potential use in gene therapy (1). Purine and pyrimidine oligonucleotides targeted to purine–pyrimidine-rich sequences form pur*pur.pyr and pyr*pur.pyr intermolecular triple helices. The oligonucleotide third strand occupies the major groove of the duplex, forming Hoogsteen hydrogen bonds with the purine bases of the duplex. Both pur*pur.pyr and mixed pur/pyr*pur.pyr triplexes can be formed at physiological pH with predominantly G*G.C triplets along with A*A.T and T*A.T triplets (2–5). Many groups have attempted to take advantage of this high degree of binding specificity in the development of ‘antigene’ TFOs to block the expression of clinically important genes (6–13). A TFO can prevent the formation of the transcription initiation complex by binding to a polypurine–polypyrimidine tract in a gene promoter; another mechanism may result from inhibiting transcription elongation (10,11,13).
TFOs have been shown to inhibit both transcription in vitro and the expression of target genes in cell culture by binding to a polypurine–polypyrimidine tract in several human gene promoters and in chromatin (6–17). There is evidence to suggest that the 3′-untranslated region (3′UTR) plays an important role in the regulation of gene expression. However, it is difficult to study the functions of the 3′UTR because it is not under the same rigid structural constraints as the 5′UTR, which has to accommodate transcriptional machinery (18–20). Triplex strategy may provide a useful approach to investigate the role of 3′UTR in the regulation of gene expression.
The Bcl-2 gene product is a 25 kDa membrane protein that functions to prevent cell apoptosis (21,22) and plays a central role in inducing resistance to radiotherapy and chemotherapy (23). This gene was first discovered by its involvement in t(14;18) translocation, commonly found in human follicular lymphoma (24). Deregulated expression of bcl-2 results from the translocation of bcl-2 to the immunoglobulin heavy chain (IgH) locus (25,26). The bcl-2 proto-oncogene has three exons with an untranslated first exon. The major transcriptional promoter, P1, is located 1386–1423 bp upstream of the translation start site. Trancripts initiating at a minor promoter, P2, located 1.3 kb downstream from P1, have been identified in some tissues (27,28). The expression of bcl-2 is regulated at both transcriptional and post-transcriptional levels (29–34). In this study, a triplex strategy was used to investigate the functions of the 3′UTR as a potential additional regulatory site of bcl-2 expression. Our results showed that the expression of bcl-2 is apparently downregulated by blocking the 3′UTR functions with TFO, suggesting that the 3′UTR of bcl-2 may represent a further target structure for modulating the expression of the bcl-2 gene. Furthermore, downregulating the anti-apoptotic Bcl-2 protein by this approach might open a new avenue for anticancer therapy.
MATERIALS AND METHODS
Oligonucleotide design
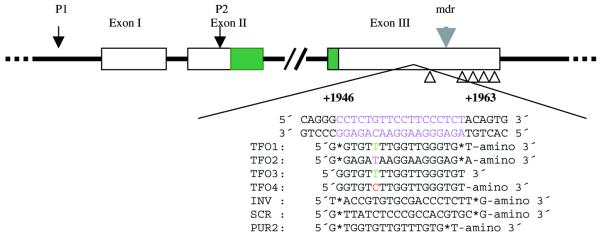
The human bcl-2 3′UTR contains an 18 bp purine–pyrimidine-rich sequence located +1946 to +1963 from the translation start site. The sequence of this region is not strictly homopurine–homopyrimidine, but contains one C.G interruption. The bcl-2 3′UTR targeted TFO, TFO1, which is a GT motif, was designed using guanine to recognize GC (G*GC triplets), thymidine to recognize AT (T*AT triplets) and thymidine along with the interruption CG (T*CG triplet). GA motif TFO2 was designed using guanine to recognize GC (G*GC triplets), adenine to recognize AT (A*AT triplets) and thymidine along with the interruption CG (T*CG triplet). TFO3 and 4 were almost the same as TFO1, except that TFO3 was not phosphorothioated at its 5′- and 3′-ends and amino-linked at its 3′-end, and TFO4 was designed using cytosine to recognize interruption CG instead of thymidine. TFO1–4 targeted to the 3′UTR of the bcl-2 gene and control oligonucleotides (INV, SCR and PUR2) were commercially obtained (Interactiva Biotech). All oligonucleotides (ODNs), except for TFO3 and 4, were phosphorothioated at their 5′- and 3′-ends and amino-linked at their 3′-ends to prevent rapid degradation. For fluorescence microscopy, ODNs were additionally labeled with FITC. All ODNs were purified by polyacrylamide gel electrophoresis and HPLC. The TFO target site within the 3′UTR of the bcl-2 gene and the sequence of oligonucleotides used in this study are shown schematically in Figure 1.
Figure 1.
