Linkage analysis demonstrates the unique contribution of chromosomal loci to the variability of total and free thyroxine in different sets of recombinant inbred mice.
Abstract
C3H/He and BALB/c mice have elevated serum thyroxine levels associated with low deiodinase type-1 activity whereas C57BL/6 (B6) mice have low thyroxine levels and elevated deiodinase type-1 activity. High-resolution genetic maps are available for four sets of recombinant inbred (RI) mice derived from B6 parents bred to C3H/He, BALB/c, DBA/2, or A strains. Total and free T4 (T-T4 and F-T4) levels in females from these RI sets (BXH, CXB, BXD, and AXBXA) were analyzed to test two hypotheses: first, serum T4 variability is linked to the deiodinase type-1 gene; second, because of their shared B6 parent, the RI sets will share linkages responsible for T-T4 or F-T4 variability. A number of chromosomes (Chr) and loci were linked to T-T4 (Chr 1, 4, 13, 11) or F-T4 (Chr 1, 6, 13, 18, 19). Linkage between T-T4 and Chr 4 was limited to CXB and BXH strains, but the locus was distinct from the deiodinase type-1 gene. Surprisingly, many linkages were unique providing “genetic signatures” for T-T4 or F-T4 in each set of RI mice. Indeed, the strongest linkage between T-T4 (or F-T4) and a Chr 2 locus (logarithm of the odds scores >4.4) was only observed in AXBXA strains. Some loci corresponded to genes/Chr associated in humans with variable TSH or T-T4 levels. Unlike inbred mice, human populations are extremely diverse. Consequently, our data suggest that the contributions of unique chromosomes/loci controlling T-T4 and F-T4 in distinct human subgroups are likely to be “buried” in genetic analyses of heterogeneous human populations.
Recombinant inbred (RI) mice are generated from the progeny of two inbred strains, such as BALB/c and C57BL/6 (B6), followed by repeated brother–sister matings of the second filial generation (F2) for 20 generations or more to establish multiple stable homozygous lines (1). The availability of high-resolution genetic maps for sets of RI mice (e.g., CXB derived from BALB/c and B6 parental strains) provides a powerful tool for mapping chromosomal loci linked with selected phenotypic traits (e.g., Refs. 2, 3). Because of our interest in autoimmune thyroid disease, we have used the RI sets CXB and BXH (the latter derived from B6 and C3H/He parents) to identify genetic loci linked to the induction of pathological thyroid stimulating antibodies and hyperthyroidism in an induced model of Graves' disease (4–6). In the course of these studies we observed differences in serum T4 levels in CXB and BXH strains (4, 5). Expanding these observations to the other two RI sets (AXBXA and BXD) is the subject of the present report.
In humans, serum T4 levels vary widely in the population but remain within a narrow range in a particular individual. Twin studies have revealed that heritability contributes up to 65% toward the serum T4 level in an individual (e.g., Refs. 7–10). Similarly, thyroid function differences among mouse strains are well known, first described in 1961 (11) with subsequent extension to a wider range of inbred mouse strains (e.g., Ref. 12). C3H/He (C3H) and B6 mice are at opposite poles, with high and low serum T4 levels, respectively (13, 14). Most T4 in serum is bound to transport proteins, with only a very small ‘free’ fraction. Because total T4 (T-T4) and free T4 (F-T4) levels in C3H and B6 strains vary in parallel (13), an inherited difference in thyroid hormone binding proteins cannot be a major factor. After transport to target tissues, iodothyronine deiodinases catalyze the conversion of T4 to the biologically active hormone tri-iodothyronine (T3) as well as to inactive forms for elimination (reviewed in Ref. 15).
Conversion of T4 to T3 in peripheral tissues is regulated by 5′-deiodinase type-1 (D1), expressed primarily in liver and kidney. Despite variation in T-T4 and F-T4 levels among mouse strains, for example in widely disparate C3H (high T4) and B6 (low T4) mice, serum T3 levels are very similar in these two strains (13). Conversely, hepatic and renal D1 activities are low in C3H mice and high in B6 mice, thereby providing an explanation for the similarity in serum T3 levels. The genetic basis for variation in hepatic D1 enzyme activity among RI mouse strains derived from C3H and B6 parents (BXH), or from BALB and B6 parents (CXB), has been assessed using restriction fragment length polymorphism and revealed segregation characteristic of a single allele difference in the D1 gene (Dio1) on chromosome (Chr) 4 (13). A corollary of these data are that alleles at the Dio1 locus may contribute to the differences in serum T4 levels (T-T4 and F-T4) in different mouse strains.
We have now used multiple sets of RI mouse strains to test the hypothesis that serum T4 variability is linked, at least in part, to the Dio1 gene. In addition, because the four RI sets share B6 as one parent strain, we hypothesized that variability in serum T-T4 and F-T4 would be linked to chromosomes or loci shared by one or more of these RI sets. Unexpectedly, neither of these hypotheses was fulfilled.
Materials and Methods
Mouse strains
The following strains of mice (females, 5–8 weeks of age) were obtained from Jackson Laboratory (Bar Harbor, ME): 1) the parental strains of the recombinant inbred sets BXH, AXBXA, and BXD, namely, C57BL/6J, C3H/HeJ, DBA/2J, and A/J, as well as the most closely related parental strains of the CXB set, C57BL/6J and BALB/cJ; 2) AXB/PgnJ 1, 2, 4, 5, 6, 8, 10, 12, 13, 15, 19a, 23, 24; BXA/PgnJ 1, 2, 4, 7, 8, 11, 12, 13, 14, 16, 24, 25, 26. We had previously obtained female mice of the following strains from Jackson Labs: CXB/ByJ 1 to 7, CXB/HiAJ 8 to 13 (4), and BXH TyJ 2, 4, 6 to 11, 14, 19, BXH KccJ 20, 22, and B6cC3–1/KccJ (5). Sera were also drawn from female mice (7–13 weeks old) of the following strains bred at the University of Tennessee, Memphis: BXD11, 12, 13, 16, 32, 40, 43, 48, 49, 65, 67, 70, 73, 75, 80, 87, 97, 99, 100, 101, 103. Mice from Jackson Laboratory were fed LabDiet 5K52/5K67 or LabDiet 5K54 (containing 2.1 and 2.2 ppm iodine, respectively). All BXD mice (Memphis) received Harlan Teklad LM-485 (2.61 ppm iodine). It should be noted that all mice of a particular RI set received the same diet.
