Abstract
Background
Histidine Hydrogen-Deuterium Exchange Mass Spectrometry (His-HDX-MS) determines the HDX rates at the imidazole C2-hydrogen of histidine residues. This method provides not only the HDX rates but also the pK a values of histidine imidazole rings. His-HDX-MS was used to probe the microenvironment of histidine residues of E. coli dihydrofolate reductase (DHFR), an enzyme proposed to undergo multiple conformational changes during catalysis.
Methodology/Principal Findings
Using His-HDX-MS, the pK a values and the half-lives (t 1/2) of HDX reactions of five histidine residues of apo-DHFR, DHFR in complex with methotrexate (DHFR-MTX), DHFR in complex with MTX and NADPH (DHFR-MTX-NADPH), and DHFR in complex with folate and NADP+ (DHFR-folate-NADP+) were determined. The results showed that the two parameters (pK a and t 1/2) are sensitive to the changes of the microenvironment around the histidine residues. Although four of the five histidine residues are located far from the active site, ligand binding affected their pK a, t 1/2 or both. This is consistent with previous observations of ligand binding-induced distal conformational changes on DHFR. Most of the observed pK a and t 1/2 changes could be rationalized using the X-ray structures of apo-DHFR, DHFR-MTX-NADPH, and DHFR-folate-NADP+. The availability of the neutron diffraction structure of DHFR-MTX enabled us to compare the protonation states of histidine imidazole rings.
Conclusions/Significance
Our results demonstrate the usefulness of His-HDX-MS in probing the microenvironments of histidine residues within proteins.
Introduction
Histidine residues can be useful probes for investigating conformational changes in proteins for two primary reasons. First, the hydrogen-deuterium exchange (HDX) rate of the imidazole C2-hydrogen varies as a function of solvent accessibility [1]. Second, the acid dissociation constant (pK a) of a histidine imidazole group changes in response to the adjacent ionizable group(s) [2], providing an indicator of the local electrostatic environment of the histidine residue. Despite having these unique properties, histidine residues have rarely been used to probe conformational changes in proteins. Presumably, this is because, until the advent of His-HDX-MS, only nuclear magnetic resonance (NMR) spectroscopy [1], [2] was able to determine the HDX rates of imidazole C2-hydrogen in proteins.
More than four decades ago, it was found that the pK a of the imidazole N-H groups can be determined by measuring the apparent HDX rates at the imidazole C2-position at various pH (pD) [3], [4], [5], thus determining both the HDX rate and pK a simultaneously. 1H NMR spectroscopy has been the analytical technique of choice to monitor the HDX reaction at the imidazole C2-position [1]. However, NMR-based methods have difficulties in assigning resonance signals to individual imidazoles and require large amounts and high concentrations of protein.
Our recent proof-of-concept study with a model protein, RNase A, demonstrated that electrospray mass spectrometry can be used to monitor the HDX reactions of histidine imidazoles in proteins [6]. The method has recently been successfully applied to investigate the structural changes in anthrax protective antigen induced by its binding to a anthrax toxin receptor [7] and in rhodopsin upon photon absorption [8]. In the present study, we demonstrate how His-HDX-MS can be used for investigating the changes in the microenvironment of histidine residues of DHFR due to ligand-induced conformational changes.
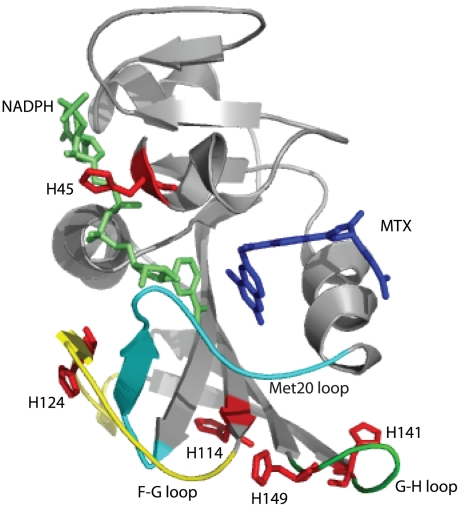
DHFR catalyzes the reduction of 7,8-dihydrofolate (DHF) through hydride transfer from reduced nicotinamide-adenine dinucleotide phosphate (NADPH) to produce 5,6,7,8-tetrahydrofolate (THF) [9]. DHFR was chosen for our study because conformational changes and long-range concerted motions are associated with catalysis [10], [11]. For example, based on an ensemble of X-ray structures, DHFR is proposed to transition between an open, occluded and closed conformation during catalysis [12]. Furthermore, single molecule fluorescence [13], and X-ray and neutron crystallography [12], [14] have observed an open and closed comformational isoforms of DHFR when bound to the chemotherapeutic drug methotrexate (MTX). The fluctuation between these different states of DHFR is due to conformational changes occurring at a regulatory loop called the Met20 loop (residues 9–24) and other loops such as the F–G (residues 116–132) and G–H loops (residues 142–150) [12], [15], [16]. Regions of these loops are far from the ligand-binding site, further demonstrating how conformational changes are propagated to disparate and distant parts of the DHFR molecule. As DHFR contains five histidine residues that are distributed close to the active site and these loops, they can be used as probes to provide insights into the conformational changes associated with ligand binding (Figure 1). In particular, His45 is located near the active site and has direct contact with the cofactor NADPH/NADP+. The other four histidines are located near or within the F–G and G–H loops (Figure 1).
Figure 1. Structure of DHFR-MTX-NADPH complex (PDB: 3DAU).
Five histidine residues are shown (red sticks) [31]. MTX and NADPH are shown in blue and lime, respectively. The Met-20 loop (residues 9–24), the F–G loop (residues 116–132) and the G–H loop (residues 142–150) are shown in cyan, yellow and green, respectively.
We report here the pK a values and half-lives (t 1/2) of HDX reactions of all five histidine residues in apo-DHFR, DHFR-MTX, DHFR -MTX-NADPH, and DHFR-folate -NADP+. These complexes provide representative snapshots of the open (apo-DHFR) and various closed forms of the enzyme [15], [17]. The present results demonstrate the utility of His-HDX-MS for probing the microenvironment of histidine residues of proteins. This method can be used for studying important biological processes such as signal transduction, receptor-drug interactions, enzyme catalysis, protein folding, and opens the door for histidine scanning for investigating protein structure-function relationships.