Schematic diagram of the human bcl-2 proto-oncogene showing the 18 bp purine–pyrimidine-rich motif at the 3′UTR, the triplex target sequence relative to the translation start site (+1946 to +1963) and the TFOs and control oligonucleotides used in this study. Except for TFO3 and 4, all ODNs are phosphorothioated at their 5′- and 3′-ends and amino-linked at their 3′-ends. TFO3 is a pure phosphodiester ODN without any modifications and TFO4 is only amino-linked at its 3′-end.
Electrophoretic mobility shift assay (EMSA)
Double-stranded oligonucleotides were prepared by heating equal amounts of complementary single strands at 75°C for 15 min and cooling slowly to room temperature. Duplex oligonucleotides were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase and purified through a Sephadex G25 column. Approximately 1 pmol of duplex oligonucleotides was incubated with increasing concentrations of specific TFOs in 10 µl of binding buffer (TFO-BB; containing 50 mM MOPS pH 7.2, 50 mM NaCl, 10 mM MgCl2) and incubated at room temperature for 2 h. Products were analyzed on a 20% non-denaturing polyacrylamide gel. To stabilize triplexes, the running buffer was the same as TFO-BB.
Plasmid construction
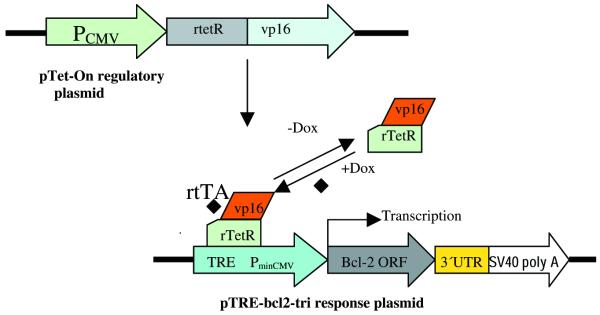
The 3′UTR of the bcl-2 gene containing the TFO binding site from +1760 to +2328 (569 bp) was amplified by reverse transcriptase-polymerase chain reaction (RT–PCR) and cloned into EcoRI and XbaI sites of vector pSPT18 to get plasmid pSPT-tri. The sequence of this cloned 3′UTR was confirmed by sequencing. Plasmid pSPT-bcl-2 containing the ORF of the bcl-2 proto-oncogene was linearized with EcoRI by partial digestion and ligated to a 3′UTR fragment cut from the EcoRI–XbaI digested plasmid pSPT-tri. After being cut with XbaI, the 4500 bp fragment was self-ligated to give the plasmid pSPT-bcl2-tri. The 1500 bp fragment containing bcl-2 ORF and 3′UTR was cut away from pSPT-bcl-2-tri and subcloned into vector pTRE (Clontech), the resulting plasmid was pTRE-bcl2-tri (Fig. 2).
Figure 2.
Schematic diagram of Tet-On expression system. The 569 bp of the 3′UTR containing the TFO binding site is located between the bcl-2 ORF and SV40 polyA of the response plasmid pTRE-bcl2-tri. The bcl-2 gene is under the control of the promoter PhCMV-1, which consists of TRE, and Pmin CMV. HeLa cells that have been stably transfected with regulatory plasmid pTet-On constitutively express rtTA which is a fusion of the wild-type Tet repressor (TetR) to the VP16 AD of herpes simplex virus. When HeLa cells are transfected with pTRE-bcl2-tri, rtTA binds to the TRE and activates transcription in the presence of Doxycyclin.
Cell line
The HeLa human cervical carcinoma cell line was obtained from Clontech. This is a Tet-On cell line, which is stably transfected with plasmid pTet-On (35). It was grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 100 µg/ml G418. Cells were cultured at 37°C with 85% humidity and 5% CO2.
Triplex formation in plasmid
32P-labeled TFO2 (1.2 pmol) was mixed with different concentrations of supercoiled plasmid pTRE-bcl2-tri in TFO-BB (containing 50 mM MOPS pH 7.2, 50 mM NaAc and 10 mM MgAc2) and incubated at room temperature for 6 h. Samples were analyzed in a 2% agarose gel using TAE buffer containing 3 mM MgAc2 as running buffer for 20 min at 80 V.
Transient transfection
Three micrograms of plasmid pTRE-bcl2-tri containing the 3′UTR sequence were incubated with 4.6 µg phosphorothioate oligonucleotide in TFO-BB at room temperature for 2 h, then diluted in 500 µl serum-free DMEM. At the same time, 12 µg of liposome (Dosper, Roche) was added to 500 µl serum-free DMEM. The DNA and liposome solutions were mixed gently and incubated at room temperature for 30 min. The day before transfection, 1 × 106 cells were plated in each well of 6-well plates. Immediately prior to transfection, cells were washed twice with Ca2+ and Mg2+ free sterile PBS. Following a 30 min incubation, the 1 ml liposome–DNA complexes were added to each well (35 mm). The plates were incubated for 6 h at 37°C, then 1 ml of DMEM containing 20% FCS and 10 µg/ml Doxycyclin was added. After 24, 48 and 72 h incubation, cells were trypsinized, washed with PBS and lysed in 200 µl cell lysis buffer (containing 150 mM NaCl, 50 mM Tris–HCl, 2 mM EDTA and 1% NP-40). To protect the protein from proteases, a tablet of protease inhibitor (Boehringer Mannheim) was dissolved in 10 ml lysis buffer. Protein concentration was determined using the Bradford method (Bio-Rad).