We restricted our study to female mice for the following reasons: 1) Graves' disease in humans occurs more frequently in women than in men; 2) T4 levels differ in male and female mice (12). In addition, because T4 levels peak earlier and are higher in DBA/2 than in B6 mice in early postnatal life (16), the mice we studied were young adults (average 5–13 weeks old). Parental strains are referred to as C3H, B6, DBA, A, and BALB; the RI set AXB/BXA as AXBXA. All studies were approved by the Institutional Animal Care and Use Committees (Los Angeles and Memphis) and performed with the highest standards of care in pathogen-free facilities.
Total and free thyroxine
T-T4 and F-T4 were measured by independent RIAs (Diagnostic Products Corporation, Los Angeles, CA). T-T4 levels were determined in 25 μl serum aliquots from individual mice for the parental strains (C3H, BALB, A, DBA, and B6) and RI strains (CXB, BXH and BXD); the numbers of mice used to obtain the mean T-T4 values are specified in the figure legends. For AXBXA mice, T-T4 was measured in duplicate aliquots (25 μl) of a serum pool for each RI strain. T-T4 levels have been reported for CXB, BXH, C3H, and B6 mice as “base-line” values before immunization with TSH receptor (TSHR) A-subunit adenovirus (4, 5). In the case of F-T4 assays, because of the requirement for a large (50 μl) volume, determinations were made on a serum pool for each RI strain. Thyroid hormone levels were computed from kit standards and expressed as μg/dl T- T4 and ng/dl F-T4.
Statistical analysis and linkage analysis
Comparisons between T-T4 and F-T4 levels in different mouse strains were performed by ANOVA (SigmaStat; Jandel Scientific Software, San Rafael, CA). Putative quantitative trait loci (QTL) involved in the variability of T-T4 and F-T4 were mapped using the genotype files for AXB/BXA, BXD, BXH and CXB strains generated by Williams et al. (17) available at http://www.nervenet.org and embedded in GeneNetwork (www.genenetwork.org). The probability of linkage between T-T4 and F-T4 and previously mapped genotypes was estimated at ∼1 centi-Morgan intervals (∼2 megabase; Mb) along the entire genome, except for the Y chromosome. To establish criteria for suggestive and significant linkage, a permutation test was performed (1000 permutations at 1-centimorgan intervals) (18). This test compares the peak likelihood ratio statistics (LRS; LRS = LOD × 4.6, where LOD is the logarithm of the odds) obtained for a given data set with the peak LRS score obtained for 1000 random permutations of the same data set. To avoid bias, analyses were performed without including data for the parental strains. It should be noted that LRS >25 is usually required to reach P < 0.05 genome-wide.
The phenotype observations have been entered into the databases in GeneNetwork (GN) (www.genenetwork.org) under the following trait accession identifiers: BXD GN 12585 (T-T4) and GN 12584 (F-T4); AXB/BXA GN 10162 (T-T4) and GN 10170 (F-T4); CXB GN 10516 (T-T4) and GN 10691 (F-T4); BXH GN 10150 (T-T4) and GN 10161 (F-T4). These data can be found by searching the CXB, BXH, AXB/BXA, or BXD databases for the name “McLachlan” (Note: T-T4 and F-T4 levels are described as “preimmunization” values to differentiate them from T-T4 values after immunization for CXB, BXH and AXB/BXA mice). These GN trait numbers are provided to enable readers to verify and extend the data analysis. Because GN interval maps connect directly to the University of California Santa Cruz Genome Browser, it is possible to explore the gene complement of chromosomal intervals together with the QTL profile. In some tables, we include information on the “Additive Effect,” which is half the difference in the mean phenotype of all cases that are homozygous for one parental allele at this marker minus the mean of all cases that are homozygous for the other parental allele at this marker.
Linkage values were combined, when appropriate, to provide data sets for two RI sets or from all four RI sets. As described for BXH and BXD RI mice (19) and CXB and BXH mice (5), we calculated the probability associated with a X2 value equal to −2(lnPBXD + lnPCXD) with four degrees of freedom where lnPBXD and lnPCXD are the natural logarithms of the probabilities derived independently for the two recombinant inbred strains in the same chromosomal interval. When combining data for four strains, the same approach was following using eight degrees of freedom. For the convenience of researchers familiar with human genetic studies, combined linkage data are provided as LOD scores (convertible to LRS values by multiplying by 4.6, as described above). In the combined analyses, “point-wise” P values are provided for the single points examined (as opposed to genome-wide tests which examine few hundred points).
Results
Serum T4 in parental strains and recombinant inbred mice
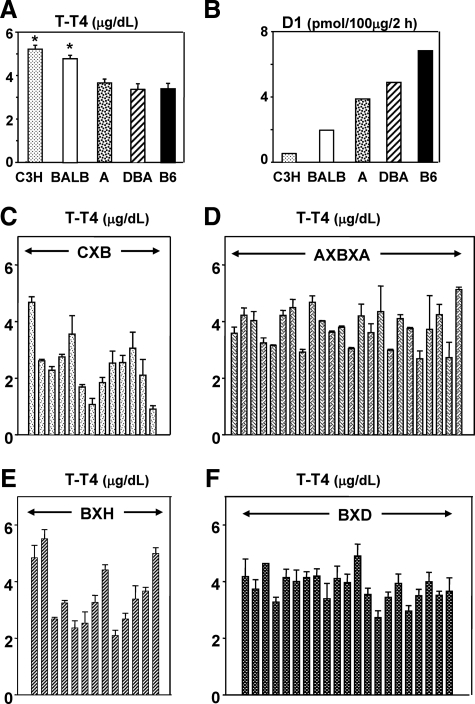
Serum T-T4 levels were significantly higher in female C3H and BALB/c (but not A and DBA/2) than in B6 mice (Fig. 1A, P < 0.05, ANOVA). Conversely, D1 activity is reported to be far lower in C3H and BALB/c than in B6 mice (Fig. 1B; data from Berry et al. (13). We next determined serum T-T4 levels in four sets of RI mice (all females) generated from the foregoing parental strains. Marked variability in T-T4 was observed in CXB and BXH (B6 × C3H) RI strains (Fig. 1, C and E), consistent with the magnitude of differences between the parental strains for these mice (Fig. 1A). Serum T-T4 levels were also variable in AXBXA (A × B6) and BXD (B6 × DBA/2) RI strains (Fig. 1, D and F), despite similar T-T4 levels in the parental strains (Fig. 1A). Although counter-intuitive, T-T4 variability in some RI strains likely arises from combinations of different genes controlling T-T4 levels in A vs. B6 mice (or DBA vs. B6 mice).
