Abstract
Purpose
Acquired somatic uniparental disomy (UPD) is commonly observed in myelodysplastic syndromes (MDS), myelodysplastic/myeloproliferative neoplasms (MDS/MPN), or secondary acute myelogenous leukemia (sAML) and may point toward genes harboring mutations. Recurrent UPD11q led to identification of homozygous mutations in c-Cbl, an E3 ubiquitin ligase involved in attenuation of proliferative signals transduced by activated receptor tyrosine kinases. We examined the role and frequency of Cbl gene family mutations in MPN and related conditions.
Methods
We applied high-density SNP-A karyotyping to identify loss of heterozygosity of 11q in 442 patients with MDS, MDS/MPN, MPN, sAML evolved from these conditions, and primary AML. We sequenced c-Cbl, Cbl-b, and Cbl-c in patients with or without corresponding UPD or deletions and correlated mutational status with clinical features and outcomes.
Results
We identified c-Cbl mutations in 5% and 9% of patients with chronic myelomonocytic leukemia (CMML) and sAML, and also in CML blast crisis and juvenile myelomonocytic leukemia (JMML). Most mutations were homozygous and affected c-Cbl; mutations in Cbl-b were also found in patients with similar clinical features. Patients with Cbl family mutations showed poor prognosis, with a median survival of 5 months. Pathomorphologic features included monocytosis, monocytoid blasts, aberrant expression of phosphoSTAT5, and c-kit overexpression. Serial studies showed acquisition of c-Cbl mutations during malignant evolution.
Conclusion
Mutations in the Cbl family RING finger domain or linker sequence constitute important pathogenic lesions associated with not only preleukemic CMML, JMML, and other MPN, but also progression to AML, suggesting that impairment of degradation of activated tyrosine kinases constitutes an important cancer mechanism.
INTRODUCTION
Mutations and genomic aberrations constitute key pathogenic lesions in myeloid malignancies. In primary acute myelogenous leukemia (pAML) and chronic myelogenous leukemia (CML), reciprocal translocations have enhanced our understanding of molecular pathogenesis, improved diagnosis and provided rational therapeutic targets. In myelodysplastic syndromes (MDS), myelodysplastic/myeloproliferative neoplasms (MDS/MPN), and AML evolved from MDS or MDS/MPN (secondary AML [sAML]), unbalanced chromosomal lesions predominate and loss of heterozygosity (LOH) is of particular importance. LOH can arise either via hemizygous deletion, where a DNA segment is lost from one homolog while the other remains at one copy per cell, or by uniparental disomy (UPD), wherein the retained homolog is duplicated to preserve two total copies per cell at the locus. Thus, analysis of recurrent regions of LOH may point toward the presence of important mutations. Mutations seen in MDS and AML affect specific classes of genes and indicate general pathways of leukemia evolution.1 For example, mutations have been found in a variety of receptor tyrosine kinases, including c-Kit, c-Mpl, and Flt-3.2–4 Mutations also affect signal transduction genes such as Jak2 and NPM-1. tP53 constitutes an example of a proapoptotic tumor suppressor gene mutated in aggressive leukemias.5–7
Until recently, metaphase cytogenetics was applied for detection of chromosomal defects including deletions resulting in LOH. Using this technique, a number of invariant chromosomal abnormalities have been described and minimal affected regions delineated, pointing towards potentially pathogenic genes. Single nucleotide polymorphism array (SNP-A) –based cytogenetic analysis allows for better resolution of chromosomal defects, with identification of previously cryptic unbalanced lesions.8 In particular, SNP-A is able to identify UPD. We and others have recently shown that somatic UPD affecting various chromosomes can be found frequently in MDS, MDS/MPN, and sAML and have identified a number of recurrent areas.9–11 Initially, UPD9p was shown to lead to homozygosity of the Jak2 V617F mutation in MPN.12–14 Using SNP-A, we have demonstrated that other areas of UPD can also be associated with homozygous mutations, including UPD13p (Flt-3 ITD) and UPD1p (c-Mpl).15,16 Based on this paradigm, we recently identified a novel recurrent area of UPD at 11q, frequently present in chronic myelomonocytic leukemia (CMML) and AML evolved from atypical MDS/MPN, and through delineation of a commonly deleted region have identified mutations in c-Cbl.15 c-Cbl is an E3 ubiquitin ligase involved in degradation of activated receptor tyrosine kinases and other tyrosine kinases, including Src kinases. Consequently, mutations affecting the RING finger domain (RFD) may have a wide range of effects on proliferation regulation, crucial to both MPN and AML. In animal studies, c-Cbl knockout led to hyper-responsiveness to ligand stimulation and expansion of them in cell pools, overall resulting in a mild proliferative phenotype.17 However, RFD mutation knock-in in a c-Cbl−/− mouse model resulted in a myeloproliferative phenotype and leukemic evolution (W.Y. Langdon, personal communication, December 2008). In a transgenic MDS NUP98/HOX13 mouse model, progression to sAML with acquisition of RAS and c-Cbl mutations occurs frequently.18
We hypothesized that mutations inactivating oncogene degradation pathways may constitute a new class of molecular lesions in myeloid malignancies modifying current paradigms of leukemogenesis. We therefore investigated the presence of mutations in the Cbl family of E3 ubiqutin ligases in selected subtypes of malignant myeloid disorders and determined the phenotypic and functional features as well as clinical outcomes. Based on the study of these features we set out to discern the pathophysiologic principles of molecular dysfunction created by E3 ubiquitin ligase lesions.