Western blot analysis
Total proteins (25 µg per lane) were resolved on a 10% SDS–polyacrylamide gel at 180 V for 60 min and transferred onto nylon membrane (Amersham) using semi-dry transferring (Bio-Rad) at 100 mA for 60 min. Thereafter, the membrane was blocked with 1% BSA in TBS for 30 min, Bcl-2 and actin proteins were detected using a 1:500 dilution of mouse monoclonal anti-Bcl-2 antibody (Dako) and a 1:5000 dilution of mouse monoclonal anti-actin antibody (Sigma). HRP-conjugated anti-mouse IgG secondary antibody (Jackson ImmunoResearch Laboratories) was used at a 1:10 000 dilution, and was visualized using the enhanced chemiluminescence (ECL) system (Pierce).
RESULTS
Triplex formation with the 3′UTR target
Triplex formation was demonstrated by EMSA (Fig. 4). Because of its decreased charge density, triplex DNA migrates more slowly than duplex DNA in gel mobility shift assay. The results indicated that TFO2 formed an extraordinarily stable triplex, with a Kd of ∼60 µM at physiological pH 7.2. TFO1, 3 and 4 also formed stable triplexes with Kds similar to that of TFO2 (data not shown). Phosphorothioated oligonucleotides are more resistant to degradation by nucleases and therefore provide advantages over phosphodiester TFOs for in vivo application. In our studies, we used TFOs phosphorothioated at both the 5′- and 3′-ends, and amino-linked at the 3′-end (Fig. 1). This phosphorothioated TFO is stable in cells at least 48 h after transfection as shown in experiments of cellular uptake of FITC-labeled TFO in HeLa cells (Fig. 3). Triplex formation in supercoiled plasmid was also confirmed by incubating TFO2 with different concentrations of supercoiled plasmid pTRE-bcl2-tri (Fig. 5). Furthermore, specific binding of TFO to the target was checked by a luciferase reporter gene system, and we found no unspecific binding of TFOs (TFO1–4) to non-target plasmid DNA (data not shown).
Figure 4.
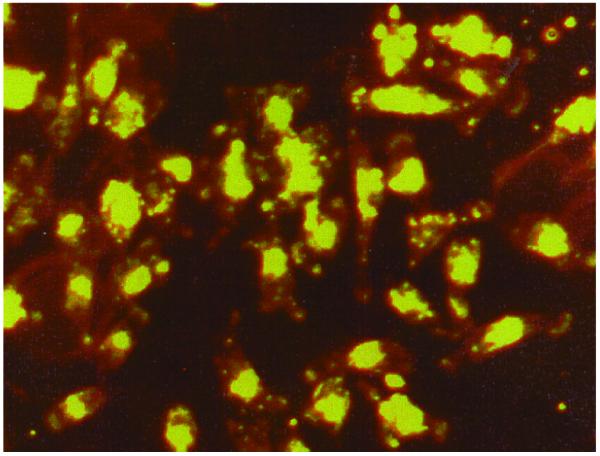
Cellular uptake of FITC-labeled ODN in HeLa cells 24 h after transfection. FITC-labeled TFO2 was transfected into HeLa cells with the help of liposome (Dosper). At different time points post-transfection cells were detected under fluorescence microscope.
Figure 3.
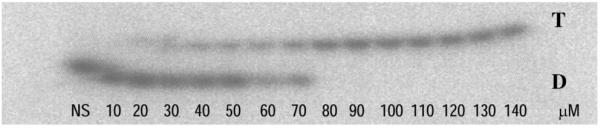
EMSA showing triplex formation of phosphorothioate ODN with a 3′UTR duplex (29 bp). The reaction was carried out in TFO-BB (see Materials and Methods); the duplex DNA concentration was 1 µM. TFO2 concentrations in each reaction are shown below the corresponding lane. NS, non-specific ODN; T, triplex; D, duplex.
Figure 5.
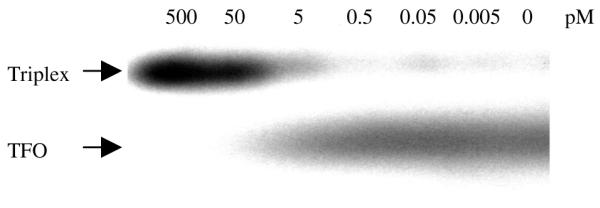
Triplex formation in supercoiled plasmid. 32P-TFO2 (1.2 pmol) and different concentrations of supercoiled plasmid pTRE-bcl2-tri (∼0–500 pM) were incubated at room temperature for 6 h. Arrows indicate positions of triplex and TFO.