Fig. 1.
T4 levels in inbred parental strains and recombinant inbred (RI) mice. A and B, Total T4 (T-T4) for C3H/He, BALB/c (abbreviated C3H and BALB, respectively), A, DBA/2 (abbreviated DBA), and B6 strains. T-T4 values are for female mice (5–13 weeks old); mean + sem for C3H (n = 15), BALB (n = 9), A (n = 14), DBA/2 (n = 14), and B6 (n = 15). *, P < 0.05; significantly different from T-T4 levels in B6 mice (ANOVA). Deiodinase type-1 (D1) activity, plotted from the data of Berry et al. (13), is for predominantly male mice. C–F, T-T4 in AXBXA, CXB, BXD, and BXH RI strains. Data are shown as the mean + sem for n = 3 or more, sd for n = 2. The numbers of sera studied were as follows: BXD n = 3–5 (except BXD 16, n = 2; BXD13, n = 1); BXH n = 6 (except BXH11 and cC3, n = 2); CXB n = 4 (except CXB5, n = 2); AXBXA n = 2. T-T4 data for CXB, BXH, C3H, and B6 strains were presented previously as “preimmunization” values (4, 5).
Serum F-T4 levels correlated with T-T4 in CXB, BXH, and AXBXA (but not BXD) strains and for the combined data of the four sets of RI mice (Supplemental Fig. 1 published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org/). The combined data for the 4 RI sets (lower panel, Supplemental Fig. 1) depict the wide range of serum T-T4 and F-T4 encompassed by females (5–13 weeks old) in CXB, BXH, AXBXA, and BXD strains. It should be noted that, as for many other mouse studies (e.g., Refs. 12, 20–23), we used a clinical assay for T-T4 and F-T4 with calibration standards in human serum. Some previous reports have used in-house assays with rodent serum standards. However, data obtained with standards in human or rodent serum provide relatively similar results (Supplemental Table 1). Moreover, because we used identical calibration standards in all assays, any bias will constant. Consequently, the T-T4 and F-T4 data we obtained provide relative values for the mouse strains under investigation. Importantly, relative values obtained by the same assay for each RI set are suitable for genetic mapping, the major goal of our study.
Whole genome interval mapping for serum T-T4 levels
Quantitative trait loci for natural endogenous variation in T-T4 were mapped as described in Materials and Methods using the database files for the AXBXA, BXD, BXH, and CXB sets of RI mice (17, 24). Surprisingly, neither of our expectations, namely shared genetic linkages between all four RI sets and/or a role for the Dio1 gene, was fulfilled.
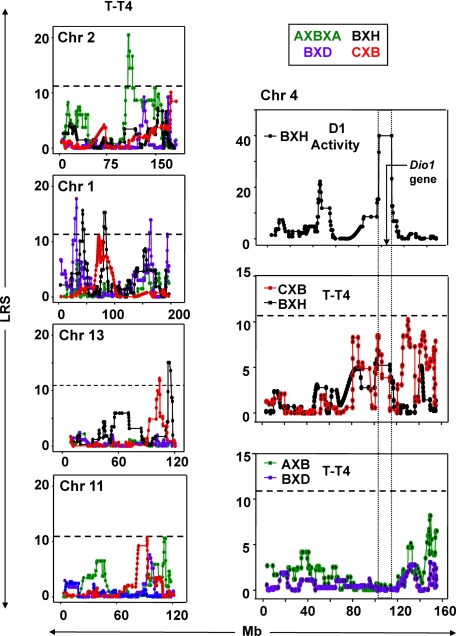
Variability in T-T4 levels was clearly linked to loci on Chrs 2, 1, 13, and (to a lesser extent) Chr 11, but not to Chr 4 bearing the Dio1 gene (Fig. 2 and Supplemental Fig. 2). The strongest linkage (LOD 4.45) for T-T4 was for a single locus on Chr 2 close to the FSHβ gene (Table 1). This linkage appeared to be unique to the AXB/BXA set, as illustrated in Fig. 2. Moreover, on performing combined linkage analysis for all four RI sets, the LOD score for T-T4 linkage was reduced to 2.69 (Supplemental Table 2), indicating that this is not a QTL for all sets. Of interest, alleles from the B6 (not the A) parental strain enhanced T-T4 levels (Supplemental Table 2).
Fig. 2.
Left, Linkage between total T4 (T-T4) and loci on Chr 2, 1, 13, and 11 in four sets of RI mice (AXBXA, green; BXD, purple; BXH, black; and CXB, red). LRS values are on the vertical axis; megabases (Mb) on the horizontal axis. The broken horizontal lines indicate the LRS value above which there is suggestive linkage for T-T4 for all 4 RI sets (mean 11.2 from BXD 10.00, AXBXA 10.87, CXB 11.81, and BXH 12.18). GeneNetwork (GN) identifications for T-T4: AXBXA (GN 10162), BXD (GN 12585), BXH (GN 10150), and CXB (GN 10516). Number of strains studied: AXB/BXA, 25; BXD, 21; CXB, 13; BXH, 13. Note: Data for all Chr (1–20, X) are provided in Supplemental Fig. 2. Right, Linkage to Chr 4 for deiodinase type-1 (D1) activity in BXH mice and for serum T-T4 in CXB, BXH, AXBXA, and BXD mice. Chromosomal distance (megabases. Mb) is on the horizontal axes and LRS values on the vertical axes. Upper panel, D1 activity in BXH mice (plotted from data in Ref. 13). Middle panel, T-T4 in CXB (GN 10516) and BXH (GN 10150). Lower panel, T-T4 in AXBXA (GN 10162) and BXD (GN 12585). The location of the Dio1 gene is indicated in the top panel. The boxed areas in all three panels define the Chr 4 region linked to Dio1 activity in BXH strains.
