Abstract
Autophagy is the mechanism by which cells consume parts of themselves to survive starvation and stress. This self-cannibalization limits cell death and tissue inflammation, recycles energy and biosynthetic substrates and removes damaged proteins and organelles, accumulation of which is toxic. In normal tissues, autophagy-mediated damage mitigation may suppress tumorigenesis, while in advanced tumors macromolecular recycling may support survival by buffering metabolic demand under stress. As a result, autophagy-activation in normal cells may suppress tumorigenesis, while autophagy inhibition may be beneficial for therapy of established tumors. The mechanisms by which autophagy supports cancer cell metabolism are slowly emerging. As cancer is being increasingly recognized as a metabolic disease, how autophagy-mediated catabolism impacts cellular and mammalian metabolism and tumor growth is of great interest. Most cancer therapeutics induce autophagy, either directly by modulating signaling pathways that control autophagy in the case of many targeted therapies, or indirectly in the case of cytotoxic therapy. However, the functional consequence of autophagy induction in the context of cancer therapy is not yet clear. A better understanding of how autophagy modulates cell metabolism under various cellular stresses and the consequences of this on tumorigenesis will help develop better therapeutic strategies against cancer prevention and treatment.
Keywords: autophagy, p62, inflammation, cancer, energy, metabolism, mitochondria
Introduction
Macroautophagy (autophagy hereafter) is a mechanism of cellular self-consumption for recycling of intracellular “cargo” such as damaged proteins and organelles. Under normal conditions, basal autophagy serves to maintain cellular homeostasis, and autophagy is induced in response to many different forms of stress including nutrient, oxygen and growth factor deprivation and chemotherapeutics [1–3].
Upon autophagy induction, the potentially toxic cargo is encapsulated by pre-autophagosomal-structures called “phagophores” that mature into double-membrane vesicles called “autophagosomes” and fuse with lysosomes where the cargo is degraded [2,4]. While short-lived proteins are degraded mainly via ubiquitin-proteasome system (UPS), autophagy is the only mechanism for degrading large protein aggregates and damaged organelles. Autophagy can non-selectively target cytoplasm for bulk degradation, or selectively target cellular components such as proteins, lipid droplets, mitochondria, peroxisomes, endoplasmic reticulum, ribosomes, and glycogen as well as invading microorganisms [1,5,6]. The mechanisms governing cargo selectivity, however, are incompletely understood at present. Given the major role of autophagy in the turnover of cellular proteins and organelles, it is not surprising that autophagy deregulation has been implicated in many disease conditions, including cancer.
Autophagy is regulated by nutrient and growth factor availability
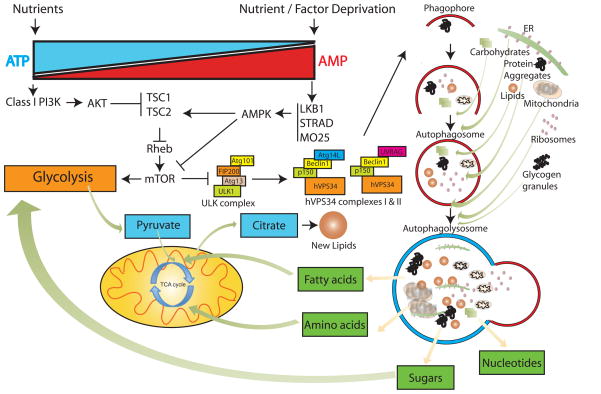
Nutrient and growth factor deprivation are major activators of autophagy [6]. Mammalian target of rapamycin (mTOR) is a protein kinase and master negative regulator of autophagy. mTOR complex 1 (mTORC1) responds to nutrient and growth factor availability by phosphorylating and inactivating two essential components of the Unc-51-like kinase (ULK1) complex, ULK1 and Atg13 [7], thereby preventing the nucleation of the phagophores (Figure 1) (reviewed in [8]). When nutrients are limiting, mTORC1 kinase activity is inhibited, de-repressing autophagy. This provides the necessary integration of growth control signals with nutrient availability and catabolism. Although starvation-induced autophagy is mainly regulated by mTOR signaling, autophagy is also regulated through mTOR-independent mechanisms (reviewed in [9–11]).
Figure 1. Regulation of autophagy under nutrient deprivation and its interaction with central carbon metabolism.