METHODS
Patients
Bone marrow aspirates were collected from 442 patients with MDS (n = 115), MDS/MPN (n = 98), MPN (n = 22), sAML (n = 110) evolved from these conditions, and pAML (n = 97) seen at Cleveland Clinic and Johns Hopkins Hospital between 2003 and 2008 (Appendix Table A1, online only). Informed consent for sample collection was obtained according to protocols approved by institutional review boards. Diagnosis was confirmed at each primary institution and assigned according to WHO classification criteria.19
Single Nuclotide Polymorphism Array Analysis
High-density Affymetrix SNP-A (250K and 6.0 arrays; Affymetrix, Santa Clara, CA) were applied as a karyotyping platform to identify LOH on chromosome 11q. Lesions identified by SNP-A were compared to the Database of Genomic Variants20 (http://projects.tcag.ca/variation/) and an internal control series (n = 1,003) to exclude known copy number variations. To confirm all regions of LOH detected by 250K SNP-A, we repeated samples when possible on 6.0 arrays and analyzed using Genotyping Console version 2.0 (Affymetrix). Signal intensity was analyzed and SNP calls determined using Gene Chip Genotyping Analysis Software version 4.0 (GTYPE, Affymetrix). Copy number and areas of UPD were investigated using a Hidden Markov Model and CN Analyzer for Affymetrix GeneChip Mapping 250K arrays (CNAG version 3.0) as previously described.21
E3 Ubiquitin Ligase Mutational Screening
To screen patients for mutations in c-Cbl, Cbl-b, Cbl-c, and Hakai, direct genomic sequencing of all exons was performed (details of primers and conditions are available on request). For sequencing, 250 ng of polymerase chain reaction (PCR) product, 3 μmol/L original forward or reverse primer, 2 μL Big Dye version 3.1 (Applied Biosystems, Foster City, CA), and 14.5 μL deionized H2O were amplified under the following conditions: 95°C (2 minutes) followed by 25 cycles of 95°C (10 seconds), 50°C (5 seconds), and 60°C (4 minutes). Sequencing was performed as previously described.15 If a mutation was intronic, RNA was extracted by TRIzol (Invitrogen, Carlsbad, CA) and reverse transcription polymerase chain reaction performed for confirmation of splice variants.
Immunohistochemical Detection of pSTAT5
Staining was performed on a Benchmark XT platform (Ventana Medical Systems, Tucson, AZ), according to the manufacturer's instructions, using mouse monoclonal antiphospho-STAT5a/b (Y694/99; Advantex BioReagents LLP, Conroe, TX) at 1:500 dilution. All stains were scored without knowledge of the clinical diagnosis or mutational status. Phospho-STAT5-positive staining (nMEG pSTAT5) was defined as previously reported.22,23 Images were obtained via digital microscopy using an Olympus BX51 microscope (Olympus America, Melville, NY) equipped with either a UPlanFl 40×/0.75 numeric aperture (NA) or a UPlanFl 100 ×/1.30 NA objective. Images were captured using a Dage-MTI Model DC330E charge-coupled device camera (Dage-MTI, Michigan City, IN) attached to the microscope with a U-TV1X-2 video adapter (Olympus America) and a 0.45× camera coupler (Diagnostic Instruments, Sterling Heights, MI).
Statistical Analysis
Overall survival was defined as the time a patient was diagnosed with a myeloid malignancy at Cleveland Clinic or Johns Hopkins to death or last known contact, and analyzed using Kaplan-Meier statistics and Cox's proportional hazards model. For comparison of the frequency of clinical features between Cbl family mutation and wild-type (WT), categoric variables were analyzed using Fisher's exact test.
RESULTS
Detection of UPD11q Using SNP-A and Detection of c-Cbl Mutation
Previously, we identified homozygous c-Cbl mutations in patients with CMML and UPD11q.15 Using SNP-A karyotyping, we studied a large cohort of patients (n = 442) with MDS and related disorders, including JMML and CML blast crisis (CML-BC), to assess the frequency of this lesion within clinical subtypes. Three hundred one and 187 cases were examined by 250K and 6.0 arrays, respectively. Forty-six cases were analyzed using both arrays, yielding identical results. Based on the analysis of 1,003 controls, we determined the average size and location of nonclonal regions of autozygosity. All of these nonclonal regions were interstitial. For the purpose of this study, we have excluded all regions of autozygosity based on the size criteria (27 Mb) derived from controls. The remaining regions were confirmed by analysis of germ-line samples (nonclonal CD3+ lymphocytes) in 45 patients (Fig 1A). Regions of homozygosity found in both bone marrow and CD3+ fractions were excluded from further analyses. We confirmed UPD11q detected on 250K arrays by repeated analyses using ultra-high density Affymetrix 6.0 arrays and Genotyping Console version 2.0 software (Fig 1B). Among a total of 133 regions of somatic UPD on multiple chromosomes including 1, 4, 17, and 21, UPD11q was most common (n = 17). LOH can also result from deletions, and deletions involving 11q23.3 were found in 29 patients (Fig 1C). Sequencing c-Cbl revealed mutations in 13 cases (76%) of UPD11q. However, among patients with deletion11q, a c-Cbl mutation was found in only one case (CMML). We also analyzed patients without LOH11q to assess the frequency of heterozygous mutations and identified five cases (1.2%).
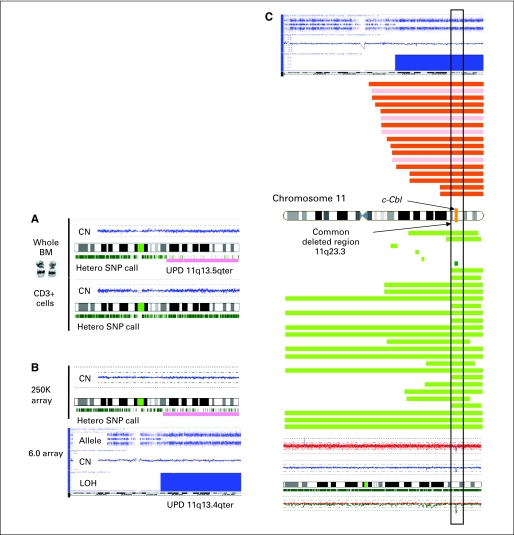
Fig 1.