Table 1.
Chromosomal (Chr) linkage for T-T4 in recombinant inbred mice (AXBXA, BXD, BXH, and CXB strains)
| Chr | Strain | LRS | LOD | Mb | Hu Chr | Gene/assoc |
|---|---|---|---|---|---|---|
| 2 | AXBXA | 20.456 | 4.45 | 107.04 | 11p13 | (FSHb) |
| 107.89 | ||||||
| 1 | BXD | 17.742 | 3.86 | 31.602 | 6q22.33 | |
| 32.031 | 6q11.1 | |||||
| BXH | 12.185 | 2.65 | 43.276 | 2q12.1 | Pou3f3a | |
| 45.530 | 2q32.2 | |||||
| CXB | 10.048 | 2.18 | 76.411 | 2q36.1 | ||
| 78.051 | 2q35-q37 | |||||
| BXH | 15.259 | 3.32 | 80.831 | 2q36.2 | For TSHb | |
| 83.218 | 2q36 | |||||
| BXD | 13.921 | 3.03 | 163.862 | 1q23 | (Fasl) | |
| 164.840 | 1q24.3 | |||||
| BXD | 11.315 | 2.46 | 193.869 | |||
| 13 | CXB | 11.814 | 2.57 | 103.77 | 5q14.3 | Pde8ba |
| 104.23 | 5q12 | |||||
| BXH | 15.047 | 3.27 | 112.89 | 5q12 | Pde4da | |
| 114.38 | 5q11.2 | |||||
| 11 | CXB | 10.048 | 2.18 | 92.865 | 17q22 | |
| 92.880 | ||||||
| AXBXA | 10.577 | 2.30 | 110.977 | 17 | Sox9 | |
| 112.542 | ||||||
| 112.773 |
Data are presented for suggestive or significant (bold) LRS values and LOD scores (where LOD = LRS/4.6). Also included are locations of loci (megabases, Mb), corresponding human Chr, and genes/associations.
Chr 1: Pou3f3, POU domain transcription factor class 3 (42.75399 Mb); Fasl, Fas ligand (163.710819 Mb). Chr 2: FSHb, follicle stimulating hormone &β (106.8963 Mb). Chr 13: Pde8b, Pde4d, phosphodiesterase 8B and 4D (95.79446 and 109.77500 Mb, respectively); Sox9, SRY-box containing gene 9 (112.6435 Mb).
TSH in humans (25).
Unlike the single locus on Chr 2, Chr 1 harbored four nonoverlapping loci associated with variation in T-T4 levels in the other three RI sets (BXD, BXH, and CXB). BXD strains had the highest score (LOD 3.9; with a peak score at ∼31.5 Mb) which was followed by the BXH scores (LOD 3.3; peak between 80.8–83.2 Mb (Table 1). These linkages appear to be specific to each RI set because combined linkage analysis for all four RI sets reduced LOD scores (Supplemental Table 2). However, as shown previously (5), there was shared linkage between serum T-T4 and genes in Chr 1 region 78–80 Mb in the BXH and CXB RI sets (Supplemental Table 2). Of interest, this region corresponds to the Chr 2q6 locus associated with serum TSH in humans (25). Two other Chr 1 linkages (163.862 and 193.869 Mb) appear to be unique to BXD family (Fig. 3, Supplemental Table 2).
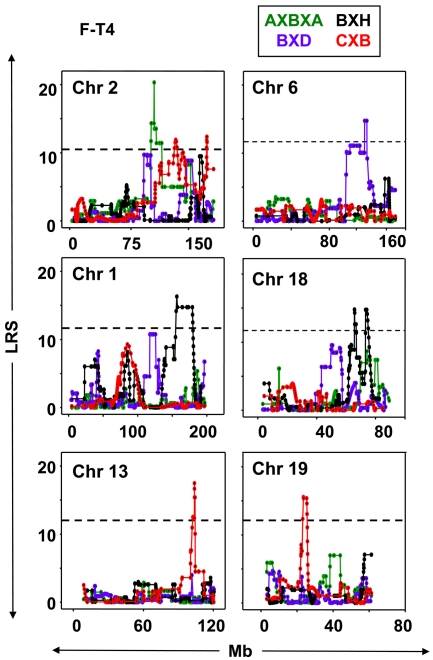
Fig. 3.
Linkage between free T4 (F-T4) and loci on Chr 2, 6, 1, 18, 13, and 19 in four sets of RI mice (AXBXA, green; BXD, purple; BXH, black; and CXB, red). LRS values are on the vertical axis; Mb on the horizontal axis. The broken horizontal lines indicate the mean LRS value for all four RI sets, above which there is suggestive linkage for T-T4 for all 4 RI sets (Mean 11.3 from BXD 10.33, AXBXA 10.87, CXB 11.81, BXH 12.18). GeneNetwork (GN) identifications for F-T4: AXBXA (GN10170), BXD (GN 12584), BXH (GN 10161), CXB (GN10691). Number of strains studied: AXBXA, 15; BXD, 21; CXB, 11; BXH, 10.
In BXH and CXB sets, T-T4 was linked to adjacent nonoverlapping loci on Chr 13 (Table 1). These Chr 13 regions include the genes for Pde8b and Pde4d which have been associated with variable TSH levels in humans (26). Lastly, in CXB and AXBXA sets, significant linkage was observed, albeit with lower LOD scores, between T-T4 and separate nonoverlapping loci on the distal end of Chr 11 (Fig. 2, Table 1).