Results and Discussion
Assignment of HDX rates to the five histidine residues of DHFR
All five histidine residues were detected in four different peptides identified by LC-MS/MS (Figure S1). Initially, the peptides were assigned to the amino acid sequence of DHFR by matching the experimentally obtained peptide masses to their expected molecular masses. The tandem mass spectra for these peptides ensured that the assignments were correct (Figure S2). The four peptides were named His45-, His114-, His124- and His141&149-peptides, based on the positions of five histidines of DHFR. Peptide ions used for calculating the rate of HDX at each of the histidine residues are shown in Table S1.
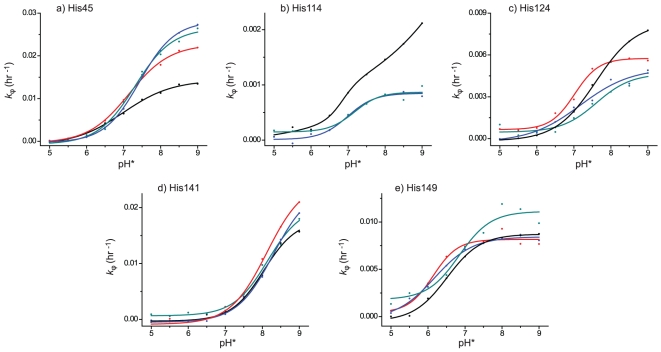
The pseudo-first-order rate constants (k ϕ) for the HDX of the imidazole C2-hydrogen of His45, His114 and His124 were calculated directly from the mass spectra of His45-, His114- and His124-peptides. As His141 and His149 occur in the same peptide, the k ϕ values for them were derived using tandem mass spectrometry where multiple product ions that contain solely His141 or His149 were produced from the His141&149-peptide (Figure S2d). Among the product ions produced, the b11 ion for His141 and y10 ion for His149 were chosen to obtain the k ϕ values, because the intensities of these two ions were among the highest observed. Thus, tandem mass spectrometry was used to provide the k ϕ values at individual histidine residues in peptides containing multiple histidine residues. The k ϕ values are plotted as a function of pH* (direct pH meter readings of the D2O buffer solutions calibrated with standard buffer solutions made with H2O) for each histidine residue in Figure 2. All of the histidine residues gave simple sigmoid curves corresponding to a single pK a except for His114 in apo-DHFR (Figure 2b). The enhanced k ϕ values above pH 8 for His114 may indicate greater solvent accessibility, which could be attributed to possible local conformational change at alkaline pH. Furthermore, we could not obtain an interpretable sigmoid curve for His114 in DHFR-MTX due to the extremely slow HDX rate. This is because based on the solvent accessible surface area His114 is more buried in the DHFR-MTX complex compared to the apo-DHFR and the other DHFR complexes.
Figure 2. pH* dependence of the k ϕ for HDX at the imidazole C2-position of five histidine residues in apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+.
The His45 (a), 114 (b), 124 (c), 141 (d) and 149 (e) from apo-DHFR (black), DHFR-MTX (red), DHFR-MTX-NADPH (blue), and DHFR-folate-NADP+ (green).
Histidine pK a values
Measured pK a values from the sigmoid curves of individual histidine residues are shown in Table 1. Interestingly, there are several ligand induced pK a changes to the histidine residues that were observed. First, the pK a of His45 increased at least 0.31 pH units upon MTX-NADPH or folate-NADP+ binding, whereas the increment was less significant (0.16 pH unit) upon MTX binding. Second, the pK a of His124 decreased at least 0.43 pH units upon MTX and MTX-NADPH binding, but not upon folate-NADP+ binding. Third, the pK a of His149 decreased at least 0.34 pH units upon MTX and MTX-NADPH binding, while the pK a changed 0.33 pH units upward upon folate-NADP+ binding. Note that we did not convert the pH* to pD to calculate the pK as, since the pK a values calculated with pH* are fairly close to those determined in H2O [18], [19]. This is because the constant term of 0.4 in pH*/pD conversion (pD = pH*+0.4) [20] is approximately canceled by a decrease of acidities of acids in D2O [18].
Table 1. pK a values of histidine residues in apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+ complex.
| pKa | ||||
| DHFR | DHFR-MTX | DHFR-MTX-NADPH | DHFR-folate-NADP+ | |
| His45 | 6.99 (±0.07) | 7.15 (±0.04) | 7.43 (±0.03) | 7.30 (±0.07) |
| His114 | 6.84 (±0.20) | N.D. | 6.99 (±0.10) | 7.17 (±0.16) |
| His124 | 7.60 (±0.03) | 7.02 (±0.06) | 7.17 (±0.20) | 7.61 (±0.19) |
| His141 | 8.04 (±0.08) | 8.12 (±0.14) | 8.29 (±0.10) | 8.14 (±0.11) |
| His149 | 6.51 (±0.04) | 6.13 (±0.13) | 6.17 (±0.10) | 6.84 (±0.17) |
Relationship between pK a and microenvironment
Histidine 45
Figure 3a shows the microenvironment of His45 in the crystal structures of apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH, and DHFR-folate-NADP+, respectively. The imidazole ring of His45 in the DHFR-MTX-NADPH and DHFR-folate-NADP+ structures is located close to the negatively charged pyrophosphoryl moiety of NADPH/NADP+; the distances between Nδ1 of His45 and the two oxygen atoms of the pyrophosphoryl moiety are within 3.5 Å. It has been well established that neighboring groups that increase the electron density of the imidazole ring (e.g., RCOO−, RO−, RS−) increase the pK a of the imidazole ring and that neighboring groups that decrease the electron density of the imidazole ring (e.g., RNH3 +, GdnH+, ImH+) decrease the pK a of the imidazole ring [21]. Thus, the increased pK a of His45 in the NADPH/NADP+ bound DHFRs are possibly due to the close proximity of the imidazole ring of His45 with the pyrophosphoryl moiety of NADPH/NADP+. The pK a of this histidine residue in DHFR-MTX is lower than those in DHFR-MTX-NADPH and DHFR-folate-NADP+, but still 0.16 pH units higher than that in apo-DHFR even though there are no NADPH/NADP+ molecules. The close distance from the carboxyl group of Glu17 to His45 (3.1 Å) may be the reason for the higher pK a in DHFR-MTX than expected. This glutamic acid residue (Glu17) is more than 13 Å away in DHFR-MTX-NADPH and DHFR-folate-NADP+, therefore it is unlikely to affect the pK a of His45. Unfortunately, Glu17 is not observed in the apo-DHFR structure, possibly due to the enhanced flexibility of the Met20 loop, destabilized by the absence of ligand binding. We predict a decreased pK a for His45 in apo-DHFR relative to binary and ternary complexes because the Met20 loop is undergoing rapid conformational switching between open and closed forms (∼200 s−1). Thus, there is no ability to form any strong binding contacts.