Effect of triplex on bcl-2 gene expression
Transient transfection experiments were done to evaluate the effects of phosphorothioate TFOs on the expression of the bcl-2 gene in the HeLa cell line. By using FITC-labeled ODNs, we could estimate a transfection efficiency of >90% in HeLa cells (Fig. 3). We used a Tet-On inducible expression system (Fig. 2) (35). The 569 bp of the 3′UTR containing a TFO binding site is located between the bcl-2 ORF and the SV40 polyA tail of the response plasmid pTRE-bcl2-tri. Bcl-2 is under the control of the promoter PhCMV-1, which consists of a Tetracyclin response element (TRE), and Pmin CMV. HeLa cells that have been stably transfected with regulatory plasmid pTet-On constitutively express Tet-controlled transcriptional activator (rtTA) which is a fusion of the wild-type Tet repressor (TetR) to the VP16 activation domain (AD) of the herpes simplex virus. When HeLa cells are transfected with pTRE-bcl2-tri, rtTA binds to the TRE and activates transcription in the presence of Doxycyclin. When TFO binds to the 3′UTR and inhibits transcription elongation, the transcribed RNA will lack the polyA tail and will be rapidly degraded in cells. When plasmid pTRE-bcl2-tri was incubated with TFO and control oligonucleotides, the amount of plasmid DNA remained constant in each triplex binding reaction to ensure that the same amounts of plasmid DNA were transfected. As shown in Figure 6, when plasmid DNA incubated with equal amounts of different oligonucleotides, GA motif TFO2 but not control ODNs, can significantly downregulate bcl-2 expression. However, the GT motif TFO1 can partially inhibit the expression of bcl-2. The data shown in Figure 7 indicate that, at low TFO2:plasmid DNA ratios, the expression of bcl-2 can be inhibited (12.5:1) or even partially repressed (1.25:1). TFO4, which recognizes CG interruption with a cytosine base also significantly downregulates bcl-2 expression and even works better than TFO2; while TFO3, which is a phosphodiester TFO, has almost no effect on bcl-2 expression (Fig. 8). Moreover, as shown in Figure 8, the effect of TFO on bcl-2 expression can last for 72 h after transfection. These results clearly demonstrate a downregulation of the 3′UTR functions of bcl-2 by TFO, and indicate that 3′UTR may play a significant role in the regulation of bcl-2 expression.
Figure 6.
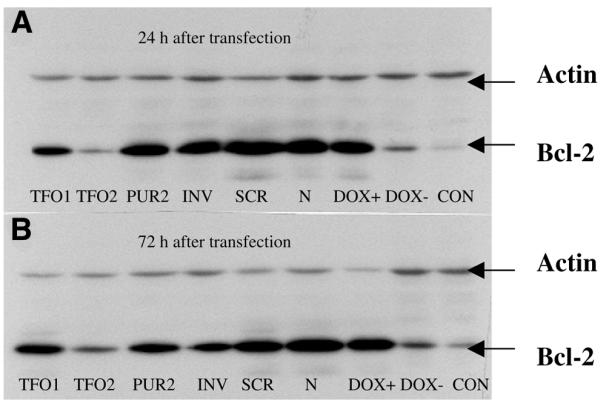
Western blot analysis showing downregulation of Bcl-2 protein expression by inhibitory effects of TFO on bcl-2 3′UTR functions. Plasmid pTRE-bcl2-tri was incubated with specific TFOs (TFO1 and 2) and controls (PUR2, INV, SCR, N) and transfected into HeLa cells that have been stably transfected with plasmid pTet-On. Cells were lysed 24 (A) and 72 h (B) post-transfection and western blots were carried out. TFO1, GT sequence; TFO2, GA sequence; PUR2, 16 nt TFO targeted to the bcl-2 promoter P2; INV, inverse oligonucleotide targeted to bcl-2 mRNA from +1 to +20; SCR, a 20 nt scrambled oligonucleotide; N, plasmid pTRE-bcl2-tri incubated with no oligonucleotide; DOX+, transfected just with plasmid pTRE-bcl2-tri; DOX–, transfected just with plasmid pTRE-bcl2-tri and without Doxycyclin in medium; CON, cells transfected without any DNA.
Figure 7.
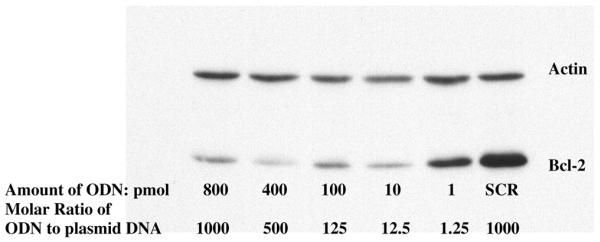
Western blot analysis showing the inhibitory effect of TFO2 on the bcl-2 gene expression at differing oligonucleotide concentrations (ODN amounts: 800, 400, 100, 10, 1 pmol). Plasmid pTRE-bcl2-tri was incubated with different concentrations of specific TFO (TFO2) and SCR, and transfected into HeLa cells that have been stably transfected with plasmid pTet-On. Cells were lysed 24 h post-transfection and western blots were carried out.
