Abstract
Conserved primers across multiple species and simultaneously specific for a certain isozyme can be rare and difficult to find. PrimerIdent was developed aiming to automate this primer design and selection process in a given nucleotide sequence alignment, providing an intuitive, easy to interpret graphical result, which offers a list of all possible primers that meet the user criteria, with a colour-code identity to each sequence in the alignment. The software here presented is a simple and intuitive web based tool that is suitable for distinguishing very similar nucleotide sequences, such as isozymescoding sequences, to enable the conserved primer design across multiple species, necessary for approaches that rely on knowing if a primer is suitable for a certain set of pre-aligned sequences, to design a specific primer to a certain sequence variation, or a combination thereof. This extremely useful software can, therefore, be used as a tool for the specific amplification of individual members of multigenic families across related species and also to evaluate the differential expression of isogenes for a given species.
Availability
Keywords: Primer design, conserved primers, multiplepurposed primers, gene-specific primers
Background
Polymerase chain reaction (PCR) is a robust, flexible and fast technique that is essential to the modern Molecular Biology and is currently used in many applications. One of the fundamental assets for a successful PCR reaction is the quality and reliability of the oligonucleotide primer design.Designing primers in conserved regions of related nucleotide sequences is of great importance for molecular biologists as it allows the usage of the same primer to successfully obtain a PCR product across multiple and/or understudied organisms. Softwares that can design conserved primers using multiple alignments have been developed, and are free to use, such as Primaclade [1].
PrimerIdent, however, uses the concept of conserved primer design for a different purpose: PCR (or RTPCR) amplification of a specific isozymecoding sequence.Isozymes are multiple forms of enzymes from the same class arising from genetically determined differences in primary structure and frequently sharing a great identity amongst them. As a consequence, it is very difficult to design primers that effectively differentiate between related genes that share a high degree of similarity. The difficulty lies on choosing primers that are specific for one sequence coding a particular isozyme and simultaneously less identical to similar sequences coding other members of the same gene family.In order to design a suitable primer for a nucleotide sequence coding a specific isozyme, it is necessary to ensure that the primer is designed for a exclusive region for that isozyme. This primer design approach makes the conserved primer design tools described above unsuitable.One way of doing this, albeit time-consuming and complex, is to find these primers using a trial/error method, with the aid of BLAST (Basic Local Alignment Search Tool ― [2]), by comparing the match score of the different published sequences, and therefore minimizing the risk of amplifying more than one nucleotide coding sequence for a type of isozyme at the same time.
Such approach proves to be even more complex if the studied organism doesn't have any nucleotide sequences published for a particular multigene family. This means that, in this case, it is necessary to find a primer that is
conserved across multiple organisms phylogenetically close to the organism studied, and simultaneously
specific for a given isozyme.
This isozyme specificity should also be consistent across those related organisms.To circumvent these complex tasks, PrimerIdent was developed aiming to automate this primer design and selection process, in a given nucleotide sequence alignment providing an intuitive, easy to interpret graphical output.
Input
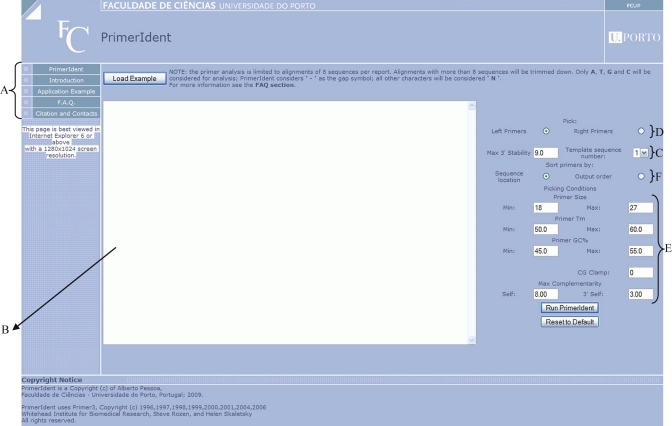
PrimerIdent was implemented as a Perl-CGI script that uses Primer3 core [3] as the primer design engine and accepts a FASTA alignment of no more than 8 sequences with a maximum length of 10 kbp at a time. Figure 1shows the web frontend. The website is located at http://primerident.up.pt, hosted by the University of Porto, Portugal. In an interactive user interface, the menu on the left
provides basic information about PrimerIdent, which includes an introduction section and application examples, as well as a F˙A˙Q. (Frequently Asked Questions) section and contact information.In order to use PrimerIdent, the user pastes the FASTA sequence alignment into the textbox
This alignment can be made in programs such as ClustalW [4] or MEGA4 [5]. Before submitting the alignment for analysis, the user has to select one of the sequences to be considered the template for primer design, normally corresponding to a cDNA of the target isozyme of the most closely related species
, and choose between left and right primer design
As the Primer3 core is used as the primer design engine, the user can also modify the different criteria that the Primer3 engine should use to pick all possible primers for that template sequence
It is also possible to sort the found primers by sequence location or output order (i.e. the same order by which the Primer3 core finds them ‐ F).