Detecting acquired, segmental uniparental disomy (UPD) using single nucleotide polymorphism array (SNP-A) technology. (A) SNP-A karyograms of both whole bone marrow (BM) cells and CD3+ lymphocytes of a patient show the somatic nature of acquired UPD in chromosome 11. The blue line represents average copy number (CN) signal intensity of SNPs on the array chip. In this instance, there are no CN variations, and thus, the blue line does not deviate from normal diploid CN. The green marks below the idiogram represent heterozygosity at particular DNA loci. In the region of UPD seen in whole BM cells, drastic reduction of heterozygous loci denotes the region of UPD (pink bar). The remaining green marks in the region of UPD delineate the presence of nonclonal cells in the sample. An abnormal chromosome 11 was not detected when metaphase cytogenetic analysis was performed on bone marrow. In CD3+ sorted lymphocytes, a normal chromosome 11 is seen. (B) Comparison of Affymetrix 250K and 6.0 arrays in the detection of UPD11q. CNAG version 3.0 analysis (top) shows clear UPD of chromosome 11 by loss of heterozygous loci. Repeated testing on the 6.0 array and analysis using Genotyping Console v2.0 software (bottom) confirms the 250K SNP-A findings. Note that the Genotyping Console output includes allele difference, loss of heterozygosity (LOH), and CN variation plots. The allele difference graph represents the genotypes for each individual SNP. Dots with a value of 1 represent SNPs with an AA genotype, whereas those with a value of −1 represent SNPs with a BB genotype. Dots at 0 represent heterozygous SNPs (AB). Complete loss of all AB SNPs indicates copy-neutral LOH. This is further shown by both the LOH and CN graphs, which show no loss in CN but clear LOH. (C) Topographic maps show regions of (red) UPD or (green) deletion in individual patients on chromosome 11q. Bars corresponding to the ideogram represent the regions affected for each patient. The c-Cbl locus was included in the common deleted region (11q23.3) among UPDs and deletions. Red and pink bars represent UPD lesions with and without c-Cbl mutations, while dark and light green bars show deletions with and without mutations. Top is 6.0 array karyogram with UPD11q and bottom is one with microdeletion 11q by 250K array analysis. c-Cbl mutations were identified in both cases.
c-Cbl Mutations and Clinical Features
We sequenced all exons of c-Cbl; all mutations, except for 1 frame shift mutation in the tyrosine kinase–binding domain, were associated with the RFD or linker sequence, which are highly conserved among species. More importantly, 12 mutations in the RFD were located at or next to a cysteine residue (63%; Fig 2). The presence of each somatic mutation was confirmed by bidirectional DNA sequencing of multiple isolates and comparison against CD3+ sorted lymphocytes when possible.
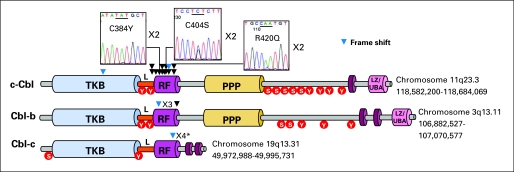
Fig 2.
Identification of variations in c-Cbl, Cbl-b and Cbl-c RING finger (RF) domain. Schematic representation shows the major domains of c-Cbl, Cbl-b, and Cbl-c, primarily the tyrosine kinase binding (TKB) domain, linker sequence (L), RF domain, proline-rich region (PPP), and leucine zipper (LZ)/ubiquitin-associated domain (UBA). Tyrosine and serine residues, represented by red circles, are phosphorylated by tyrosine kinases. Genomic DNA sequencing of all exons in c-Cbl, Cbl-b, and Cbl-c revealed the presence of (black arrow) missense and (blue arrow) frame shift mutations or frame shift polymorphisms in L or RF domain, except for a case with a mutation in the TKB domain. In c-Cbl, some basepair changes occured in a homozygous state because of UPD and resulted in the substitution of cysteine or arginine residues at positions 384 shared in two patients (C384Y), 404 in three patients (C404S/Y), and 420 in three patients (R420Q/P). (*) Frame shift polymorphism.
In one case of a patient with MDS (refractory anemia subtype) and monosomy 7 who transformed to AML, SNP-A karyotyping revealed the UPD11q, yet after transformation to AML sequencing identified a c-Cbl mutation creating a novel splice site resulting in a longer transcript (Appendix Fig A1, online only). We also found a hemizygous mutation in the linker sequence in a CMML patient with a microdeletion of 11q23.3 previously undetected by metaphase cytogenetics (Appendix Fig A2, online only). Eleven mutations were found in CMML and similar forms of MDS/MPN unclassifiable either at or before the time of testing; 6 of these patients progressed to sAML (55%). In total, two (5%) of 38 patients with CMML, 10 (9%) of 110 with sAML, and one (1%) of 115 patients with MDS carried mutant c-Cbl (Appendix Table A2, online only). In addition, c-Cbl mutations were found in four (19%) of 21 patients with JMML and one (10%) of 10 with CML-BC.
Other Mutations and Nonsynonimous SNPs in Related E3 Ubiquitin Ligases
The Cbl family contains several other E3 ligases, including Cbl-b (3q) and Cbl-c (19q) as well as a novel member with high homology called Hakai (7q). Based on structural and functional similarities we hypothesized that these genes can also harbor mutations associated with similar clinical phenotypes. When we sequenced 12 patients with corresponding UPDs, no mutations were found. However, when we sequenced all patients in our cohort without c-Cbl mutations, we identified three with a heterozygous and one with a hemizygous Cbl-b mutation (Appendix Fig A3, online only) and three patients (one cell line) with a Cbl-c frame shift polymorphism with a single base insertion, all affecting the RFD (Fig 2). In addition, we identified 12 (7%) of 167 patients with myeloid malignancies harboring another rare nonsynonymous SNP in Cbl-c, also affecting the RFD (H405Y). The frequency of this SNP in the general population is less than 1% (data not shown).