Figure 3. Microenvironment of histidine residues in apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+ structures. a) His45, b) His114, c) His124, d) His141, and e) His149.
The carbon atoms of apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+ are shown in yellow, lightblue, green, and cyan, respectively.
Histidine 114
With the exception of the DHFR-MTX complex, the pK a values of His114 were almost the same in apo-DHFR and the other complexes, and the values were slightly higher than the intrinsic pK a value of a fully solvent exposed histidine residue (pK a≈6.5) [6]. As discussed previously, the pK a of His114 was unable to be measured in the DHFR-MTX complex due to slow exchange rates as the imidazole side chain is more buried relative to apo-DHFR and the other complexes. There is no electron-donating or electron-withdrawing group found near His114 in apo-DHFR. The closest one is the side-chain carboxyl group of Glu154 that is 7.8 Å away (Figure 3b). Whereas in other three structures, the Oε1 of Glu154 is localized close proximity of the Nε2 of His114 (within 3 Å). Despite its proximity to Glu154 the pK a values of this histidine residue are not elevated large extent as is seen on His141 (see below). We hypothesize that a hydrogen bonding network exists among His114, Glu154 and Ser135, in which Oγ of Ser135 donates a hydrogen to Oε2 of Glu154 and then Oε1 of Glu154 donates a hydrogen to Nε1 of His114 to form the hydrogen bonds; consequently, there is no electron-donating effect of the carboxyl group of Glu154 to His114.
Histidine 124
The pK a values of His124 in DHFR-MTX and DHFR-MTX-NADPH were lower than apo-DHFR, while the pK a remained the same when folate-NADP+ was bound. His124 is surrounded by the backbone carbonyl oxygen atoms of Asp11, Arg12, Gly121 and Asp122 in all the four structures (Figure 3c). These are the only electron-donating atoms around His124 and no electron-withdrawing groups were found. Based on the crystal structures we are unable to explain why the pK a of His124 in DHFR-MTX and DHFR-MTX-NADPH are lower than those in apo-DHFR and DHFR-folate-NADP+.
Histidine 141
The pK a of His141 in all the DHFR samples were high (≥8) compared to the intrinsic pK a value of a fully solvent exposed histidine residue (pK a≈6.5) [6], indicating there is a negatively charged group or groups in close proximity to this histidine. As expected, in all structures the imidazole ring of His141 is in close proximity to Glu139 (Figure 3d), suggesting that it forms a salt-bridge with this presumably charged glutamic acid residue at neutral pH.
Histidine 149
The pK a of His149 in DHFR-MTX and DHFR-MTX-NADPH were significantly lower than in apo-DHFR, while the pK a increased upon folate-NADP+ binding. As the electron density on the imidazole ring is affected by surrounding electronegative atoms, their distances to the Nδ1 atom may correlate with pK a. In fact, the combined distances to the carbonyl oxygen atoms of Asp116 and Ser148 from the Nδ1 of His149 appear to correlate well with the observed pK a of His149 (Figure 3e). The order of distances are as follows: DHFR-MTX (9.6 Å)>DHFR-MTX-NADPH (9.3 Å)>apo-DHFR (8.0 Å)>DHFR-folate-NADP+ (6.3 Å). The longer combined distances correlate well with lower pK a values. These results suggest that the electron-donating effects by the backbone carbonyl oxygens of Asp116 and Ser148 to His149 is the determinant factor of the pK a of the side chain of His149.
Histidine t 1/2 values
The t 1/2 values for the 5 histidine residues of apo-DHFR, and the DHFR binary and ternary complexes are shown in Table 2. The significant changes in t 1/2 due to ligand binding are: 1) the t 1/2 of His45 decreased greater than 1.7-fold upon MTX, MTX-NADPH and folate-NADP+ binding, 2) the t 1/2 of His114 increased at least 2-fold upon MTX, MTX-NADPH and folate-NADP+ binding, and 3) the t 1/2 of His124 increased at least 1.4-fold upon MTX, MTX-NADPH and folate-NADP+ binding.
Table 2. Half-lives of H-D exchange at histidine residues in apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+ complex.