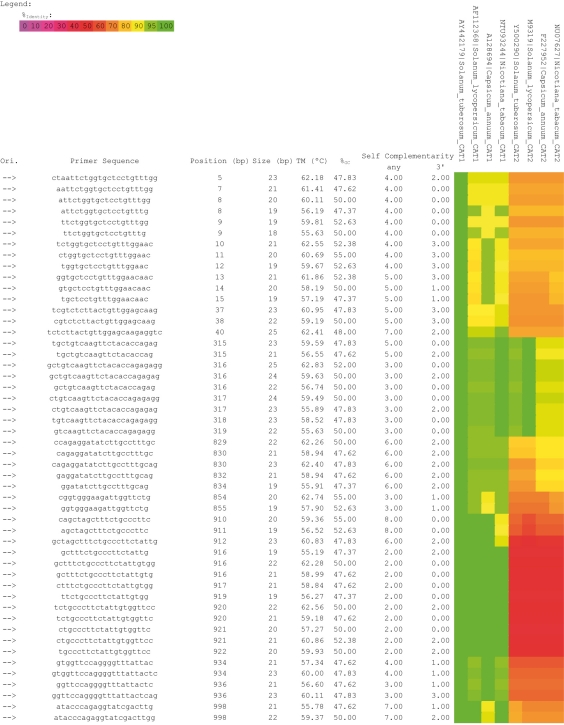
Figure 1.
PrimerIdent sample report for left primer analysis of Solanaceae Catalase (CAT) isozymes sequences. This sample report was trimmed in order to fit the page. Along with Solanum tuberosum (potato) CAT1 (AY442179 ― used as template sequence) the alignment also includes Solanum lycopersicum (tomato) CAT1 (AF112368), Capsicum annuum (chilli-pepper) CAT1 (AY128694), Nicotiana tabacum (tobacco) CAT1 (NTU93244), S. tuberosum CAT2 (AY500290), S. lycopersicum CAT2 (M93719), C. annum CAT2 (AB007190) and N. tabacum CAT2 (NTU07627). In this example it is possible to find primers with a high identity to CAT1 sequences and simultaneously an identity as low as possible for CAT2 sequences.
Output
PrimerIdent runs the Primer3 core in the background against the template sequence assigned in the web form, listing all possible primers for the template sequence, using the criteria submitted in the web interface by the user.
PrimerIdent will then calculate the identity of all the primers from that list to each sequence included in the alignment and renders a final HTML report with the list of all primers depicting for each one the identities calculated for each sequence and a colour encoded pattern for easy interpretation (Figure 2).The colour code reflects the degree of identity of each primer to the aligned sequences. The colour variation is a function of the identity match using the following references: 0% identity corresponds to the colour pink, 50% to red, 90% to yellow and 100% to green.The resulting PrimerIdent report can then be used to easily find a primer with a specific identity pattern that will suit the needs of the researcher. In the case of the sample report shown in Figure 2, PrimerIdent easily depicts primers with a high identity to Catalase 1 (CAT1) sequences and simultaneously an identity as low as possible for Catalase 2 (CAT2).
Figure 2.
PrimerIdent Web Interface. Although best viewed in Microsoft Internet Explorer 6 or above, PrimerIdent also works in other mainstream browsers, such as Mozilla FireFox, Apple Safari and Opera.
The HTML report includes useful information about the designed primers: their position in the alignment, size, melting temperature, GC percentage, 3' selfcomplementarity and any selfcomplementarity. This additional information helps researchers to choose the best primer for a certain application if more than one primer with the identity pattern that suits their needs is found. If a primer pair is required, then the researcher will have to get separate reports for left and right primers, see what primers have the identity pattern needed and choose the pair based on the primer position included in the report.
Acknowledgments
The authors wish to thank the Systems Management and Computation Unit, (Digital University Department, Reitoria ― University of Porto), especially to José António Sousa and Anabela Vieira for providing and supporting the web hosting facilities for http://primerident.up.pt.
Footnotes
Citation:Pessoa et al, Bioinformation 5(2): 52-54 (2010)
References
- 1.Gadberry MD, et al. Bioinformatics. 2005;21:1263–1264. doi: 10.1093/bioinformatics/bti134. [DOI] [PubMed] [Google Scholar]
- 2.Altschul SF, et al. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Rozen S, Skaletsky H. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 4.Thompson JD, et al. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamura K, et al. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]