Figure 8.
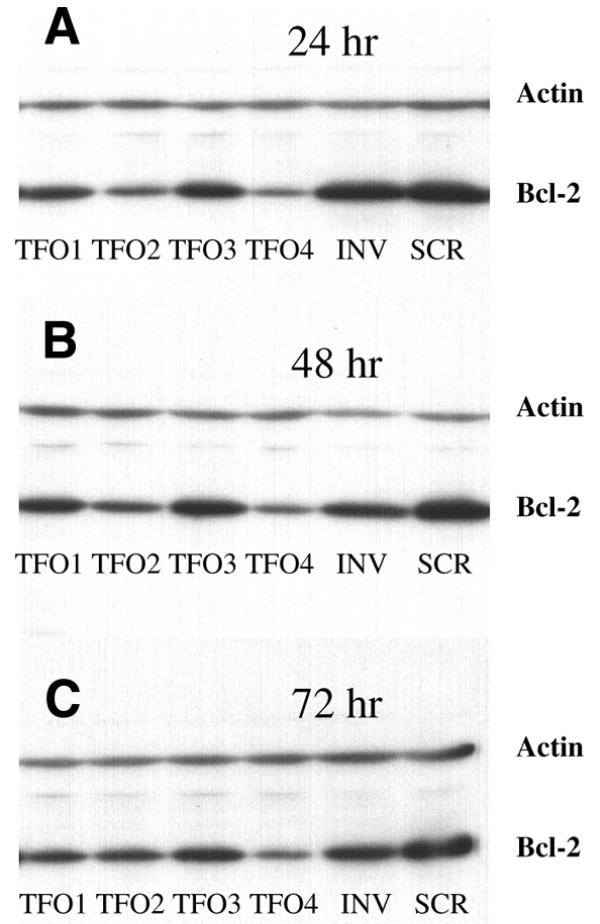
Effects of different kinds of TFOs on the expression of the Bcl-2 protein at different time points. Plasmid pTRE-bcl2-tri was incubated with different TFOs (TFO1–4) and controls (INV, SCR), and transfected into HeLa cells that had been stably transfected with plasmid pTet-On. Cells were lysed 24 (A), 48 (B) and 72 h (C) post-transfection and western blots were carried out.
DISCUSSION
The overall objective of this study was to develop a TFO-based gene silencing strategy that could downregulate the expression of the bcl-2 proto-oncogene. The choice of 3′UTR as a triplex target was based on the following considerations. (i) The 3′UTR may play a significant role in the regulation of gene expression; (ii) the triplex may inhibit the transcriptional elongation and the truncated mRNA lacking a polyA tail is unstable and is rapidly degraded in cells; (iii) we aimed to expand TFO target sites to the transcribed and 3′UTR regions of genomes.
Triplex formation in the promoter or coding sequences of several genes have been described previously (6–17). However, none has been used in 3′UTR sequences. We have found an 18 bp polypurine–polypyrimidine sequence at the 3′UTR of the bcl-2 gene. Our data demonstrated that the triplex structure of the pur*pur.pyr motif can be formed at the bcl-2 3′UTR with the third strand being antiparalleled to the purine-rich strand of the duplex (Fig. 4). The ability of this oligonucleotide to inhibit expression of bcl-2 (Figs 6–8) suggests that the plasmid is incorporated into the nucleus and, when formed, the triplex is stable in the nucleus. To our knowledge, this is the first example of using an oligonucleotide targeted to the 3′UTR of a gene.
We formed the triplex in vitro by incubating supercoiled plasmid DNA with TFO (Fig. 5), and then the entire DNA complex was transfected into HeLa cells. As we expected, none of the controls was able to decrease the Bcl-2 protein level. However, TFO2 specifically decreased the Bcl-2 protein level as shown by western blotting. The GT sequence TFO1 could partially inhibit the expression of bcl-2 (Fig. 6), this is in agreement with the previous findings that AG TFO forms extraordinarily stable triplexes (8). Kim et al. (10) observed some non-specific inhibition with control oligonucleotides unless excess oligonucleotide was removed after incubation with plasmid. In the present study, no significant non-specific effects of sequence unrelated TFOs were observed even without purification after incubation.
Beal and Dervan (3) have previously shown that G*G.C, A*A.T and T*A.T triplets stabilize a triple helix to a greater extent than the other 13 natural triplets in a thymidine–EDTA moiety. They also found, while oligonucleotides containing A, C or G opposite a single C.G base pair did not provide cleavage, T-substituted oligonucleotides gave weak but better cleavage than other oligonucleotides (3). However, in our experiments, the C-substituted oligonucleotide (TFO4) shows equal or stronger inhibition of Bcl-2 protein expression than T-substituted oligonucleotides (TFO1 and 2) (Fig. 8).