Possible contribution of the Dio1 gene to serum T-T4 levels
To assess further a possible contribution of Dio1 allelic variants to variability in serum T4 levels, we first used GeneNetwork to map the data on hepatic D1 activities in BXH strains reported by Berry et al. (13). As expected, linkage was highly significant (LOD 8.7) between D1 activity and the Chr 4 region from 101 to 114.0 Mb within which the Dio1 gene is located (Fig. 2, top right panel). These data validate the approach used in the present study.
Turning to the relationship between the Dio1 gene and serum T-T4, whole genome mapping did not overtly indicate linkage between T-T4 and Chr 4. A more detailed focus on this chromosome suggested that some loci might be involved in T-T4 variability in CXB and BXH sets, but not in BXD or AXBXA sets (Fig. 2, middle and lower right panels, respectively). Moreover, further analysis clearly excluded any strong linkage between T-T4 and the Dio1 gene. Thus, combined (but not individual) linkage analysis in CXB and BXH sets for the interval 106 to 114 Mb containing the Dio1 gene gave a low but statistically significant LOD score (1.69, point-wise P = 0.02) (Table 2). However, a higher LOD score (2.20, point-wise P = 0.0063) was obtained using a combined analysis of CXB and BXH data for linkage between T-T4 and a region upstream of Dio1 (102.5–102.8 Mb), near the Pde4b gene (Table 2). For both of these Chr 4 intervals (106.7–114.0 and 102.5–102.8 Mb), C3H alleles (in BXH) and BALB/c alleles (in CXB) contributed to the increase in serum T-T4 levels (Table 2). In addition, an increased LOD score (2.65, point-wise P = 0.002) was obtained from combined analysis in all four RI sets for an upstream Chr 4 region (130–147 Mb) (Table 2). Overall, these data call into question a role for Dio1 in serum T-T4 variability, at least in the RI sets that we have studied, and raise the possibility that variants in neighboring genes on Chr 4 may be of greater significance.
Table 2.
Combined linkage analysis for T-T4 (Chr 4) and F-T4 (Chr 1)
| Chr | Strain | LOD | Additivea | Mb | Interval | χ2 | P | LODCom |
|---|---|---|---|---|---|---|---|---|
| T-T4 | ||||||||
| 4 | CXB | 1.84 | 0.690 | 102.810b | 100.928 | 14.34 | 0.006299 | 2.20 |
| BXH | 1.27 | 0.705 | 102.347 | to | (2 RI) | |||
| AXBXA | 0.13 | 0.103 | 102.547 | 102.81 | ||||
| BXD | 0.08 | −0.074 | 100.928 | |||||
| 4 | CXB | 1.39 | 0.606 | 106.724c | 106.724 | 11.61 | 0.020508 | 1.69 |
| CXH | 1.14 | 0.662 | 114.011 | to | (2 RI) | |||
| BXD | 0.03 | 0.042 | 106.724 | 114.01 | ||||
| AXBXA | 0.11 | −0.092 | 105.207 | |||||
| 4 | CXB | 2.22 | 0.801 | 130.145 | 130.15 | 24.06 | 0.002238 | 2.65 |
| AXBXA | 1.28 | −0.289 | 146.990 | to | (4 RI) | |||
| BXH | 1.11 | −0.651 | 142.331 | 146.99 | ||||
| BXD | 0.61 | 0.194 | 132.126 | |||||
| F-T4 | ||||||||
| 1 | BXD | 1.53 | −0.100 | 42.104 | 37.75 to 42.104 | 15.29 | 0.0041344 | 2.38 |
| BXH | 1.79 | −0.058 | 37.775 | (2 RI) | ||||
| 1 | CXB | 2.01 | −0.120 | 84.893 | 83.218 to 84.893 | 24.55 | 0.0039205 | 2.41 |
| BXH | 1.34 | −0.093 | 83.218 | (2 RI) |
Chr 4: combined LOD scores (LODCom) were computed in 2 RI sets for BXH and CXB strains (one parental strain has defective D1 activity) or in 4 RI sets. Chr 1: LODCom scores were computed for F-T4 in 2 RI strains. LOD scores and additive effects (see below) for individual RI sets are included.
Additive Effect: in BXH strains, a positive additive effect indicates that C3H alleles increase trait values, and a negative additive effect indicates that B6 alleles increase trait values.
Pde4b: phosphodiesterase 4B, cAMP specific (101.927543 Mb).
Dio1: deiodinase type-I gene (106.964069 Mb); human Chr 1p33-p32.
Whole genome interval mapping for F-T4
F-T4 was linked to loci on six chromosomes (Fig. 3), most of which were specific for particular RI sets. Three linkages for F-T4 (Table 3) coincide with those for T-T4 (Table 1). In particular, the same Chr 2 locus (peak at ∼108 Mb) is linked to F-T4 and T-T4 in AXBXA sets. Other shared T-T4 and F-T4 linkages included the following: 1) the Chr 13 locus at ∼104 Mb in CXB mice; 2) the Chr 1 locus with a peak at ∼165 Mb for T-T4 in BXD (Table 1) and for F-T4 in BXH mice (Table 3). F-T4 was not linked to Chr 4 in any RI sets (Supplemental Fig. 3).
Table 3.