Clinical Characteristics in Patients With Cbl Family Mutation
In order to identify the pathologic subtypes in which Cbl family member mutations may play a role, we systematically investigated a wide range of myeloid malignancies. They were most commonly associated with MDS/MPN subentities, including CMML and atypical MDS/MPN, and some cases of typical MDS. In addition, Cbl family mutations appear to be present in sAML with an antecedent history of MDS/MPN. Based on this distribution of c-Cbl mutations, we analyzed other related disease entities within MDS/MPN and MPN; we also identified c-Cbl mutations in JMML and in CML-BC (Appendix Table A2). This pattern suggests that c-Cbl mutations can be a characteristic feature of atypical MDS/MPN syndromes or serve as a second genetic hit facilitating malignant evolution. Our study demonstrates the ubiquitous nature of Cbl family mutations, in particular in the context of very circumscribed phenotypes of myelomonocytic neoplasms.
Several common clinical features were identified among 27 patients affected by the Cbl family of variants, including monoblast or monocyte proliferation (74%), splenomegaly (81%), and surface expression of c-kit on malignant cells (93%). Characteristic nuclear features included abnormal lobulation and hyperchromatic and raisinoid nuclei of megakaryocytes seen in patients with CMML (Appendix Fig A4, online only). By immunohistochemistry, c-Cbl mutant megakaryocyte nuclei displayed aberrant pSTAT5 staining (86%; Appendix Fig A4), while pSTAT5 was not expressed in patients without c-Cbl mutations. In three cases, pSTAT5 expression was not detected in specimens obtained before the mutation was present (Appendix Fig A4). The clinical phenotype of patients with Cbl-b mutations was not distinguishable from that of patients with c-Cbl mutations. The allelic pattern of Cbl family mutations plays an important role in disease phenotype in patients; in sAML eight (62%) of 13 c-Cbl family mutations were homozygous (Fig 3A).
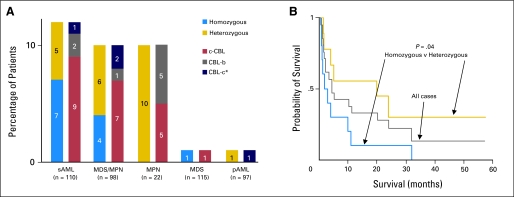
Fig 3.
Cbl family mutations in myeloid malignancies and unique clinical characteristics of patients with mutations. (A) Cbl family mutations are frequently observed in secondary acute myelogenous leukemia (sAML; 12%), myelodysplastic/myeloproliferative neoplasms (MDS/MPN; 10%), and MPN (10%). Homozygous mutations of c-Cbl are more frequent in sAML (7%) than MDS/MPN (4%). Cbl-b mutations and/or Cbl-c frame shift polymorphism (*) are seen in all disease phenotypes. (B) Kaplan-Meier analysis shows overall survival in all cases (n = 20; gray line). Median survival is 5 months and the survival rate is 10%. By comparing patients homozygous for Cbl family mutation (n = 10; blue line) to those heterozygous (n = 10; gold line), there is a statistically significant difference in overall survival between these two cohorts regardless of treatment or remission/relapse.
To evaluate the impact of Cbl family mutations, we first compared mutant and WT cases on the basis of various clinical parameters (Table 1). In mutant cases, monocyte counts were significantly higher as compared with patients with WT Cbl family genes. A higher proportion of patients with c-Cbl mutations were treated with intense chemotherapy or stem cell transplantation, suggesting that these therapies were more frequently selected because of the aggressive biology of the Cbl mutation-associated disease. When we performed univariate analysis of survival impact of various clinical variables, significant differences in WBC, monocyte counts, disease risk, and the presence of Cbl family gene mutations (Table 2) were found. Chemotherapy was an adverse risk factor for survival, likely as it correlated with more advanced disease in multivariate analyses. The prognosis of patients with Cbl family mutations was poor, especially in those with homozygous mutations. The median overall survival was 5 months for all patients, with a significant difference of 1.7 months versus 20 months for patients with homozygous and heterozygous mutations, respectively (P = .04; Fig 3B). In multivariate analyses advanced disease (hazard ratio [HR], 5.64; 95% CI, 3.8 to 8.36) and Cbl family mutations (HR, 2.17; 95% CI, 1.18 to 4.02; Table 3) were shown to be independent adverse factors for overall survival.
Table 1.
Comparison of Clinical Characteristics Between Wild-Type and Mutant Cbl Family Genes
| Variable* | Cbl Family Mutant (n = 17) | Cbl Family Wild Type (n = 307) | P |
|---|---|---|---|
| Age, years | |||
| ≥ 60 | 10 | 224 | .21 |
| < 60 | 6 | 62 | |
| Sex | |||
| Male | 8 | 183 | .2 |
| Female | 9 | 106 | |
| WBC count, ×109/L | |||
| ≥ 10 | 9 | 93 | .11 |
| < 10 | 8 | 197 | |
| Monocyte count, ×109/L | |||
| ≥ 1 | 13 | 53 | < .0001 |
| < 1 | 4 | 237 | |
| Metaphase cytogetetics | |||
| Abnormal | 10 | 133 | .62 |
| Normal | 7 | 139 | |
| Disease risk† | |||
| Advanced grade | 12 | 141 | .7 |
| Low grade | 4 | 142 | |
| Therapy and response | |||
| Chemotherapy‡ | 14 | 130 | .001 |
| Stem cell transplantation | 4 | 21 | .035 |
| Complete remission | 3 | 22 | .73 |
Some clinical data are not available.