Our results also showed that expression of bcl-2 can be repressed at the TFO:plasmid DNA ratio of 12.5:1 or even partially inhibited at the ratio of 1.25:1 (Fig. 7). Moreover, data presented here agree with the suggestion that the formation of a highly stable triple helix may be capable of preventing transcription elongation independent of the formation of a covalent bond with triplex target, though modified or chemically reactive oligonucleotides may enhance the ability of TFOs to inhibit transcription elongation (36).
Several reports have demonstrated that TFOs targeted to positive regulatory factor binding sites inhibit transcription in cells. McShan et al. (37) demonstrated that a TFO targeted to the Sp1 binding sites in the long terminal repeats of human immunodeficiency virus inhibits viral transcription in infected cells. It has been demonstrated that TFOs targeted to the human Ki-Ras or Her-2/Neu promoter inhibit the binding of a protein in HeLa nuclear extracts (38,39). Triplex with the human c-myc P2 promoter also inhibits binding of the important regulatory factor, MAZ, and blocks transcription of c-myc in a cell free in vitro transcription system (40). It has also been demonstrated that a triplex can prevent the binding of recombinant Sp1 and inhibits transcription of the cyclin D1 promoter in stable transfected HeLa cells (9). Triplex-mediated repression of transcription may also result from the inhibition of formation of the proximal initiation complex assembly. It has been shown by Maher et al. (41) that triple helical complexes bound to the promoter inhibit in vitro transcription primarily by blocking assembly of the initiation complexes rather than by occluding the positive regulatory factor.
Coding sequences, such as the 3′UTR, of genes remain potential targets for triplex formation and including the coding sequence will expand the number of triplex targets available in genes of interest. A potential mechanism by which TFOs may lead to a reduction in gene expression is by inhibiting transcription elongation. The oligonucleotide is designed to form a triple helix in the coding sequence, such as 3′UTR, of the gene of interest in order to prevent the progression of RNA polymerase in its synthesis of the mRNA. One advantage of this approach is to increase the number of potential triplex target sequences since the transcribed region of a gene is generally much larger than the cis-regulatory elements of the promoter. Another possible advantage of a target in the coding region of a gene is a somewhat greater accessibility of the coding sequence to triplex formation since the TFO will not be required to displace a previously bound transcription factor (7). As our data show, TFOs targeted to the 3′UTR can effectively downregulate the expression of the gene of interest. The mechanism of gene expression inhibition by TFO targeting to the 3′UTR may lie in the fact that the triplex inhibits transcription elongation and the truncated mRNA (lacking a polyA tail) is very unstable and is rapidly degraded in cells. This study provides an example for expanding the target sites of oligonucleotide-based gene therapy to 3′UTR.
It has been reported that phosphorothioated TFOs with 3′-terminal amino groups are still able to form triplexes and partially block c-pim-1 promoter activity (8). This is also confirmed by our data shown in Figure 6, that TFO2 repressed the expression of the bcl-2 gene. Moreover, our results further showed that an amino linkage at the 3′-end alone is enough to prevent ODN from nucleases in vivo as TFO4 can significantly downregulate the Bcl-2 protein level for up to 72 h after transfection (Fig. 8).
It is well known that deregulated expression of some proto-oncogenes, such as bcl-2 and c-myc, results from the translocation of these genes to the IgH locus and the promoters of these genes are under control of the IgH enhancer (25,26). However, our data imply that deregulated expression of such translocated genes may also be influenced by the regulation of the 3′UTR of the hybrid, such as Bcl-2–IgH, which functions to stablize mRNA more efficiently than that of untranslocated counterpart genes. Further experiments are needed to investigate if TFO targeted to the 3′UTR of the bcl-2 gene can downregulate the expression of bcl-2 in bcl-2-overexpressing cells such as B cell lymphoma cell lines.
In summary, our studies expanded the TFO target site to the 3′UTR of a gene of interest and demonstrated that TFOs targeted to the bcl-2 3′UTR downregulate the expression of the bcl-2 proto-oncogene in transiently transfected HeLa cells, and suggest that the TFO strategy is a promising approach for treating diseases related to bcl-2 overexpression.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to Dr Marcel Pilarts for the construction of plasmid pTRE-bcl2-tri. We also thank Schemmel Elke for technical assistance.