| t1/2 (day) | ||||
| apo-DHFR | DHFR-MTX | DHFR-MTX-NADPH | DHFR-folate-NADP+ | |
| His45 | 2.21 (±0.07) | 1.27 (±0.05) | 1.02 (±0.01) | 1.09 (±0.04) |
| His114 | 16.57* | >50 | 34.07 (±1.64) | 33.22 (±2.31) |
| His124 | 3.46 (±0.06) | 5.01 (±0.14) | 5.71 (±0.61) | 6.20 (±0.67) |
| His141 | 1.62 (±0.10) | 1.17 (±0.12) | 1.22 (±0.09) | 1.42 (±0.12) |
| His149 | 3.31 (±0.05) | 3.54 (±0.13) | 3.42 (±0.08) | 2.60 (±0.17) |
Histidine 45
The t 1/2 values of His45 in DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+ were lower than apo-DHFR, suggesting that His45 has greater solvent accessibility (ability of bulk solvent to access the particular histidine imidazole ring) in the binary and tertiary complexes than in apo-DHFR. His45 is located close to the negatively charged pyrophosphoryl moiety of NADP+/NADPH in the crystal structures of DHFR-MTX-NADPH and DHFR-folate-NADP+ (Figure 3a). The negatively charged pyrophosphoryl moiety is expected to stabilize the cationic imidazolium of His45. Since the rate of the HDX reaction at the imidazole C2-position is directly proportional to the concentration of cationic imidazolium [4], the close proximity of the pyrophosphoryl moiety of NADPH/NADP+ to His45 is likely to be the contributing factor for the higher exchange rates of the ligand bound ternary complexes compared to apo-DHFR. The higher exchange rate of His45 in DHFR-MTX compared to apo-DHFR may be its close proximity to the carboxyl group of Glu17. However, since this glutamic acid residue cannot be seen in apo-DHFR structure, no conclusion can be made.
Histidine 114 and 124
In contrast to His45, the t 1/2 of His114 and His124 increased upon MTX, MTX-NADPH and folate-NADP+ binding, suggesting that the solvent accessibilities of these histidine residues decreased. Examination of the crystal structures of apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH, and DHFR-folate-NADP+ demonstrated that upon ligand binding the side-chain of His114 underwent a conformational change (Figure 4a), that resulted in the formation of a new hydrogen-bond between the imidazole Nε2 atom and the Oε1 of Glu154 and a contact with the side chain methylene group of Cys152. These interactions appear to contribute to the slower HDX in the ligand bound structures. In apo-DHFR, His114's imidazole side-chain faces the solvent. A similar trend was noticed for His124, where upon ligand binding there is a conformational change of the imidazole side-chain leading to contacts with residues 121-123 (Figure 4b). It should be noted that solvent permeation and local fluctuation events are assumed to be important determinants of HDX of proteins [22], the contribution of which cannot always be predicted from the structural data.
Figure 4. Ligand-induced conformational changes of His114 (a) and 124 (b).
The carbon atoms of apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+ are shown in yellow, lightblue, green, and cyan, respectively.
Histidine 141 and 149
The t 1/2 of His141 and 149 were almost the same in apo-DHFR and the other complexes. The side chain of His141 is exposed to the bulk solvent and no notable differences are observed in its microenvironment among the four crystal structures (Figure 3d), thus consistent with our observations on its comparable pK a and t 1/2 values in the apo-DHFR and the other complexes. On the other hand, as discussed previously, we observed subtle differences in the electrostatic environment around His149 between structures (Figure 3e) that were sufficient to cause the changes in its pK a but not its t 1/2 values. Thus, the results show that the subtle differences in the electrostatic environment did not alter the solvent accessibilities of His149.
Comparison of His-HDX-MS with NMR and neutron crystallography data
We are able to compare our findings for the DHFR-MTX complex with NMR and neutron crystallography studies. Poe and co-workers determined the pK a of five histidine residues in E. coli DHFR complex with MTX using 1H NMR [23]. The assignments of the five histidine C2-NMR resonances were done based on the local electrostatic environments of the five histidine residues in the crystal structure of DHFR-MTX [24]. The pK a values assigned to the five histidine residues are not consistent with those determined by His-HDX-MS. This might be because the pK a assignments in the NMR study were made based on the electrostatic environment of the five histidine residues derived from the DHFR-MTX crystal structure. Since the assignments of pK a values to the individual histidine residues by His-HDX-MS is straightforward and no assumptions are involved [6], we are confident that the pK a assignments in our experiments are correct. Other than E. coli DHFR, numerous NMR studies have been performed on Lactobacillus casei DHFR including a study determining the pK a of seven histidine residues in this enzyme [25]. Unfortunately we are unable to do a suitable comparison with this work due to considerable sequence variability between the two species.
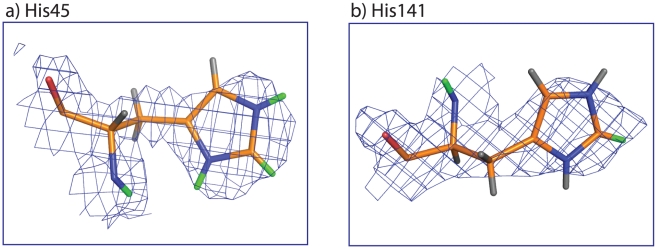
Since neutron diffraction provides an experimental method of directly locating deuterium atoms in proteins including the C2-deuteron of histidine [26], we analyzed the neutron diffraction data of DHFR-MTX complex [14]. As we expected, from the nuclear density maps, it was evident that the C2-position of His45 and 141, which were the two residues that showed the fastest HDX (Table 2), undergo HDX (Figure 5a and b). The other residues did not show significant nuclear densities at the C2-positions, which correlates well with the slow-exchanging histidines in our study. Based on the His-HDX-MS experiments conducted at 37°C over 3 days, His114 hardly undergoes HDX, while His45, 124, 141 and His149 approximately undergo 80, 34, 83, 44% (calculated from the k ϕ max values of these residues), respectively. The crystals in the neutron crystallography study underwent HDX at 4°C for 4 weeks. Since the rate of HDX at 4°C is predicted to be much slower than the rate at 37°C, the extent of deuteration in the crystal is expected to be lower than that at 37°C for 3 days. Hence, it is not surprising that poor nuclear density was observed for His114, His124 and His149.
Figure 5. Nuclear density map of His45 and 141 in DHFR-MTX taken from PDB (ID: 2INQ).
Deuterium and hydrogen atoms are shown as green and gray sticks, respectively. Other atoms are carbon (orange), nitogen (blue) and oxygen (red).