Chromosal linkages for F-T4 in recombinant inbred mice (AXBXA, BXD, BXH, and CXB strains)
| Chr | Strain | LRS | LOD | Mb | Hu Chr | Gene/assoc |
|---|---|---|---|---|---|---|
| 2 | AXBXA | 20.301 | 4.41 | 106.692 | 11p13 | (FSHb) |
| 1 | BXD | 10.762 | 2.34 | 117.558 to | 2q14.2 | (Ptpn4) |
| 125.82 | 2q14.1 | |||||
| 157.504 | 1q25.2 | |||||
| 164.317 | 1q23 | |||||
| 164.587 | 1q24.3 | a | ||||
| 171.142 to | 1q23.3 | |||||
| BXH | 14.729 | 3.20 | 179.106 | 1q43-q44 | (Akt3) | |
| 117.066 | 10q11.2 | (Ret) | ||||
| 119.083 | 12p13.33 | |||||
| 6 | BXD | 14.735 | 3.20 | 119.377 | 12p13.31 | |
| 103.772 | 5q14.3 | Pde8bb | ||||
| 13 | CXB | 15.37 | 3.34 | 104.23 | 5q12 | |
| 69.158 | 18q21.2 | (Rab27b) | ||||
| 77.031 | 18q21.1 | Ref.c | ||||
| 18 | BXH | 14.73 | 3.20 | 77.841 | 18q21.1 | |
| 23.654 | 9q21.12 | |||||
| 19 | CXB | 15.37 | 3.34 | 25.403 | 9p24.3 |
As in Table 1, data are presented for suggestive LRS values or LOD scores (LOD = LRS/4.6). Also included are locations of loci (megabases, Mb), corresponding human Chr, and the closest genes or previously observed associations.
Chr 1: Akt3, thymoma viral proto-oncogene 3 (178.952245 Mb); Ptpn4, protein tyrosine phosphatase nonreceptor type 4 (121.554669 Mb). Chr 2: FSHb, follicle stimulating hormone &β (106.8963 Mb). Chr 6: Ret, ret proto-oncogene (118.101765 Mb). Chr 13: Pde8b and Pde4d, phosphodiesterase 8B and 4D (95.79446 Mb, 109.77500 Mb). Chr 18: Rab27b: RAS oncogene family (70.13878 Mb).
As for BXD T-T4, Chr 1 (Table 1).
TSH in humans (26).
F-T4 in humans (25).
The following chromosomal linkages with F-T4 (Table 3) were novel and did not coincide with T-T4 loci: 1) Chr 6 (117–119 Mb) in BXD strains; 2) Chr 18 (69–78 Mb) in BXH mice (one locus corresponds to human Chr 18q21.1, associated with free T4 in humans) (25); 3) Chr 19 loci (∼24 Mb) in CXB strains. Loci on Chr 1 linked to T-T4 included 118–126 Mb (BXD strains), 171–179 Mb (BXH strains) (Table 3), ∼38–42 Mb (shared by BXD and BXH), and 83–84 Mb shared by CXB and BXH (Table 2).
Discussion
We have jointly and separately analyzed total and free serum thyroxine levels in four families of RI mice (17). We examined serum T4 levels in the five parental lines used to generate these sets. Serum T-T4 levels were higher in parental C3H and BALB mice than in B6, A, or DBA/2 mice, in agreement with data from other studies (e.g., Ref. 13). These trends for T-T4 are the reverse of those for hepatic D1 activity (13). Turning to the RI strains themselves, T-T4 and F-T4 levels vary markedly among strains within each set, particularly among the BXH and CXB sets, consistent with elevated T-T4 levels in the parental strains (C3H or BALB/c) and low levels in the common B6 parent. We restricted our studies to females because variability in thyroid hormone levels and TSH was reported for male vs. female mice (12). In addition, a study of quantitative trait loci for several hormones including T-T4 in young vs. old UM-HET3 mice (4 or 15 months) demonstrated age-dependent effects (27). All mice in our study (5–13 weeks) correspond to the group of young adults.
T-T4 and F-T4 levels were examined in four RI sets to test two hypotheses. The first hypothesis was that serum T4 variability is linked, at least in part, to the Dio1 gene. Combined analysis for the CXB and BXH (but not AXB/BXA or BXD) sets revealed linkage between serum T-T4 variability and Chr 4 but the loci appears to be distinct from Dio1. A similar Chr 4 locus was linked to T-T4 levels in genetically heterogeneous mice (27). These “UM-HET3” mice are of interest because they are derived from four of the parental strains used to generate the RI sets in our study, namely by crossing F1 offspring of (BALB/c × B6) with the F1 of (C3H/He × DBA/2) (27). Consistent with our finding for RI sets, the Chr 4 locus in UM-HET3 mice is closer to Pde4b (a cAMP-specific phosphodiesterase) than to Dio1. Overall, data from both studies support linkage between T-T4 and a Chr 4 locus distinct from the Dio1 gene.
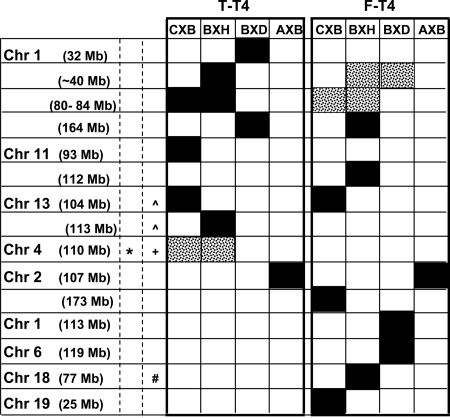
The second hypothesis that we addressed was that, because all four RI sets share B6 as one parental strain, variability in serum T-T4 and/or F-T4 would be linked to chromosomes or loci shared by one or more of these RI sets. Again, our findings were contrary to expectation: most linkages were distinct and provided “genetic signatures” for the individual RI sets (Fig. 4). Some chromosomal linkages were similar for T-T4 and F-T4, and a few Chr/loci were shared by two RI sets. Importantly, however, the linkage (with Chr 2) which gave the highest LOD scores (4.5 for T-T4 and 4.4 for F-T4) was unique to the AXBXA set. This Chr 2 locus is very close to the FSHβ gene. Although another gene may be involved, it is possible that FSH contributes to male:female thyroid hormone differences, as observed for other mouse strains (12).
Fig. 4.