References
- 1.Chan P.P. and Glazer,P.M. (1997) Triplex DNA: fundamentals, advances, and potential applications for gene therapy. J. Mol. Med., 75, 267–282. [DOI] [PubMed] [Google Scholar]
- 2.Frank-Kamenetskii M.D. and Mirkin,S.M. (1995) Triplex DNA structures. Annu. Rev. Biochem., 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 3.Beal P.A. and Dervan,P.B. (1991) Second structural motif for recognition of DNA by oligonuleotide-directed triple-helix formation. Science, 251, 1360–1363. [DOI] [PubMed] [Google Scholar]
- 4.Mergny J.L., Sun,J.S., Rougee,M., Montenary-Garestier,T., Barcelo,F., Chomilier,J. and Helene,C. (1991) Sequence specificity in triple-helix formation: Experimental and theoretical studies of the effect of mismatches on triplex stability. Biochemistry, 30, 9791–9798. [DOI] [PubMed] [Google Scholar]
- 5.Durland R.H., Kessler,D.J., Gunnell,S., Duvic,M., Pettitt,B.M. and Hogan,M.E. (1991) Binding of triple helix forming oligonucleotides to sites in gene promoters. Biochemistry, 30, 9246–9255. [DOI] [PubMed] [Google Scholar]
- 6.Olivas W.M. and Maher,L.J.,III (1996) Binding of DNA oligonucleotides to sequences in the promoter of the human bcl-2 gene. Nucleic Acids Res., 24, 1758–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebbinghaus S.W., Fortinberry,H. and Gamper,H.B.,Jr (1999) Inhibition of transcription elongation in the HER-2/neu coding sequence by triplex-directed covalent modification of the template strand. Biochemistry, 38, 619–628. [DOI] [PubMed] [Google Scholar]
- 8.Svinarchuk F., Debin,A., Bertrand,J.-R. and Malvy,C. (1996) Investigation of the intracellular stability and formation of a triple helix formed with a short purine oligonucleotide targeted to the murine c-pim-1 proto-oncogene promoter. Nucleic Acids Res., 24, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H.G. and Miller,D.M. (1998) A novel triplex-forming oligonucleotide targeted to human Cyclin D1 (bcl-1, proto-oncogene) promoter inhibits transcription in HeLa cells. Biochemistry, 37, 2666–2672. [DOI] [PubMed] [Google Scholar]
- 10.Kim H.G., Reddoch,J.F., Mayfield,C., Ebbinghaus,S., Vigneswaran,N., Thomas,S., Jones,D.E.,Jr and Miller,D.M. (1998) Inhibition of transcription of the human c-myc protooncogene by intermolecular triplex. Biochemistry, 37, 2299–2304. [DOI] [PubMed] [Google Scholar]
- 11.Joseph J., Kandala,J.C., Veerapanane,D., Weber,K.T. and Guntaka,R.V. (1997) Antiparallel polypurine phosphorothioate oligonucleotides form stable triplexes with the rat α1(I) collagen gene promoter and inhibit transcription in cultured rat fibroblasts. Nucleic Acids Res., 25, 2182–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kochetkova M., Iversen,P.O., Lopez,A.F. and Shannon,M.F. (1997) Deoxyribonucleic acid triplex formation inhibits granulocyte macrophage colony-stimulating factor gene expression and suppression growth in juvenile myelomonocytic leukemic cells. J. Clin. Invest., 99, 3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rininsland F., Johnson,T.R., Chernicky,C.L., Schulze,E., Burfeind,P. and Ilan,J. (1997) Suppression of insulin-like growth factor type I receptor by a triple-helix strategy inhibits IGF-I transcription and tumorigenic potential of rat C6 glioblastoma cells. Proc. Natl Acad. Sci. USA, 94, 5854–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giovannangeli C., Diviacco,S., Labrousse,V., Gryaznov,S., Charneau,P. and Helene,C. (1997) Accessibility of nuclear DNA to triplex-forming oligonucleotides: The integrated HIV-1 provirus as a target. Proc. Natl Acad. Sci. USA, 94, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belousov E.S., Afonina,I.A., Kutyavin,I.V., Gall,A.A., Reed,M.W., Gamper,H.B., Wydro,R.M. and Meyer,R.B.(1998) Triplex targeting of a native gene in permeabilized intact cells: covalent modification of the gene for the chemokine receptor CCR5. Nucleic Acids Res., 26, 1324–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guieysse A.L., Praseuth,D., Grigoriev,M., Harel-Bellan,A. and Helene,C. (1996) Detection of covalent triplex within human cells. Nucleic Acids Res., 24, 4210–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G., Seidman,M.M. and Glazer,P.M. (1996) Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science, 271, 802–805. [DOI] [PubMed] [Google Scholar]
- 18.Lewis J.D. and Tollervey,D. (2000) Like attracts like: Getting RNA processing together in the nucleus. Science, 288, 1385–1389. [DOI] [PubMed] [Google Scholar]
- 19.Conne B., Stutz,A. and Vassalli,J.-D. (2000) The 3′-untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nature Med., 6, 637–641. [DOI] [PubMed] [Google Scholar]