Global conformational changes upon ligand binding
It has been proposed that long-range molecular dynamics can provide energy for enzymatic reactions. DHFR has been proposed to be such an enzyme [9], [27]. Studies such as X-ray crystallography [12], NMR relaxation experiments [17] and amide HDX mass spectrometry [28], have indicated that the conformation of DHFR changes continuously throughout the course of the enzymatic reaction. As mentioned before, the major sites of conformational change include the active site loop (residues 9-24, termed the Met-20 loop), the F-G loop (residues 116-132) and the G-H loop (residues 142-150). These loop regions are distant from the active site. Except for His45, which has direct contact with the cofactor, the other histidine residues are at least 10 Å away from the active site (see Figure 1). His124 is in the F-G loop and His114 is at the base of the F-G loop. His149 is in the G-H loop and His141 is at the base of the G-H loop. We observed dramatic effects on either pK a or t 1/2 or both on the five histidine residues upon ligand binding. Our results, therefore, indicate that even though the Met20 loop in all the ligand-bound complexes analyzed here adapt a closed conformation, considerable differences in their electrostatic environment and/or solvent accessibilities exists.
We have also compared the HDX rates at C2-position of the five histidine residues with the amide HDX rates on the peptides that contain those histidine residues [29]. No correlation was found between the results. This is not surprising because His-HDX rates are specific to the side chain of histidine residues, whereas amide HDX rates are summed HDX rates on all the backbone amides in the peptides that contain one or more of those histidine residues.
His-HDX-MS for structural biology
Our results demonstrate that with only sub-nanomole amounts of sample, His-HDX-MS can provide unambiguous assignments of pK a and t 1/2 for individual histidine residues in proteins, and the two parameters (pK a and t 1/2) are extremely sensitive to changes in the microenvironment around histidine residues. Therefore, this method should be useful to study the changes in the microenvironment of histidine residues due to conformational changes induced by ligand binding and protein-protein interactions. We postulate that this method is applicable to proteins that are not amenable to analysis by NMR spectroscopy such as large proteins/protein complexes (MW>100 kDa), complex protein mixtures like whole cell lysates, and proteins that are available in limited amount. An obvious limitation of His-HDX-MS is that the technique relies on the presence of histidine residues at sites or near the sites of interest. Many histidine residues may be surface exposed and “uninteresting” (i.e., show no change in their pK a and t 1/2 values upon ligand binding), or simply the frequency of histidine residues in proteins of interest may be too low for the technique to describe global structural changes. We think the limitation of the technique can be overcome by introducing histidine residues at the sites of interest using site-directed mutagenesis. Such a ‘His-scanning’ approach will be tested in the future. Another limitation is the long incubation time required for sufficient HDX (at least several hours [6]). This means that His-HDX-MS cannot detect dynamical changes that occur on a short time-scale. This is an inherent limitation of the method. Moreover, the requirement of long incubation times will be problematic for proteins that are not stable during the incubation. Further improvement of mass spectrometry instrumentation that allows us to measure smaller changes in the M and M+1 ratio of peptide peaks will be needed in order to shorten the required incubation time.
Materials and Methods
Material
Deuterium oxide (D2O, 99.9%) was purchased from Cambridge Isotope Laboratories (Andover, MA), and deuterium chloride (DCl) and sodium deuteroxide (NaOD) were purchased from Sigma-Aldrich (St. Louis, MO). Folate and methotrexate (MTX) were purchased from Sigma-Aldrich. Immobilized chymotrypsin was purchased from Princeton Separations (Adelphia, NJ) and Staphylococcus aureus V-8 protease (Glu-C) was purchased from Thermo Fisher Scientific (Waltham, MA). All other chemicals and materials used were either reagent grade or were of the highest quality that was commercially available.
Preparation of E. coli DHFR and DHFR-ligand complexes
E. coli DHFR was expressed and purified using a similar protocol for expression and purification of B. anthracis DHFR, as we described previously in [30]. Briefly, the cDNA was cloned into the Champion pET-Sumo vector (Invitrogen, Carlsbad, CA) and then transformed into chemically competent BL21(DE3) E. coli cells (Novagen-EMD Biosciences, Gibbstown, NJ) for IPTG-inducible protein expression. The overexpressed protein was purified using an immobilized Nickel affinity column (Ni-NTA; Qiagen, Valencia, CA), and the 6xHis-SUMO tag attached to the N-terminus of DHFR was cleaved by ULP1 protease in the presence of a surfactant, IGEPAL CA-630 (Sigma-Aldrich). The cleaved DHFR protein was separated from the non-cleaved DHFR by a subsequent pass over immobilized Nickel affinity resin, and the IGEPAL was removed from the protein by weak anion exchange chromatography (DEAE2; Bio-Rad, Hercules, CA). As a polishing step, concentrated protein was injected onto a Superdex-75 gel filtration column (GE Healthcare, Piscataway, NJ) and the middle peak fractions were pooled for the HDX-MS experiments. DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+ complexes were prepared as follows. Due to the poor solubilities of MTX and folate in aqueous solvent, a five-fold molar excess of these ligands were added as a solid to the apoenzyme while the DHFR concentration was relatively dilute, 0.75–1.5 mg/mL. After a short incubation with the ligands, the DHFR-ligand complexes were then concentrated 10-fold with a YM10 Centricon (Millipore, MA, USA) device. The cofactors (NADP+ or NADPH at a five-fold molar excess) were added directly to the concentrated protein.
Buffer solutions in D2O
Buffers, 50 mM MES (pH 5.0–7.5) and 50 mM HEPES (pH 8.0 - 9.0), were made with 99.9% D2O. All buffers contained 50 mM NaCl. The pH of the buffers was adjusted with diluted DCl or NaOD and measured with a Solution Analyzer Model 4603 (Amber Science, Eugene, Oregon) equipped with a glass/AgCl electrode (Model 476086, Nova Analytics, Woburn, MA). The final D2O content of the buffers were assumed to be 99.9%, neglecting the exchangeable hydrogens attached to heteroatoms of MES and HEPES that remained in the buffers. The reported pH* values are direct pH meter readings of the D2O buffer solutions calibrated with standard buffer solutions made with H2O and are uncorrected for the isotope effect at the glass electrode.