The major extra-cellular nutrient sensing pathways, controlled by PI3K-I and adenosine monophosphate activated kinase (AMPK) tightly regulate autophagy through mTOR signaling, although other mTOR independent mechanisms exist [9–11]. Under nutrient-replete conditions, autophagy is inhibited by mTOR and inactivation of the ULK1 complex. Metabolic stress relieves this inhibition to activate autophagy, and activates AMPK. AMPK inhibits mTOR by activating its negative regulator, the tuberous sclerosis protein 2 (TSC2) and inhibiting its positive-regulatory subunit, Raptor. The ULK1 complex activates the vacuolar sorting protein 34 (hVPS34)- and Beclin1-containing complexes (complex I and complex II), to initiate phagophore formation (Figure 1) [9]. Phagophores nucleate and expand around the cargo encapsulating it and targeting it to lysosomes for degradation. Degradation products of autophagy substrates may re-enter glycolysis and the TCA cycle for anabolic as well as catabolic processes leading to generation of energy and biomass production. For an extensive review of other forms of autophagy regulation, please see [11,12,55].
Functional autophagy involves the coordinate action of multiple autophagy-related (Atg) gene products that regulate nucleation, elongation and maturation of the phagophore membranes around the cargo (autophagosomes), and their fusion with lysosomes forming autophagolysosomes (Figure 1) [12]. Cargo selection is achieved, in part, by ubiquitin modifications of targets, which are recognized by ubiquitin-interacting domains (UBA) of the autophagy cargo receptor proteins such as p62/SQSTM1 and neighbor of BRCA1 gene 1 (NBR1), and are targeted to autophagosomes via their LC3-interacting regions (LIR) [13]. Recent studies have identified a phosphatidylinositol 3-phosphate (PI3P) binding protein, autophagy-linked FYVE protein (Alfy) that orchestrates assembly of autophagy machinery around p62-captured cargo, aiding in their capture and sequestration in autophagosomes [14]. p62 and NBR1 themselves are autophagy substrates and are degraded along with the cargo in normal cells, and accumulate under autophagy-deficiency thus serving as potential biomarkers for defective autophagy.
Autophagy promotes stress survival and maintains tissue homeostasis
Evidence from mouse models with deficiency in genes essential for autophagosome formation (Figure 1) suggests that autophagy is involved in stress survival, early embryonic development, and prevention of neurodegeneration, Crohn’s disease (CD) and tumorigenesis [2,6]. While atg6/beclin1−/− mice die early during embryogenesis (E7.5-E8.5), beclin1+/− mice with a partial autophagy defect are developmentally normal although tumor prone (see below) [15,16]. Atg5 is required for pre-implantation development in mice although dispensable for later embryonic development [17,18]. Newborn pups with maternally inherited autophagy proteins but deficient for atg5, atg7 and atg3, fail to survive neonatal starvation and display signs of bioenergetic impairment [19–21]. Atg4c knockout mice have increased propensity for carcinogen-induced tumorigenesis (see below) [22] while Atg4b knockout mice show impaired inner ear development and balance functions [23]. Central nervous system-targeted deficiency in atg5 or atg7 in mice causes the accumulation of toxic protein and organelles and neuronal degeneration [24,25]. Liver-specific Atg7-deficiency causes accumulation of p62-containing protein aggregates and p62-mediated liver damage [26]. Deficiency in atg5 and atg16L1 causes tissue injury in intestinal Paneth cells leading to CD, a known risk factor for colorectal cancer in humans [27]. Atg7−/− mice show impaired white adipocyte differentiation and are predisposed to premature death when fed a high-fat diet [28]. These observations suggest that autophagy sustains cellular and mammalian viability through damage mitigation and maintenance of metabolic homeostasis.
Autophagy defects reduce viability, but promote tumorigenesis
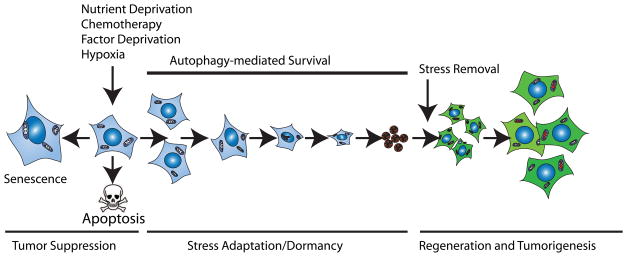
Autophagy is induced in cells under metabolic stress, but in apoptosis-competent cells, apoptosis is also activated as a tumor suppressor mechanism. In apoptosis-deficient cells, autophagy promotes remarkable long-term stress survival leading to dormancy and then regeneration upon stress removal (Figure 2). Autophagy-defective cells, however, can eventually succumb to prolonged metabolic stress and undergo cell death by necrosis when apoptosis is disabled [29].