In advanced group, secondary acute myelogenous leukemia, refractory anemia with excess blasts, and chronic myelomonocytic leukemia 2. The others are in low grade group
Chemotherapy includes mitoxantrone, idarubicin, daunorubicin, cytarabine, etoposide, hydroxyurea, fludarabine gemtuzumab, 5-azacitidine, decitabine, lenalidomide, arsenic trioxide, and valproic acid.
Table 2.
Univariate Analysis of Overall Survival in Clinical Variables
| Variable* | No. of Patients | Overall Survival |
|||
|---|---|---|---|---|---|
| Mean (months) | Hazard Ratio | 95% CI | P | ||
| Age, years | |||||
| ≥ 60 | 220 | 32 | 1.23 | 0.83 to 1.82 | .31 |
| < 60 | 64 | 36 | |||
| Sex | |||||
| Male | 176 | 31 | 1.37 | 0.98 to 1.91 | .07 |
| Female | 108 | 36 | |||
| WBC count, ×109/L | |||||
| ≥ 10 | 79 | 25 | 1.83 | 1.30 to 2.57 | .001 |
| < 10 | 205 | 36 | |||
| Monocyte count | |||||
| ≥ 1 | 48 | 24 | 1.79 | 1.22 to 2.64 | .003 |
| < 1 | 236 | 35 | |||
| Metaphase cytogetetics | |||||
| Abnormal | 124 | 30 | 1.37 | 0.98 to 1.90 | .063 |
| Normal | 140 | 35 | |||
| Disease risk† | |||||
| Advanced grade | 141 | 18 | 6.25 | 4.27 to 9.15 | < .0001 |
| Low grade | 142 | 46 | |||
| Cbl family gene | |||||
| Mutation | 16 | 9 | 4.64 | 2.69 to 8.00 | < .0001 |
| Wild type | 268 | 34 | |||
| Chemotherapy | |||||
| Treatment | 131 | 26 | 2.13 | 1.54 to 2.93 | < .0001 |
| No treatment | 153 | 40 | |||
| Stem cell transplantation | |||||
| Treatment | 24 | 34 | 0.87 | 0.49 to 1.54 | .63 |
| No treatment | 260 | 33 | |||
| Complete remission | |||||
| Yes | 23 | 30 | 0.70 | 0.40 to 1.23 | .72 |
| No | 95 | 25 | |||
Some clinical data are not available.
In advanced group, secondary acute myelogenous leukemia, RAEB, and chronic myelomonocytic leukemia 2. The others are in low-grade group.
Table 3.
Multivariate Analysis of Overall Survival in Clinical Variables
| Variable* | Hazard Ratio | 95% CI | P |
|---|---|---|---|
| Disease risk (advanced grade/low grade) | 5.64 | 3.8 to 8.36 | < .0001 |
| Cbl family gene (mutation/wild type) | 2.17 | 1.18 to 4.02 | .013 |
WBC, monocyte count, disease risk, chemotherapy, and Cbl family mutation were included in multivariate analysis.
DISCUSSION
In leukemia, cytogenetic abnormalities identify underlying pathophysiology and carry enormous prognostic and therapeutic significance. Various examples of gain of function and inactivating mutations show that homologous recombination may lead to duplication of affected alleles.24 Consequently, areas of somatic UPD may point toward genes carrying putative pathogenic mutations. When high density SNP-A was applied as a karyotyping platform in a large number of patients with MDS and related disorders, we noted recurrent somatic UPD at chromosome 11q, particularly frequent in MDS/MPN. Commonly deleted region mapping and analysis of genes located within this region led us to hypothesize that c-Cbl may contain mutations. Sequencing of c-Cbl in patients affected by somatic UPD11q revealed RFD and linker sequence mutations present in patients with CMML, MDS/ MPN unclassifiable, and sAML derived from these conditions or MDS. We also found new mutations in Cbl-b with a clinical phenotype similar to that seen with c-Cbl RFD mutations. Consequently, our results imply that E3 ubiquitin ligases constitute a novel class of genes in whom mutations reflect a novel general mechanism of leukemogenesis. This notion is supported by the variety of pathomorphologic subentities of myeloid malignancies affected by mutations of the Cbl family. Moreover, a novel frame shift polymorphism was found in Cbl-c in patients with MDS/MPN. However, the relevance of this otherwise extremely rare polymorphism is not clear as the corresponding gene does not show significant expression in myeloid cells.