Assessment of stability of DHFR and its complexes by circular dichroism spectroscopy
We have studied the stability of Apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+. The proteins were incubated at 37°C for 72 hrs. Then, the CD spectra were monitored at the far-UV wavelengths. Far-UV CD spectra of apo DHFR and its complexes display a minimum between 215–220 nm, indicating β-sheet structure. A representation of the CD spectra at pH 7.0 is shown in Supporting Information Figure S3, which demonstrates that DHFR retains its predominantly β-structure over the 72 hour time course.
Deuteration and digestion of DHFR
Apo-DHFR, DHFR-MTX, DHFR-MTX-NADPH and DHFR-folate-NADP+ (650 pmol in H2O) were diluted 10-times with D2O, of which 4 µL (260 pmol) were mixed with 36 µL of buffer with different pH* values (pH 5.0–9.0), and incubated at 37°C for 72 hr. The final D2O content in the reaction mixture was approximately 99%, assuming no HDX through moisture in the atmosphere. The duration of incubation was set to be sufficiently long for determining the HDX rate constant (k ϕ) of His114, which is the slowest exchanging histidine residue in DHFR. We have demonstrated that the HDX reaction follows pseudo-first-order kinetics, and a linear relationship between the HDX rate and the incubation time can be obtained with a model peptide [6]. The reaction was stopped by mixing with 5 µL formic acid, and the protein was freed from the buffer salts using an Ultra Micro Spin C18 column (Nest Group, Southboro, MA) according to the manufacturer's instructions and dried in a Speed Vac. The protein was redissolved in 20 µL 0.1 M ammonium bicarbonate and digested with 0.25 µg immobilized chymotrypsin at 25°C for 1 hr. After the digestion the solution was centrifuged at 3,000× g for 1 min in a tabletop centrifuge and the supernatant was recovered. The recovered chymotryptic peptides were further digested by 1 µg of V8 protease for another 1 hr at 25°C. The resulting digest solution was dried in a Speed Vac and redissolved in 800 µL 0.1% TFA and analyzed by LC-MS/MS as described below.
LC-MS/MS
The digests prepared above were analyzed by LC-MS/MS using a UltiMate 3000 LC systems (Dionex, San Francisco, CA) interfaced to a LTQ-Orbitrap XL mass spectrometer (Thermo-Finnigan, Breman, Germany). The platform was operated in the nano-LC mode using the standard nano-ESI API stack fitted with a picotip emitter (uncoated fitting, 10 µm spray orifice, New Objective Inc., Woburn, MA). The solvent flow rate through the column was maintained at 300 nL/min using a 1∶1000 splitter system. The protein digests (5 µL) were injected into a reversed-phase C18 PepMap trapping column (0.3×5 mm, 5 µm particle size, Dionex) equilibrated with 0.1% TFA/2% acetonitrile (v/v) and washed for 5 min with the equilibration solvent at a flow rate of 25 µL/min, using an isocratic loading pump operated through an autosampler. Note that the use of 0.1% TFA instead of 0.1% formic acid was required to retain one of the histidine containing peptides (His45 peptide) onto the trapping column. After the washing step, the trapping column was switched in-line with a reversed-phase C18 Acclaim PepMap 100 column (0.075×150 mm, Dionex) and the peptides were chromatographed using a linear gradient of acetonitrile from 2% to 50% in aqueous 0.1% formic acid over a period of 40 min at 300 nL/min and the eluate was directly introduced into the mass spectrometer. The mass spectrometer was operated in a data-dependent MS to MS/MS switching mode, with the two most intense ions in each MS scan subjected to MS/MS analysis. The full MS scan was performed at 60000 resolution and the subsequent MS/MS analysis was performed at 30000 resolution. The total scan cycle frequency was approximately 1 sec. The precursor ion isolation width was set to be m/z±2.0 that allowed to transmit the M and M+2 isotopic ions of the peptide for CID. The threshold intensity for the MS/MS trigger was set at 2000 and the fragmentation was carried out using the CID mode using a normalized collision energy (NCE) of 35. The data was entirely collected in the profile mode. Dynamic exclusion function for previously selected precursor ions was not enabled during the analysis. Xcalibur software (version 2.0.5, build 0704, Thermo-Finnigan) was used for instrument control, data acquisition, and data processing.
Measurement of pK a and half-life (t 1/2) of HDX reaction
The pseudo-first-order rate constant (k ϕ) of HDX reaction was determined by monitoring the changes in the ratios of M+1/M isotopic peak of a given peptide before (time = 0) and after (time = t) the HDX reaction. The intensities of the M and M+1 peaks at time t = 0 are represented as I M(0) and I M+1(0), respectively, and the intensities of the same peaks at time t are represented as I M(t) and I M+1(t). Since the HDX reaction follows pseudo-first-order kinetics, the intensities of I M(t) and I M+1(t) can be expressed by equation 1 and 2, respectively.
| (1) |
 |
(2) |
where P is the fractional D2O content in the solvent (P = 1 when D2O content is 100%). By taking the ratio I M+1(t)/I M(t) and arranging the equation for k ϕ, the equation 3 is derived.
| (3) |
where R(0) = I M+1(0)/I M(0) and R(t) = I M+1(t)/I M(t), and t is the incubation time (hr). Note that only a single histidine residue is allowed in the peptide to determine k ϕ at the peptide level. The equation is fundamentally the same as the equation 4 in our previous report [6], but the term P (fractional D2O content) is incorporated into the equation to correct the k ϕ value upward when P<1.
The pKa value was obtained from the sigmoid titration curve plotted k ϕ versus pH* using Origin Graphing software (Ver. 8.0, OriginLab, Northampton, MA) using the following equation.
where A1 is the minimum rate constant at the lowest pH, A2 is the maximum rate constant at the highest pH, x 0 is the point of inflection, and δx is the change in x corresponding to the most significant change in y values.
The half-life (t 1/2, day) of the exchange reaction was estimated using the equation: t 1/2 (day) = ln2/k ϕ max/24, where k ϕ max (hr−1) is the maximum rate constant obtained from the plateau to the alkaline side of the sigmoid titration curve.