Figure 2. Mechanism of autophagy-mediated stress survival and tumor cell dormancy.
Metabolic stress is a potent trigger for senescence or apoptosis as two important tumor suppression mechanisms. Autophagy is induced under metabolic stress as a survival mechanism that delays apoptosis [29,56]. Apoptosis-defective cells survive metabolic stress through autophagy-mediated stress-adaptation. Upon prolonged stress, cells exit the cell cycle, and alter gene expression and metabolism to conserve energy leading to senescence, dormancy or quiescence [29,31]. Upon removal of stress, dormant cells re-enter the cell cycle and resume proliferation that can potentially lead to regeneration and tumor relapse. Deciphering the molecular mechanisms of this stress-adaptation is important to prevent tumor dormancy to improve cancer therapy.
Contrary to the survival function of autophagy, autophagy defects that impair survival can promote tumorigenesis, and mice with allelic loss of beclin1 are prone to hepatocellular carcinoma (HCC), lung adenocarcinomas, lymphomas, and mammary hyperplasia [15,16]. Moreover, oncogenic insults such as constitutive activation of the class-I phosphatidylinositol 3-kinase (PI3K-I) pathway inhibit autophagy by activating mTOR, suggesting a role for autophagy in tumor suppression [6]. The apparent paradox that the loss of a survival pathway leads to tumorigenesis can be reconciled by the damage-mitigation and metabolism-supporting functions of autophagy with different functional consequences at various stages during the process of cancer development. Thus, the role of autophagy in cancer is context-dependent.
Autophagy suppresses tumorigenesis by limiting necrosis and inflammation
Regions of chronic tissue inflammation are breeding grounds for cancer-causing mutations [3,30]. In mammals, chronic tissue damage and inflammation can be caused by pathogen infections or exposure to toxins, and lead to tumorigenesis [3]. Autophagy-deficiency can recreate this condition by preventing the normal adaptation to metabolic stress, without the need for external inducers of cell death, tissue damage, and chronic inflammation. Genetic inhibition of autophagy indeed leads to cell death, tissue damage, and chronic inflammation due to a mal-adaptation to normal stress [29,31]. Furthermore, accumulation of cytotoxic “cellular garbage” under autophagy-deficiency may provide the perfect recipe for a damaging microenvironment and local inflammation conducive for tumor growth [3,32]. However, the exact mechanism by which autophagy-defects lead to inflammation and tumor promotion can be tissue dependent.
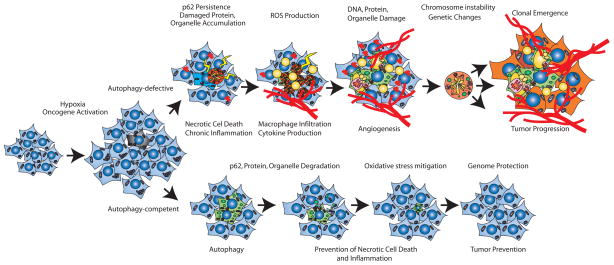
Liver-specific deficiency in atg7 causes accumulation of p62 and poly-ubiquitinated protein aggregates inducing p62-dependent liver damage [33]. Allelic loss of beclin 1 causes p62-aggregate accumulation and inhibition of canonical nuclear factor-κB (NF-κB) pathway leading to hepatocellular carcinoma (HCC) [32], which phenocopies hepatocyte-targeted deficiency in the NF-κB activators IKK-β or NEMO [34,35]. It is known that inhibition of NF-κB survival signaling in the liver causes apoptosis, and recruitment of liver-specific macrophages (Kupffer cells) producing inflammation and compensatory-proliferation in the surviving hepatocytes leading to HCC [36]. Thus autophagy functions to limit chronic cell death and inflammation to suppress tumorigenesis (Figure 3). It will be of interest to establish whether tumors arising in other tissues are linked to chronic tissue damage due to suppression of autophagy, either through direct inactivation of essential autophagy genes or indirect activation of signaling pathways that suppress autophagy.