Cas-Br-M, a retrovirus, contains v-Cbl which corresponds to about one third of the murine c-Cbl gene and contains only the murine phosphotyrosine binding domain.25 This virus consistently induces a type of pre-B cell lymphoma in infected mice. The importance of c-Cbl in hematopoiesis has been previously demonstrated in knockout mice that show hyper-responsiveness to hematopoietic growth factors, expansion of the progenitor and stem cell pool, and mild myeloproliferative features.17 However, recent results obtained with an RFD knock-in in a c-Cbl−/− mouse model parallels the phenotype observed in patients; the mutant mouse demonstrated a severe myeloproliferative phenotype (W.Y. Langdon, personal communication). Indeed, in patients mutations were predominantly located in the RFD and affected structurally essential cysteines, possibly led to inactivation of RFD function by frame shift, or created novel splicing sites resulting in larger transcripts. In addition, c-Cbl mutations were homozygous or hemizygous, implying that the presence of a WT allele is protective. It is likely that mutations do not lead to the simple knockout of c-Cbl function. Rather, by affecting the RFD, they render it a proto-oncogene, consistent with the oncogenic properties of v-Cbl. Previously, mutations of c-Cbl have been described in a limited number of patients with AML, but neither their function nor their clinical phenotype could be delineated without a comprehensive study of corresponding karyotypes and clinical outcomes.26–28
c-Cbl is a member of the Cbl family of E3 ubiquitin ligases, which poly- or monoubiquitinate a number of important tyrosine kinases serving as important transduction elements of proliferative signals and activated tyrosine kinase receptors, including Flt-3, c-kit, and M-CSF.29,30 Consequently, inactivation of ubiquitination may lead to enhanced and prolonged signaling, a function which can explain the phenotype in patients (Fig 4). Based on this essential role of E3 ligases, we hypothesized that another Cbl family member, Cbl-b, may also be affected by mutations in myeloid malignancies. Sequencing of these genes in patients who did not harbor c-Cbl mutations revealed that these genes can also be affected by mutations leading to the inactivation of the RFD. These patients displayed a clinical phenotype analogous to those with c-Cbl mutations. The clinical features corresponding to c-Cbl mutations included monocytic features, aberrant and increased phosphorylation of pSTAT5, and monocytoid blasts. An increased frequency of mutations in patients with frank AML may argue either that c-Cbl mutations lead to an invariant progression to an aggressive phenotype, or that they constitute a second hit event frequently occurring in the context of atypical myeloproliferative disorders. In fact, unlike other reports that looked at patients with inv16, we found no mutation of Cbl family genes in de novo AML, including French-American-British type M4 or M5.28 Both theories are supported by the dismal prognosis of patients with Cbl family mutations. The close association of c-Cbl mutations with monocyte expansion, such as that seen in JMML, CMML, or sAML with monocytoid features, suggests a primary role of c-Cbl mutations in the pathogenesis of these diseases, while occurrence of c-Cbl mutations during evolution to AML in serially studied patients and the high proportion of cases with advanced leukemia affected by c-Cbl mutations argues for its auxiliary facilitator role.
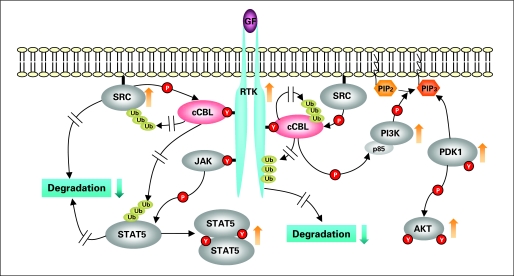
Fig 4.
Potential intracellular consequences of c-Cbl mutations. c-Cbl is a member of the E3 ubiquitin ligase Cbl family, which poly- or monoubiquitinate a number of important receptor tyrosine kinases (RTK), including Flt-3, c-kit, and CSF-1, for degradation. Inactivation of ubiquitination activity through mutations occurring in RING finger domain may lead to enhanced and/or prolonged hematopoietic growth factor signaling, a function which can contribute to the clinical phenotype of patients with c-Cbl mutation. However, in addition to RTK, knockout of c-Cbl ubiquitination activity may result in enhanced phosphorylation of SRC kinase, STAT5, and also c-Cbl itself. Ultimately, the elevated level of important transduction factors such as STAT5 and PI3K as well as increased SRC kinase activity can lead to aberrant proliferative responses.
Taken together, our data suggests that Cbl family mutations constitute a novel class of pathogenic molecular lesions associated with a spectrum of myeloid malignancies characterized by myeloproliferative features and poor prognosis. Inactivation of the RFD, and thereby ubiquitination involved in downmodulation of proliferative signaling, constitutes a general mechanism of leukemogenesis likely present in a variety of malignancies.
Appendix
Fig A1.
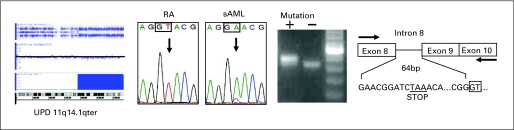
In a patient with secondary acute myelogenous leukemia (sAML) transformed from myelodysplastic syndrome refractory anemia with monosomy 7, uniparental disomy (UPD) 11q14.1qter was detected only after AML evolution by single nucleotide polymorphism assay analysis, at which time a homozygous mutation was seen in the intron 8 splice donor site of c-Cbl. This mutation results in a splice variant, leading to a longer transcript with a frame shift in RF domain.
Fig A2.
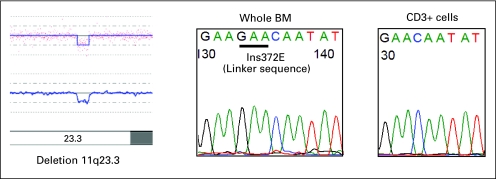
A microdeletion 11q23.3 is detected in c-Cbl in a patient with chronic myelomonocytic leukemia 1, leading to a hemizygous insertion mutation in the linker sequence. BM, bone marrow.
Fig A3.
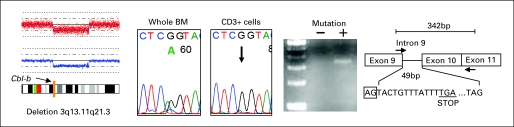
In a patient with sAML evolved from myelodysplastic/myeloproliferative neoplasms, a chromosome 3 abnormality was seen by metaphase cytogenetics and single nucleotide polymorphism array analysis revealed a deletion in the region of Cbl-b. Direct sequencing of whole bone marrow (BM) showed a hemizygous mutation (1204-51 G>A) in intron 9, whereas this mutation is not detected in CD3+ lymphocytes. Reverse transcription polymerase chain reaction revealed the utilization of a novel splice acceptor site and mutant-specific amplification, resulting in a frame shift in the RF domain.
Fig A4.