Structural Analysis
The comparison of protein structures was performed using the program PyMOL (Molecular Graphics System software, DeLano Scientific, Palo Alto, CA). The structural data of apo-DHFR (PDB code number: 5DFR), DHFR-MTX (PDB code number: 2INQ), DHFR-MTX-NADPH (PDB code number: 3DAU) and DHFR-folate-NADP+ (PDB code number: 1RX2) as deposited in the Protein Data Bank were used in the comparison.
Supporting Information
Representative LC-MS/MS total ion current chromatogram of peptides derived from DHFR. The protein digest (3.1 pmol) was injected into a reversed-phase C18 PepMap trapping column (0.3×5 mm, 5 µm particle size, Dionex) equilibrated with 0.1% TFA/2% acetonitrile (v/v) and washed for 5 min with the equilibration solvent at a flow rate of 25 µL/min. After the washing step, the trapping column was switched in-line with a reversed-phase C18 Acclaim PepMap 100 column (0.075×150 mm, Dionex) and the peptides were chromatographed using a linear gradient of acetonitrile from 2% to 50% in aqueous 0.1% formic acid over a period of 40 min at 300 nL/min and the eluate was directly introduced into the mass spectrometer. The full MS scan was performed at 60000 resolution and the subsequent MS/MS analysis was performed at 30000 resolution. The precursor ion isolation width was set to be m/z±2.0 that allowed to transmit the M and M+2 isotopic ions of the peptide for CID.
(EPS)
Tandem mass spectra of His45- (a), His114- (b), His124- (c) and His141&149-peptide (d). Precursor ions were m/z 328.7 (z = 2), 513.8 (z = 2), 1028.9 (z = 2) and 804.3 (z = 2) for His45-, His114-, His124- and His141&149-peptide, respectively. Only b and y series product ions are labeled.
(EPS)
Circular dichroism (CD) spectra of apo-DHFR (A), DHFR-MTX (B), DHFR-MTX-NADPH (C), and DHFR-folate-NADP+ (D). The spectra were measured after a 72 hr incubation period in D2O buffer at 37°C. Black line represents CD spectra at 0 hr time point whereas red line represents CD spectra after incubation at 37°C for 72 hr.
(EPS)
Histidine-containing peptides/peptide fragments monitored. *His141&149-peptide (Ser138 - Tyr151) contains His141 and 149. The amino acid sequences, positions and monitored ions shown for His141 and 149 are for the fragment ions (b11 and y10, respectively) produced from the peptide by collision-induced dissociation.
(XLS)
Acknowledgments
The authors wish to thank Matthias Buck for assisting circular dichroism spectroscopy analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by National Institutes of Health (NIH) grant P30EY-11373 (Visual Sciences Research Center of Case Western Reserve University) and funds from Case Western Reserve University and the Cleveland Foundation. QW and CD were funded by a Department of Defense (DOD) Laboratory Directed Research and Development award (20070131ER). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Markley JL, Cheung SM. Differential exchange of the C2-hydrogens of histidine side chains in native proteins: proposed general technique for the assignment of histidine NMR peaks in proteins. Proc Int Conf Stable Isot Chem Biol Med, 1st. 1973:103–118. [Google Scholar]
- 2.Sachs DH, Schechter AN, Cohen JS. Nuclear magnetic resonance titration curves of histidine ring protons. I. Influence of neighboring charged groups. J Biol Chem. 1971;246:6576–6580. [PubMed] [Google Scholar]
- 3.Harris TM, Randall JC. Deuterium exchange reactions at the C2-position of imidazoles. Chem Ind. 1965;41:1728–1729. [PubMed] [Google Scholar]
- 4.Vaughan J, Mughrabi ZE, Wu C. The kinetics of deuteration of imidazole. J Org Chem. 1970;35:1141–1145. [Google Scholar]
- 5.Amyes TL, Diver ST, Richard JP, Rivas FM, Toth K. Formation and stability of N-heterocyclic carbenes in water: the carbon acid pKa of imidazolium cations in aqueous solution. J Am Chem Soc. 2004;126:4366–4374. doi: 10.1021/ja039890j. [DOI] [PubMed] [Google Scholar]
- 6.Miyagi M, Nakazawa T. Determination of pKa values of individual histidine residues in proteins using mass spectrometry. Anal Chem. 2008;80:6481–6487. doi: 10.1021/ac8009643. [DOI] [PubMed] [Google Scholar]
- 7.Wimalasena DS, Janowiak BE, Lovell S, Miyagi M, Sun J, et al. Evidence that histidine protonation of receptor-bound anthrax protective antigen is a trigger for pore formation. Biochemistry. 2010;49:6973–6983. doi: 10.1021/bi100647z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodowski DT, Palczewski K, Miyagi M. Conformational changes in the G protein-coupled receptor rhodopsin revealed by His-hydrogen-deuterium exchange. Biochemistry. 2010. [DOI] [PMC free article] [PubMed]
- 9.Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal PK, Billeter SR, Rajagopalan PT, Benkovic SJ, Hammes-Schiffer S. Network of coupled promoting motions in enzyme catalysis. Proc Natl Acad Sci U S A. 2002;99:2794–2799. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horiuchi Y, Ohmae E, Tate S, Gekko K. Coupling effects of distal loops on structural stability and enzymatic activity of Escherichia coli dihydrofolate reductase revealed by deletion mutants. Biochim Biophys Acta. 2010;1804:846–855. doi: 10.1016/j.bbapap.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopalan PT, Zhang Z, McCourt L, Dwyer M, Benkovic SJ, et al. Interaction of dihydrofolate reductase with methotrexate: ensemble and single-molecule kinetics. Proc Natl Acad Sci U S A. 2002;99:13481–13486. doi: 10.1073/pnas.172501499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett B, Langan P, Coates L, Mustyakimov M, Schoenborn B, et al. Neutron diffraction studies of Escherichia coli dihydrofolate reductase complexed with methotrexate. Proc Natl Acad Sci U S A. 2006;103:18493–18498. doi: 10.1073/pnas.0604977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McElheny D, Schnell JR, Lansing JC, Dyson HJ, Wright PE. Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc Natl Acad Sci U S A. 2005;102:5032–5037. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong KF, Selzer T, Benkovic SJ, Hammes-Schiffer S. Impact of distal mutations on the network of coupled motions correlated to hydride transfer in dihydrofolate reductase. Proc Natl Acad Sci U S A. 2005;102:6807–6812. doi: 10.1073/pnas.0408343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 18.Martin RB. Deuterated Water Effects on Acid Ionization Constants. Science. 1963;139:1198–1203. doi: 10.1126/science.139.3560.1198. [DOI] [PubMed] [Google Scholar]