Figure 3. Mechanisms by which autophagy defects promote tumorigenesis.
Metabolic stress due to hypoxia, nutrient deprivation and oncogene activation trigger proliferation but also induce apoptosis, which limits tumor growth. Metabolic stress activates autophagy as a protective mechanism in metabolically stressed tumor regions. Autophagy-competent cells survive, while autophagy-defective cells succumb to cell death in hypoxic regions, promoting a chronic inflammatory state that can favor tumor growth [3,29,31]. Thus by limiting tissue damage, autophagy may suppress tumorigenesis. At the subcellular level, metabolic stress also induces oxidative damage and p62-accumulation. In normal cells, coordinate induction of autophagy degrades damaged proteins, lipids and organelles, preventing toxic buildup of “cellular garbage” in a p62-dependent manner, thus protecting the genome. In autophagy-defective cells, the degradation of toxic material is prevented, leading to persistently high levels of p62 and the formation of p62-containing protein aggregates, accumulation of damaged mitochondria and ROS production, leading to DNA damage and chromosomal instability that can promote tumorigenesis [3,32].
Autophagy limits genome instability
Another important mechanism of oncogenesis is the accumulation of tumor-promoting DNA alterations through exposure to oxidative stress. Metabolic stress in normal tissues can trigger oxidative stress through accumulation of unfolded proteins and damaged organelles, which are normally eliminated by autophagy. Stress that upregulates autophagy also induces the autophagy cargo receptor p62, which binds and aggregates poly-ubiquitinated proteins tagged for degradation, and delivers them to the autophagy pathway for degradation [32]. In autophagy-competent cells [37], the p62-dependent recycling and clearance of cellular garbage functions to mitigate oxidative stress. When autophagy is defective, however, p62 and cargo are not degraded and persistence of high levels of p62 and associated cargo elicits cytotoxic response. Consistent with this, p62-deficiency delays mortality in mice with liver-specific atg7-deficiency [26]. This cytotoxicity includes activation of the DNA damage response, alteration in gene expression, and elevated chromosome gains and losses that may accelerate DNA mutations that potentially contribute to tumorigenesis (Figure 3) [32]. Moreover, persistent ectopic expression of p62 reconstitutes cytotoxic response and tumorigenesis, and knocking down p62 rescues the oxidative stress phenotype suggesting that clearance of p62 and p62-containing aggregates is one mechanism by which autophagy prevents tumorigenesis [32].
Recent evidence suggest that p62 directly mediates oxidative stress by sequestering the kelch-like ECH-associated protein 1 (Keap1), an adaptor of the E3-ligase for the degradation of the NF-E2-related factor 2 (Nrf2) transcription factor, promoting Nrf2-mediated stress response [38,39]. Additionally, persistent Nrf2 activation due to liver-specific deletion of Keap1 exacerbates the hepatotoxicity in autophagy-deficient mice, which is suppressed by loss of Nrf2 suggesting that p62-dependent deregulation of Nrf2-Keap1 system at least in part is responsible for the tissue damage observed under autophagy-deficiency [38]. p62 in turn, is induced by Nrf2 in response to oxidative stress creating a positive feedback loop [40]. This mechanism of autophagy-mediated tumor suppression by cellular garbage disposal suggests that stimulation of autophagy may be an important approach to cancer prevention.
Autophagy and cancer metabolism: The nightlife of autophagy
Human cancers are known to have altered metabolism tailored to suit their specific growth requirements. Cancer cells preferentially utilize glucose through aerobic glycolysis (Warburg effect). A recent metabolomic analysis of human colon and gastric cancers showed low glucose and high glycolytic intermediates and lactate levels. They also displayed signs of excessive glutamine consumption and enhanced degradation and recycling of proteins proposed to be through autophagy [41] although other mechanisms are possible. It is well appreciated that cancer cells have an increased need for energy and a carbon/nitrogen source for biomass production due to hyper-proliferation, both of which are achieved primarily through metabolism. As a major provider of energy substrates under stress [11,19,42], autophagy can support metabolism under these conditions of extreme demand.