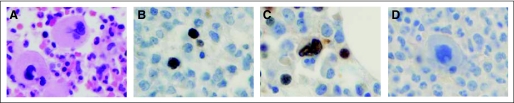
Characteristic features of megakaryocytes in patients with c-Cbl mutations. (A) A patient with chronic myelomonocytic leukemia (CMML) with bone marrow fibrosis and c-Cbl mutation shows abnormal megakaryocytes (abnormal lobulation, hyperchromatic and raisinoid nucleus). pSTAT5 staining was positive in the nuclei of megakaryocytes and erythroid precursors in a patient with secondary acute myelogenous leukemia (B), and refractory anemia with excess blasts 1 (C); both had a c-Cbl mutation. (D) In comparison, pSTAT5 staining was negative in megakaryocytes of a patient in refractory anemia phase before acquiring c-Cbl mutation. Original magnification, ×100.
Table A1.
Diagnosis and Metaphase Cytogenetics Findings of Enrolled Patients (n = 442)
| Diagnosis by Subgroup | No. | Abnormal Metaphase Cytogenetics (%) |
|---|---|---|
| MDS | ||
| RA/RARS/RCMD/RCMD-RS/5q/MDS-U | 77 | 48 |
| RAEB I/II | 38 | 42 |
| MDS/MPN | ||
| MDS/MPNu | 39 | 31 |
| CMML I/II | 38 | 42 |
| JMML | 21 | 19 |
| MPN | ||
| PV/PMF/ET | 12 | 17 |
| CML blast crisis | 10 | 100 |
| AML | ||
| pAML | 97 | 56 |
| sAML | ||
| From MDS | 64 | 39 |
| From MDS/MPN | 36 | 48 |
| From MPN | 10 | 20 |
Abbreviations: sAML, secondary acute myelogenous leukemia; RA, refractory anemia; RARS, refractory anemia with ringed sideroblasts; RCMD, refractory cytopenia with multilineage dysplasia; RCMD-RS, RCMD with ringed sideroblasts; RAEB, refractory anemia with excess blasts; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasms; MPNu, MPN unclassifiable; CMML, chronic myelomonocytic leukemia; JMML, juvenile myelomonocytic leukemia; PV, polycythemia vera; PMF, primary myelofibrosis; ET, essential thrombocytopenia; CML, chronic myelogenous leukemia; pAML, primary acute myelogenous leukemia.
Table A2.
Disease Phenotype of Patients With Cbl Family Mutations
| Diagnosis |
Cbl Family Variants |
|||
|---|---|---|---|---|
| All Cbl Family | c-Cbl | Cbl-b | Cbl-c* | |
| MDS/MPN to sAML | 6 of 36 | 5 | 1 | 0 |
| MDS/MPNu | 3 of 39 | 1 | 1 | 1 |
| CMML | 2 of 38 | 2 | 0 | 0 |
| JMML | 5 of 21 | 4 | 0 | 1 |
| MDS to sAML | 6 of 64 | 4 | 1 | 1† |
| RAEB | 1 of 38 | 1 | 0 | 0 |
| Low-grade MDS | 0 of 77 | 0 | 0 | 0 |
| MPN to sAML | 1 of 10 | 1 | 0 | 0 |
| MPN | 0 of 12 | 0 | 0 | 0 |
| CML blast crisis | 2 of 10 | 1 | 1 | 0 |
| CBF pAML | 0 of 27 | 0 | 0 | 0 |
| Non-CBF pAML | 1 of 70 | 0 | 0 | 1 |
| Total | 27 of 442 | 19 | 4 | 4 |
Abbreviations: MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasms; sAML, secondary acute myelogenous leukemia; MPNu, MPN unclassifiable; CMML, chronic myelomonocytic leukemia; JMML, juvenile myelomonocytic leukemia; RAEB, refractory anemia with excess blasts; CBF, core binding factor; pAML, primary acute myelogenous leukemia.
Cbl-c frame shift polymorphism. Cbl-c gene is not known to be expressed in myeloid malignancies.
Cell line from a patient with secondary AML (sAML).
Footnotes
Supported by in part by Grants No. RO1HL-082983, U54 RR019391 (J.P.M., M.A.S.), K24 HL-077522, DOD-MP048018 (M.A.M.), and by a grant from Aplastic Anemia & MDS International Foundation and Robert Duggan Charitable Fund (J.P.M.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Mikkael A. Sekeres, Celgene (C), Pharmion (C); Jaroslaw P. Maciejewski, Eisai (C) Stock Ownership: None Honoraria: None Research Funding: Mikkael A. Sekeres, Celgene; Jaroslaw P. Maciejewski, Celgene, Genzyme Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Hideki Makishima, Seiji Kojima, Alan F. List, Michael A. McDevitt, Jaroslaw P. Maciejewski
Financial support: Mikkael A. Sekeres, Michael A. McDevitt, Jaroslaw P. Maciejewski
Administrative support: Christine O'Keefe, Michael A. McDevitt, Jaroslaw P. Maciejewski
Provision of study materials or patients: Hideki Makishima, Heather Cazzolli, Andrew Dunbar, Hideki Muramatsu, Ronald L. Paquette, Seiji Kojima, Alan F. List, Mikkael A. Sekeres, Michael A. McDevitt, Jaroslaw P. Maciejewski
Collection and assembly of data: Hideki Makishima, Heather Cazzolli, Andrew Dunbar, Ramon Tiu, Jungwon Huh, Hideki Muramatsu, Christine O'Keefe, Eric Hsi, Ronald L. Paquette, Alan F. List, Michael A. McDevitt, Jaroslaw P. Maciejewski
Data analysis and interpretation: Hideki Makishima, Hadrian Szpurka, Ramon Tiu, Jungwon Huh, Christine O'Keefe, Eric Hsi, Ronald L. Paquette, Michael A. McDevitt, Jaroslaw P. Maciejewski
Manuscript writing: Hideki Makishima, Hadrian Szpurka, Ramon Tiu, Christine O'Keefe, Ronald L. Paquette, Alan F. List, Mikkael A. Sekeres, Michael A. McDevitt, Jaroslaw P. Maciejewski
Final approval of manuscript: Hideki Makishima, Heather Cazzolli, Hadrian Szpurka, Andrew Dunbar, Hideki Muramatsu, Christine O'Keefe, Eric Hsi, Ronald L. Paquette, Seiji Kojima, Alan F. List, Mikkael A. Sekeres, Michael A. McDevitt, Jaroslaw P. Maciejewski
REFERENCES
- 1.Gilliland DG. Hematologic malignancies. Curr Opin Hematol. 2001;8:189–191. doi: 10.1097/00062752-200107000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Schnittger S, Kohl TM, Haferlach T, et al. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood. 2006;107:1791–1799. doi: 10.1182/blood-2005-04-1466. [DOI] [PubMed] [Google Scholar]