Genetic signatures: chromosomal loci linked to T-T4 and F-T4 in RI mouse strains aged 5–13 weeks (CXB, BXH, BXD, and AXBXA, the latter abbreviated AXB). For simplification, approximate locations are provided for loci (precise locations are in Tables 1–3; Supplemental Table 2). Solid bars, Significant or suggestive linkage. Speckled bars, Significant linkage from combined analysis. *, Loci identified in UM-HET3 mice (27). Similar to loci identified in humans: ^, for TSH (26); #, for F-T4 (25); +, Dio1 (humans and mice). Note: In UM-HET3 mice, the Chr 4 locus (D4Mit155, 102.069572 Mb) (27) is part of the broader region (100–114 Mb) linked to deiodinase activity in BXH mice and, albeit weakly, to T-T4 variability in CXB and BXH strains. This D4Mit155 locus is closer to Pde4b (101.92 Mb), a cAMP-specific phosphodiesterase, rather than to Dio1 (106.96 Mb).
Elucidating the genetic basis for variability in thyroid function is currently an important topic in humans to determine whether individual responses to treatment relate to polymorphisms in candidate traits (28, 29), with the long-term goal of individualizing thyroid hormone replacement therapy (30). In this light, we compared our observations in RI mice with susceptibility genes associated with thyroid function traits in humans. We obtained no evidence in mice for a contribution by TSHR or thyroglobulin gene polymorphisms to T-T4 or F-T4 levels, as observed in humans for serum TSH levels (26, 31). It is possible that the parental mouse strains lack polymorphisms for the TSHR or thyroglobulin genes. The absence of linkage suggests a limitation of these RI sets in providing a “snapshot” of susceptibility genes controlling T-T4 or F-T4 levels. However, some linkages in RI mice correspond to Chr loci or genes observed in humans: 1) mouse Chr 1 (∼80 Mb) and Chr 2q6, associated with serum TSH in humans (25); 2) mouse Chr 13 (∼104 Mb) and Chr 5q 14.3, 5q12, and 5q11.2, harboring the genes for Pde8b and Pde4d which are associated with TSH levels in humans (26); and 3) mouse Chr 18 (∼77 Mb) and Chr 18q21.1 and Chr 18q21.1, loci which are associated with F-T4 in humans (25).
In summary, consistent with multiple processes contributing to serum T-T4 and F-T4 levels, we observed that a number of chromosomal loci play a role in the variability of these traits in mice. Surprisingly, we found that linkage with Chr 4 was distinct from Dio1, the gene responsible for D1 activity. Also unexpectedly, four recombinant inbred sets of mice derived from one shared parental strain had distinct “genetic signatures,” with limited sharing of chromosomal loci linked to T-T4 (or F-T4) levels in serum. Unlike inbred mouse strains, human populations are extremely diverse. Consequently, our data suggest that the contributions of unique chromosomes/loci controlling T-T4 and F-T4 in distinct human subgroups may be “buried” in genetic analyses of heterogeneous human populations.
Acknowledgments
We thank Boris Catz for support of this study.
This work was supported by National Institutes of Health Grants DK54684 and DK82390 (to S.M.M.), National Institute on Alcohol Abuse and Alcoholism Grant U01AA014425 (to L.L.), DK19289 (to B.R), U01AA13499, U24AA13513, P20-DA-21131, U01CA105417, and U24RR021760 (to R.W.W.).
Disclosure Summary: The authors have nothing to declare.
Footnotes
- B6
- C57BL/6
- Chr
- chromosome
- D1
- deiodinase type-1
- Dio1
- D1 gene
- F-T4
- free T4
- LOD
- logarithm of the odds
- LRS
- likelihood ratio statistic
- QTL
- quantitative trait loci
- RI
- recombinant inbred
- T-T4
- total T4
- T3
- tri-iodothyronine
- TSHR
- TSH receptor.
References
- 1. Bailey DW. 1971. Recombinant-inbred strains. An aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation 11:325–327 [DOI] [PubMed] [Google Scholar]
- 2. Airey DC, Lu L, Williams RW. 2001. Genetic control of the mouse cerebellum: identification of quantitative trait loci modulating size and architecture. J Neurosci 21:5099–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mountz JD, Yang P, Wu Q, Zhou J, Tousson A, Fitzgerald A, Allen J, Wang X, Cartner S, Grizzle WE, Yi N, Lu L, Williams RW, Hsu HC. 2005. Genetic segregation of spontaneous erosive arthritis and generalized autoimmune disease in the BXD2 recombinant inbred strain of mice. Scand J Immunol 61:128–138 [DOI] [PubMed] [Google Scholar]
- 4. Aliesky HA, Pichurin PN, Chen CR, Williams RW, Rapoport B, McLachlan SM. 2006. Probing the genetic basis for thyrotropin receptor antibodies and hyperthyroidism in immunized CXB recombinant inbred mice. Endocrinology 147:2789–2800 [DOI] [PubMed] [Google Scholar]
- 5. McLachlan SM, Aliesky HA, Pichurin PN, Chen C-R, Williams RW, Rapoport B. 2008. Shared and unique susceptibility genes in a mouse model of Graves' disease determined in BXH and CXB recombinant inbred mice. Endocrinology 149:2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rapoport B, Williams RW, Chen C-R, McLachlan SM. 2010. Immunoglobulin heavy chain variable region genes contribute to the induction of thyroid stimulating antibodies in recombinant inbred mice. Genes Immun 3:254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meikle AW, Stringham JD, Woodward MG, Nelson JC. 1988. Hereditary and environmental influences on the variation of thyroid hormones in normal male twins. J Clin Endocrinol Metab 66:588–592 [DOI] [PubMed] [Google Scholar]
- 8. Andersen S, Pedersen KM, Bruun NH, Laurberg P. 2002. Narrow individual variations in serum T(4) and T(3) in normal subjects: a clue to the understanding of subclinical thyroid disease. J Clin Endocrinol Metab 87:1068–1072 [DOI] [PubMed] [Google Scholar]