Mutations in genes encoding mitochondrial enzymes, such as succinate dehydrogenase (SD), fumarate hydratase (FH) and isocitrate dehydrogenase (IDH), are observed in certain types of human cancers and are linked to activation of hypoxia inducible factors (Hifs) [43,44]. Activation of oncogenes such as Hif1-α and c-Myc transcriptionally upregulate glycolosis genes, promote glycolysis and anaerobic breakdown of pyruvate to lactate [44,45]. Similarly, Myc activation promotes dependence on glutamine as a carbon source for the tricarboxylic acid (TCA) cycle (glutaminolysis) [46,47]. Activation of Hifs induces transcription of the Bcl-2/adenovirus E1B 19K-interacting protein 3 (Bnip3) gene to induce mitochondrial-selective autophagy, potentially elevating autophagy-dependence. Indeed, autophagy is required to access intracellular fat storage for free fatty acids to support metabolism and autophagy defects cause accumulation of lipid droplets [48]. Lipophagy (autophagy of lipids) provides fatty acids for β-oxidation that generates acetyl-CoA in mitochondria to support the TCA cycle. Autophagic degradation of preexisting intracellular proteins is a major source of free amino acid pools in stressed cells [11,42], and is shown to increase the free amino acid pools in C. elegans [49]. Autophagy-defects prevent degradation of both long-lived as well as short-lived proteins [50,51], and plasma amino acid levels rapidly decrease in atg5−/− and atg7−/− neonates after birth [19,20]. The free amino acid pool feeds into central carbon metabolism at multiple points [11] that include pyruvate, acetyl-CoA and TCA intermediates such as α-ketoglutarate (α-KG) (from glutamine) and oxaloacetate (from aspartate). Given the role of autophagy in the generation of these substrates through macromolecular recycling, defects in autophagy may have a profound impact on mitochondrial substrate availability and function. The metabolism-supporting role of autophagy may have dramatic consequences to established tumors with increased metabolic demand. In this setting, it will be interesting to examine the role of autophagy-defects in tumors with constitutive activation of oncogenes such as myc and ras where elevated metabolism may require autophagy to meet the elevated metabolic demand of deregulated tumor growth.
Autophagy, mitochondrial health and energy metabolism
How exactly defects in autophagy may impinge altered cancer metabolism and potentially support tumor growth is currently unclear, but most likely it involves autophagy’s role in maintaining mitochondrial health under stress. Damaged mitochondria are a major source of reactive oxygen species (ROS) production in cells, and mitophagy is the mechanism that clears depolarized mitochondria to maintain cellular homeostasis [52]. Mitochondria undergo multiple rounds of fusion-fission cycles to maintain the pool of healthy mitochondria. Without autophagy, the mitochondrial pool becomes progressively contaminated by abnormal and dysfunctional mitochondria [53,54]. Autophagy clears depolarized-mitochondria through the PTEN-induced putative kinase 1 (PINK1)- and Parkin1-dependent ubiquitination of the mitochondrial proteins, including the voltage-dependent anion channel (VDAC1) and the recruitment of p62 that targets them to autophagosomes [53]. Failed mitochondrial quality control may be compounded by substrate limitation due to autophagy-defects, especially under metabolic stress. Additionally, impaired mitochondrial metabolism and consequent impairment in energy production may potentially select for increased glycolysis and lactate production contributing to the Warburg effect. Thus failure of mitochondrial quality control in autophagy-defective cells may be an alternative route to less dependence on oxidative metabolism, promoting the Warburg effect, although more detailed investigation is necessary to address this issue.
Conclusion
The role of autophagy in cancer is complex and context-dependent. Autophagy is an important tumor suppressor pathway that limits oxidative stress and tissue damage that can promote cancer initiation. However, autophagy also supports cellular metabolism that has the potential to aid the growth of advanced tumors with elevated metabolic demand. Although recent studies have contributed tremendously to our understanding of the role and relevance of autophagy in cancer, there are many areas of the basic biology of autophagy that are not yet understood. For example, how does autophagy selectively access major energy sources such as lipids and carbohydrates, and to what extent do these sources contribute to stress survival? Although the role of autophagy in degrading macromolecules is well established, how the autophagy pathway is integrated with central carbon metabolism and possibly the Warburg effect is not known. Is activation of autophagy by tumors under metabolic stress merely survival promoting, or can it sustain production of new biomass? If and how autophagy provides net flux into total biomass for biosynthetic pathways and how defects in autophagy might impact these processes in advanced tumors are worth detailed investigation. Answers to these pertinent questions will also help to reveal how autophagy-defects alter mitochondrial function. Another interesting and yet unanswered question is how tumors with autophagy defect, and presumably mitochondrial and TCA cycle impairment, sustain tumor growth. With a new appreciation of the role of metabolism in cancer, it will be of great interest to elucidate the role of cellular catabolism by autophagy in this context.