- 19.Krezel A, Bal W. A formula for correlating pKa values determined in D2O and H2O. J Inorg Biochem. 2004;98:161–166. doi: 10.1016/j.jinorgbio.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Glasoe PK, Long FA. Use of glass electrodes to measure acidities in deuterium oxide Journal of Physical Chemistry. 1960;64:188–190. [Google Scholar]
- 21.Barlin GB, Perrin DD. Dissociation constants in the elucidation of structure. In: Bentley KW, Kirby GW, editors. Elucidation of Organic Structures by Physical and Chemical Methods Part I. New York: Wiley; 1972. pp. 611–677. [Google Scholar]
- 22.Bennett BC, Gardberg AS, Blair MD, Dealwis CG. On the determinants of amide backbone exchange in proteins: a neutron crystallographic comparative study. Acta Crystallogr D Biol Crystallogr. 2008;D64:764–783. doi: 10.1107/S0907444908012845. [DOI] [PubMed] [Google Scholar]
- 23.Poe M, Hoogsteen K, Matthews DA. Proton magnetic resonance studies on Escherichia coli dihydrofolate reductase. Assignment of histidine C-2 protons in binary complexes with folates on the basis of the crystal structure with methotrexate and on chemical modifications. J Biol Chem. 1979;254:8143–8152. [PubMed] [Google Scholar]
- 24.Matthews DA, Alden RA, Bolin JT, Freer ST, Hamlin R, et al. Dihydrofolate reductase: x-ray structure of the binary complex with methotrexate. Science. 1977;197:452–455. doi: 10.1126/science.17920. [DOI] [PubMed] [Google Scholar]
- 25.Wyeth P, Gronenborn A, Birdsall B, Roberts GC, Feeney J, et al. Histidine residues of Lactobacillus casei dihydrofolate reductase: paramagnetic relaxation and deuterium-exchange studies and partial assignments. Biochemistry. 1980;19:2608–2615. doi: 10.1021/bi00553a012. [DOI] [PubMed] [Google Scholar]
- 26.Niimura N, Chatake T, Kurihara K, Maeda M. Hydrogen and hydration in proteins. Cell Biochem Biophys. 2004;40:351–369. doi: 10.1385/CBB:40:3:351. [DOI] [PubMed] [Google Scholar]
- 27.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto T, Izumi S, Gekko K. Mass spectrometry on hydrogen/deuterium exchange of dihydrofolate reductase: effects of ligand binding. J Biochem. 2004;135:663–671. doi: 10.1093/jb/mvh080. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto T, Izumi S, Gekko K. Mass spectrometry on segment-specific hydrogen exchange of dihydrofolate reductase. J Biochem. 2004;135:17–24. doi: 10.1093/jb/mvh002. [DOI] [PubMed] [Google Scholar]
- 30.Bennett BC, Xu H, Simmerman RF, Lee RE, Dealwis CG. Crystal structure of the anthrax drug target, Bacillus anthracis dihydrofolate reductase. J Med Chem. 2007;50:4374–4381. doi: 10.1021/jm070319v. [DOI] [PubMed] [Google Scholar]
- 31.Bennett BC, Wan Q, Ahmad MF, Langan P, Dealwis CG. X-ray structure of the ternary MTX.NADPH complex of the anthrax dihydrofolate reductase: a pharmacophore for dual-site inhibitor design. J Struct Biol. 2009;166:162–171. doi: 10.1016/j.jsb.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative LC-MS/MS total ion current chromatogram of peptides derived from DHFR. The protein digest (3.1 pmol) was injected into a reversed-phase C18 PepMap trapping column (0.3×5 mm, 5 µm particle size, Dionex) equilibrated with 0.1% TFA/2% acetonitrile (v/v) and washed for 5 min with the equilibration solvent at a flow rate of 25 µL/min. After the washing step, the trapping column was switched in-line with a reversed-phase C18 Acclaim PepMap 100 column (0.075×150 mm, Dionex) and the peptides were chromatographed using a linear gradient of acetonitrile from 2% to 50% in aqueous 0.1% formic acid over a period of 40 min at 300 nL/min and the eluate was directly introduced into the mass spectrometer. The full MS scan was performed at 60000 resolution and the subsequent MS/MS analysis was performed at 30000 resolution. The precursor ion isolation width was set to be m/z±2.0 that allowed to transmit the M and M+2 isotopic ions of the peptide for CID.
(EPS)
Tandem mass spectra of His45- (a), His114- (b), His124- (c) and His141&149-peptide (d). Precursor ions were m/z 328.7 (z = 2), 513.8 (z = 2), 1028.9 (z = 2) and 804.3 (z = 2) for His45-, His114-, His124- and His141&149-peptide, respectively. Only b and y series product ions are labeled.
(EPS)
Circular dichroism (CD) spectra of apo-DHFR (A), DHFR-MTX (B), DHFR-MTX-NADPH (C), and DHFR-folate-NADP+ (D). The spectra were measured after a 72 hr incubation period in D2O buffer at 37°C. Black line represents CD spectra at 0 hr time point whereas red line represents CD spectra after incubation at 37°C for 72 hr.
(EPS)
Histidine-containing peptides/peptide fragments monitored. *His141&149-peptide (Ser138 - Tyr151) contains His141 and 149. The amino acid sequences, positions and monitored ions shown for His141 and 149 are for the fragment ions (b11 and y10, respectively) produced from the peptide by collision-induced dissociation.
(XLS)