- 9. Samollow PB, Perez G, Kammerer CM, Finegold D, Zwartjes PW, Havill LM, Comuzzie AG, Mahaney MC, Goring HH, Blangero J, Foley TP, Barmada MM. 2004. Genetic and environmental influences on thyroid hormone variation in Mexican Americans. J Clin Endocrinol Metab 89:3276–3284 [DOI] [PubMed] [Google Scholar]
- 10. Hansen PS, Brix TH, Sorensen TI, Kyvik KO, Hegedus L. 2004. Major genetic influence on the regulation of the pituitary-thyroid axis: a study of healthy Danish twins. J Clin Endocrinol Metab 89:1181–1187 [DOI] [PubMed] [Google Scholar]
- 11. Van Heyniningen HE. 1961. Differences in thyroid function of several strains of mice. Proc Soc Exp Biol Med 106:37–40 [DOI] [PubMed] [Google Scholar]
- 12. Pohlenz J, Maqueem A, Cua K, Weiss RE, Van Sande J, Refetoff S. 1999. Improved radioimmunoassay for measurement of mouse thyrotropin in serum: strain differences in thyrotropin concentration and thyrotroph sensitivity to thyroid hormone. Thyroid 9:1265–1271 [DOI] [PubMed] [Google Scholar]
- 13. Berry MJ, Grieco D, Taylor BA, Maia AL, Kieffer JD, Beamer W, Glover E, Poland A, Larsen PR. 1993. Physiological and genetic analyses of inbred mouse strains with a type I iodothyronine 5′ deiodinase deficiency. J Clin Invest 92:1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schoenmakers CH, Pigmans IG, Poland A, Visser TJ. 1993. Impairment of the selenoenzyme type I iodothyronine deiodinase in C3H/He mice. Endocrinology 132:357–361 [DOI] [PubMed] [Google Scholar]
- 15. St Germain DL, Galton VA, Hernandez A. 2009. Minireview: Defining the roles of the iodothyronine deiodinases: current concepts and challenges. Endocrinology 150:1097–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seyfried TN, Glaser GH, Yu RK. 1979. Thyroid hormone influence on the susceptibility of mice to audiogenic seizures. Science 205:598–600 [DOI] [PubMed] [Google Scholar]
- 17. Williams RW, Gu L, Qi S, Lu L. 2001. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol 2:Research0046.1-0046.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Churchill GA, Doerge RW. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williams RW, Strom RC, Goldowitz D. 1998. Natural variation in neuron number in mice is linked to a major quantitative trait locus on Chr 11. J Neurosci 18:138–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. East-Palmer J, Szkudlinski MW, Lee J, Thotakura NR, Weintraub BD. 1995. A novel, nonradioactive in vivo bioassay of thyrotropin (TSH). Thyroid 5:55–59 [DOI] [PubMed] [Google Scholar]
- 21. Costagliola S, Many MC, Denef JF, Pohlenz J, Refetoff S, Vassart G. 2000. Genetic immunization of outbred mice with thyrotropin receptor cDNA provides a model of Graves' disease. J Clin Invest 105:803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagayama Y, Kita-Furuyama M, Ando T, Nakao K, Mizuguchi H, Hayakawa T, Eguchi K, Niwa M. 2002. A novel murine model of Graves' hyperthyroidism with intramuscular injection of adenovirus expressing the thyrotropin receptor. J Immunol 168:2789–2794 [DOI] [PubMed] [Google Scholar]
- 23. Chen C-R, Pichurin P, Nagayama Y, Latrofa F, Rapoport B, McLachlan SM. 2003. The thyrotropin receptor autoantigen in Graves' disease is the culprit as well as the victim. J Clin Invest 111:1897–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, Mott R, Flint J. 2006. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol 4:e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panicker V, Wilson SG, Spector TD, Brown SJ, Kato BS, Reed PW, Falchi M, Richards JB, Surdulescu GL, Lim EM, Fletcher SJ, Walsh JP. 2008. Genetic loci linked to pituitary-thyroid axis set points: a genome-wide scan of a large twin cohort. J Clin Endocrinol Metab 93:3519–3523 [DOI] [PubMed] [Google Scholar]
- 26. Arnaud-Lopez L, Usala G, Ceresini G, Mitchell BD, Pilia MG, Piras MG, Sestu N, Maschio A, Busonero F, Albai G, Dei M, Lai S, Mulas A, Crisponi L, Tanaka T, Bandinelli S, Guralnik JM, Loi A, Balaci L, Sole G, Prinzis A, Mariotti S, Shuldiner AR, Cao A, Schlessinger D, Uda M, Abecasis GR, Nagaraja R, Sanna S, Naitza S. 2008. Phosphodiesterase 8B gene variants are associated with serum TSH levels and thyroid function. Am J Hum Genet 82:1270–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harper JM, Galecki AT, Burke DT, Pinkosky SL, Miller RA. 2003. Quantitative trait loci for insulin-like growth factor I, leptin, thyroxine, and corticosterone in genetically heterogeneous mice. Physiol Genomics 15:44–51 [DOI] [PubMed] [Google Scholar]
- 28. Heemstra KA, Hoftijzer HC, van der Deure WM, Peeters RP, Fliers E, Appelhof BC, Wiersinga WM, Corssmit EP, Visser TJ, Smit JW. 2009. Thr92Ala polymorphism in the type 2 deiodinase is not associated with T4 dose in athyroid patients or patients with Hashimoto thyroiditis. Clin Endocrinol (Oxf) 71:279–283 [DOI] [PubMed] [Google Scholar]
- 29. Cooper-Kazaz R, van der Deure WM, Medici M, Visser TJ, Alkelai A, Glaser B, Peeters RP, Lerer B. 2009. Preliminary evidence that a functional polymorphism in type 1 deiodinase is associated with enhanced potentiation of the antidepressant effect of sertraline by triiodothyronine. J Affect Disord 116:113–116 [DOI] [PubMed] [Google Scholar]
- 30. Dayan CM, Panicker V. 2009. Novel insights into thyroid hormones from the study of common genetic variation. Nat Rev Endocrinol 5:211–218 [DOI] [PubMed] [Google Scholar]
- 31. Peeters RP, van Toor H, Klootwijk W, de Rijke YB, Kuiper GG, Uitterlinden AG, Visser TJ. 2003. Polymorphisms in thyroid hormone pathway genes are associated with plasma TSH and iodothyronine levels in healthy subjects. J Clin Endocrinol Metab 88:2880–2888 [DOI] [PubMed] [Google Scholar]