Acknowledgments
We thank the members of the laboratories of Drs. Joshua D. Rabinowitz (Lewis-Sigler Institute for Integrative Genomics, Princeton University), and Eileen White for comments and helpful discussions. E. W. acknowledges support from the NIH (R37 CA53370, RO1 CA130893, RC1 CA147961, and the New Jersey Commission on Cancer Research (09-1083-CCR-EO) and the DOD (W81XWH06-1-0514 and W81XWH05).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levine B, Kroemer G. Autophagy in the Pathogenesis of Disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 22:212–217. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 6.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al. Regulation of Mammalian autophagy in physiology and pathophysiology. Physiol Rev. 90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- **7.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. This paper reports that mTORC1 suppresses autophagy through direct regulation of the ULK1-Atg13-FIP200 complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meijer AJ, Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol Aspects Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- *11.Rabinowitz JD, White E. Autophagy and Metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. This review discusses the role of autophagy in metabolism, its regulation, and its implications for cancer and degenerative diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vadlamudi RK, Shin J. Genomic structure and promoter analysis of the p62 gene encoding a non-proteasomal multiubiquitin chain binding protein. FEBS Lett. 1998;435:138–142. doi: 10.1016/s0014-5793(98)01021-7. [DOI] [PubMed] [Google Scholar]
- 14.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. 15. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukamoto S, Kuma A, Mizushima N. The role of autophagy during the oocyte-to- embryo transition. Autophagy. 2008;4:1076–1078. doi: 10.4161/auto.7065. [DOI] [PubMed] [Google Scholar]
- **18.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for pre-implantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. This paper provides the first evidence that autophagy is required for preimplantation development in mice although dispensable for later embryonic development. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425– 434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 21.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 23.Marino G, Fernandez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodriguez F, Salvador-Montoliu N, Vega JA, Germana A, Fueyo A, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 120:2331–2344. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 25.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 27.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. 16. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 31.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **32.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. This paper discusses the aberrant p62 accumulation in autophagy-defective tumor cells and its role in tissue damage, inflammation and cancer induction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata JI, Ezaki J, Murata S, et al. Homeostatic Levels of p62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 34.Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, Pasparakis M. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119–132. doi: 10.1016/j.ccr.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 35.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 36.Sun B, Karin M. NF-kappaB signaling, liver disease and hepatoprotective agents. Oncogene. 2008;27:6228–6244. doi: 10.1038/onc.2008.300. [DOI] [PubMed] [Google Scholar]
- 37.Mathew R, Degenhardt K, Haramaty L, Karp CM, White E. Immortalized mouse epithelial cell models to study the role of apoptosis in cancer. Methods Enzymol. 2008;446:77–106. doi: 10.1016/S0076-6879(08)01605-4. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 39.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *41.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et al. Quantitative metabolome profiling of 17 colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. This paper reports direct and simultaneous measurements of global metabolite levels in clinical tumors and adjacent normal tissues using capillary electrophoresis coupled to mass spectrometry-based metabolomics. [DOI] [PubMed] [Google Scholar]
- 42.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 43.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 44.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. N Engl J Med. 2009;360:813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis BC, Prescott JE, Campbell SE, Shim H, Orlowski RZ, Dang CV. Tumor induction by the c-Myc target genes rcl and lactate dehydrogenase A. Cancer Res. 2000;60:6178–6183. [PubMed] [Google Scholar]
- 46.Dang CV. Glutaminolysis: Supplying carbon or nitrogen or both for cancer cells? Cell Cycle. 9:3884–3886. doi: 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- 47.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. This paper discusses lipophagy as a form of autophagy to access fat stores in lipid droplets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 50.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 52.Wild P, Dikic I. Mitochondria get a Parkin' ticket. Nat Cell Biol. 12:104–106. doi: 10.1038/ncb0210-104. [DOI] [PubMed] [Google Scholar]
- **53.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. This paper reports that mitochondrial fission followed by selective fusion segregates dysfunctional mitochondria and permits their removal by autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira, Gutkind S, Daniels MP, Komatsu M, et al. Mitochondrial dysfunction and oxidative 18 stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]