- 20.Marx J. (2000) Interfering with gene expression. Science, 288, 1370–1372. [DOI] [PubMed] [Google Scholar]
- 21.Hockenberry D., Zutter,M., Hickey,W., Nahm,M. and Korsmeyer,S.J. (1991) BCL2 protein is topographically restricted in tissues characterized by apoptotic cell death. Proc. Natl Acad. Sci. USA, 88, 6961–6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaux D., Cory,S. and Adams,J.M. (1988) Bcl-2 gene promoters haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature, 335, 440–442. [DOI] [PubMed] [Google Scholar]
- 23.Reed J.C. (1997) Bcl-2 family proteins: regulators of apoptosis and chemoresistance in hematologic malignancies. Semin. Hematol., 34, 9–19. [PubMed] [Google Scholar]
- 24.Yunis J.J. (1983) The chromosomal basis of human neoplasia. Science, 221, 227–236. [DOI] [PubMed] [Google Scholar]
- 25.Cleary M., Smith,S.D. and Sklar,J. (1986) Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell, 47, 19–28. [DOI] [PubMed] [Google Scholar]
- 26.Graninger W., Seto,M., Boutain,B., Goldman,P. and Korsmeyer,S.J. (1987) Expression of Bcl-2 and Bcl-2-Ig fusion transcripts in normal and neoplastic cells. J. Clin. Invest., 80, 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsujimoto Y. and Croce,C.M. (1986) Analysis of the structure, transcripts, and protein products of bcl-2, the gene involved in human follicular lymphoma. Proc. Natl Acad. Sci. USA, 83, 5214–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seto M., Jaeger,U., Hockett,R.D., Graninger,W., Bennett,S., Goldman,P. and Korsmeyer,S.J. (1988) Alternative promoters and exons, somatic mutation and deregulation of the Bcl-2-Ig fusion gene in lymphoma. EMBO J., 7, 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H.M. and Boxer,L.M. (1995) π1 binding sites are negative regulators of bcl-2 expression in pre-B cells. Mol. Cell. Biol., 15, 3840–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harigai M., Miyashita,T., Hanada,M. and Reed,J.C. (1996) A cis-acting element in the BCL-2 gene controls expression through translational mechanisms. Oncogene, 12, 1369–1374. [PubMed] [Google Scholar]
- 31.Wilson B.E., Mochon,E. and Boxer,L.M. (1996) Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol. Cell. Biol., 16, 5546–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zauli G., Gibellini,D., Caputo,A., Bassini,A., Negrini,M., Monne,M., Mazzoni,M. and Capitani,S. (1995) The human immunodeficiency virus type-1 Tat protein upregulates Bcl-2 gene expression in Jurkat T-cell lines and primary peripheral blood mononuclear cells. Blood, 86, 3823–3834. [PubMed] [Google Scholar]
- 33.Mayo M.W., Wang,C.Y., Drouin,S.S., Madrid,L.V., Marshall,A.F., Reed,J.C., Weissman,B.E. and Baldwin,A. (1999) WT1 modulates apoptosis by transcriptionally upregulating the bcl-2 proto-oncogene. EMBO J., 18, 3990–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhlmann E.J., Subramanian,T., Vater,C.A., Lutz,R. and Chinnadurai,G. (1998) A potent cell death activity associated with transient high level expression of BCL-2. J. Biol. Chem., 273, 17926–17932. [DOI] [PubMed] [Google Scholar]
- 35.Yin D.X., Zhu,L. and Schimke,R.T. (1996) Tetracycline-controlled gene expression system achieves high-level and quantitative control of gene expression. Anal. Biochem., 235, 195–201. [DOI] [PubMed] [Google Scholar]
- 36.Escude C., Giovanangeli,C., Sun,J.-S., Lloyd,D.H., Chen,J.-K., Gryaznov,S.M., Garestier,T. and Helene,C. (1996) Stable triple helices formed by oligonucleotide N3′→P5′ phosphoramidates inhibit transcription elongation. Proc. Natl Acad. Sci. USA, 93, 4365–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McShan W.M., Rossen,R.D., Laughter,A.H., Trial,J., Kessler,D.J., Zendegui,J.G., Hogan,M.E. and Orson,F.M. (1992) Inhibition of transcription of HIV-1 in infected human cells by oligodeoxynucleotides designed to form DNA triple helices. J. Biol. Chem., 267, 5712–5721. [PubMed] [Google Scholar]
- 38.Mayfield C., Squibb,M. and Miller,D. (1994) Inhibition of nuclear protein binding to the human Ki-ras promoter by triplex-forming oligonucleotides. Biochemistry, 33, 3358–3363. [DOI] [PubMed] [Google Scholar]
- 39.Ebbinghaus S.W., Gee,J.E., Rodu,B., Mayfield,C.A., Sanders,G. and Miller,D.M. (1993) Triplex formation inhibits HER-2/neu transcription in vitro. J. Clin. Invest., 92, 2433–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim H.G. and Miller,D.M. (1995) Inhibition of in vitro transcription by a triplex-forming oligonuleotide targeted to human c-myc P2 promoter. Biochemistry, 34, 8165–8171. [DOI] [PubMed] [Google Scholar]
- 41.Maher L.J.,III, Wold,B. and Dervan,P.B. (1989) Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science, 245, 725–730. [DOI] [PubMed] [Google Scholar]