- 3.Pikman Y, Lee BH, Mercher T, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3:e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mrozek K. Cytogenetic, molecular genetic, and clinical characteristics of acute myeloid leukemia with a complex karyotype. Semin Oncol. 2008;35:365–377. doi: 10.1053/j.seminoncol.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frohling S, Lipka DB, Kayser S, et al. Rare occurrence of the JAK2 V617F mutation in AML subtypes M5, M6, and M7. Blood. 2006;107:1242–1243. doi: 10.1182/blood-2005-09-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 7.Haferlach C, Dicker F, Herholz H, et al. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22:1539–1541. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]
- 8.Maciejewski JP, Mufti GJ. Whole genome scanning as a cytogenetic tool in hematologic malignancies. Blood. 2008;112:965–974. doi: 10.1182/blood-2008-02-130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohamedali A, Gaken J, Twine NA, et al. Prevalence and prognostic significance of allelic imbalance by single-nucleotide polymorphism analysis in low-risk myelodysplastic syndromes. Blood. 2007;110:3365–3373. doi: 10.1182/blood-2007-03-079673. [DOI] [PubMed] [Google Scholar]
- 10.Gondek LP, Dunbar AJ, Szpurka H, et al. SNP array karyotyping allows for the detection of uniparental disomy and cryptic chromosomal abnormalities in MDS/MPD-U and MPD. PLoS ONE. 2007;2:e1225. doi: 10.1371/journal.pone.0001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gondek LP, Tiu R, O'Keefe CL, et al. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008;111:1534–1542. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 13.Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 14.Baxter EJ, Scott LM, Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 15.Dunbar AJ, Gondek LP, O'Keefe CL, et al. 250K SNP array karyotyping identifies acquired uniparental disomy and homozygous mutation, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;15:10349–10357. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szpurka H, Gondek LP, Mohan SR, et al. UPD1p indicates the presence of MPL W515L mutation in RARS-T, a mechanism analogous to UPD9p and JAK2 V617F mutation. Leukemia. 2009;23:610–614. doi: 10.1038/leu.2008.249. [DOI] [PubMed] [Google Scholar]
- 17.Murphy MA, Schnall RG, Venter DJ, et al. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol Cell Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slape C, Liu LY, Beachy S, et al. Leukemic transformation in mice expressing a NUP98-HOXD13 transgene is accompanied by spontaneous mutations in Nras, Kras, and Cbl. Blood. 2008;112:2017–2019. doi: 10.1182/blood-2008-01-135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swerdlow SH, Campo E, Harris NL, et al. ed 4. Lyon, France: IARC Press; 2008. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 20.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 21.Gondek LP, Tiu R, Haddad AS, et al. Single nucleotide polymorphism arrays complement metaphase cytogenetics in detection of new chromosomal lesions in MDS. Leukemia. 2007;21:2058–2061. doi: 10.1038/sj.leu.2404745. [DOI] [PubMed] [Google Scholar]
- 22.Szpurka H, Tiu R, Murugesan G, et al. Refractory anemia with ringed sideroblasts associated with marked thrombocytosis (RARS-T), another myeloproliferative condition characterized by JAK2 V617F mutation. Blood. 2006;108:2173–2181. doi: 10.1182/blood-2006-02-005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aboudola S, Murugesan G, Szpurka H, et al. Bone marrow phospho-STAT5 expression in non-CML chronic myeloproliferative disorders correlates with JAK2 V617F mutation and provides evidence of in vivo JAK2 activation. Am J Surg Pathol. 2007;31:233–239. doi: 10.1097/01.pas.0000213338.25111.d3. [DOI] [PubMed] [Google Scholar]
- 24.Lupski JR, Stankiewicz P. Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blake TJ, Shapiro M, Morse HC, III, et al. The sequences of the human and mouse c-cbl proto-oncogenes show v-cbl was generated by a large truncation encompassing a proline-rich domain and a leucine zipper-like motif. Oncogene. 1991;6:653–657. [PubMed] [Google Scholar]
- 26.Caligiuri MA, Briesewitz R, Yu J, et al. Novel c-CBL and CBL-b ubiquitin ligase mutations in human acute myeloid leukemia. Blood. 2007;110:1022–1024. doi: 10.1182/blood-2006-12-061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sargin B, Choudhary C, Crosetto N, et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007;110:1004–1012. doi: 10.1182/blood-2007-01-066076. [DOI] [PubMed] [Google Scholar]
- 28.Abbas S, Rotmans G, Lowenberg B, et al. Exon 8 splice site mutations in the gene encoding the E3-ligase CBL are associated with core binding factor acute myeloid leukemias. Haematologica. 2008;93:1595–1597. doi: 10.3324/haematol.13187. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005;6:907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- 30.Swaminathan G, Tsygankov AY. The Cbl family proteins: Ring leaders in regulation of cell signaling. J Cell Physiol. 2006;209:21–43. doi: 10.1002/jcp.20694. [DOI] [PubMed] [Google Scholar]