Abstract
Real-time quantitative PCR assays, targeting part of the ammonia-monooxygenase (amoA), nitrous oxide reductase (nosZ), and 16S rRNA genes were coupled with 15N pool dilution techniques to investigate the effects of long-term agricultural management practices on potential gross N mineralization and nitrification rates, as well as ammonia-oxidizing bacteria (AOB), denitrifier, and total bacterial community sizes within different soil microenvironments. Three soil microenvironments [coarse particulate organic matter (cPOM; >250 μm), microaggregate (53–250 μm), and silt-and-clay fraction (<53 μm)] were physically isolated from soil samples collected across the cropping season from conventional, low-input, and organic maize-tomato systems (Zea mays L.- Lycopersicum esculentum L.). We hypothesized that (i) the higher N inputs and soil N content of the organic system foster larger AOB and denitrifier communities than in the conventional and low-input systems, (ii) differences in potential gross N mineralization and nitrification rates across the systems correspond with AOB and denitrifier abundances, and (iii) amoA, nosZ, and 16S rRNA gene abundances are higher in the microaggregates than in the cPOM and silt-and-clay microenvironments. Despite 13 years of different soil management and greater soil C and N content in the organic compared to the conventional and low-input systems, total bacterial communities within the whole soil were similar in size across the three systems (~5.15×108 copies g−1 soil). However, amoA gene densities were ~2 times higher in the organic (1.75×108 copies g−1 soil) than the other systems at the start of the season and nosZ gene abundances were ~2 times greater in the conventional (7.65×107 copies g−1 soil) than in the other systems by the end of the season. Because organic management did not consistently lead to larger AOB and denitrifier communities than the other two systems, our first hypothesis was not corroborated. Our second hypothesis was also not corroborated because canonical correspondence analyses revealed that AOB and denitrifier abundances were decoupled from potential gross N mineralization and nitrification rates and from inorganic N concentrations. Our third hypothesis was supported by the overall larger nitrifier, denitrifier, and total bacterial communities measured in the soil microaggregates compared to the cPOM and silt-and-clay. These results suggest that the microaggregates are microenvironments that preferentially stabilize C, and concomitantly promote the growth of nitrifier and denitrifier communities, thereby serving as potential hotspots for N2O losses.
Keywords: amoA, nosZ, soil microenvironments, N transformation rates, cropping system
1. Introduction
Managed ecosystems have received much attention for their role in nitrogen (N) cycling as the increased rate of mineral N fertilizer application to crop production systems significantly contributes to the global increase of nitrous oxide (N2O) emissions (Mosier et al., 1998) and N leaching (Foster et al., 1986). A better mechanistic understanding of nutrient cycling within cropping systems – both conventional and alternative – is necessary to promote long-term crop management practices that enhance agroecosystem services, such as, carbon (C) storage, N retention, and mitigation of greenhouse gas emissions.
Agricultural management practices (e.g., tillage, organic amendment addition, cropping rotation, and irrigation) can strongly influence the size, composition, and activity of the microbial community in soils (Beare et al., 1992; Bossio et al., 1998; Frey et al., 1999). For example, agricultural practices, such as mineral N fertilizer application, significantly alter ammonia-oxidizing bacteria (AOB) populations, which subsequently impact nitrification and production rates of nitrous oxide (N2O), a potent greenhouse gas, in crop production systems (e.g., Kowalchuk and Stephen, 2001). Cropping systems that receive high organic matter inputs have been characterized by greater microbial activity as well as greater N mineralization compared to systems receiving only mineral fertilizer-N (Gunapala and Scow, 1998; Kramer et al., 2002). However, if organic matter-C additions occur prior to the peak in N demand by the crops, then this can lead to increased microbial immobilization of NO3− (Recous et al., 1990; Sakala et al., 2000), which can reduce N losses from the system via leaching and/or denitrification (Robertson, 1997; Scow, 1997). Consequently, the interactions between nutrient management practices and microbial-mediated N dynamics need to be better understood in order to optimize trade-offs between greater soil organic matter levels and potential N losses in agroecosystems.
Characterizing microbial communities associated with physico-chemically and spatially heterogeneous soil microenvironments may reveal the microbial communities responsible for important ecosystem processes and elucidate how microbial processes scale up from the soil to the ecosystem level. In a comparison between aggregated and unaggregated soils, Sexstone et al. (1988) suggested that the spatial separation of available C, NO3−, and denitrifying bacteria was the major factor limiting denitrification in macroaggregated soil. Additionally, Seech and Beauchamp (1988) have shown that denitrifier population sizes and activity increase as the size of aggregates decrease. Contrastingly, others have shown that denitrification (i.e., N2O production) is positively correlated with intact aggregate size (Drury et al., 2004; Miller et al., 2009). Molecular analyses have shown that the microaggregate environment selects for a microbial community dominated by Actinobacteria and higher bacterial abundance than the communities found on the outside of the microaggregate and those associated with macroaggregates (Ranjard et al., 2000; Mummey et al., 2006).
Recent advances in environmental molecular biology techniques have enabled researchers to investigate the relationship between soil management, microbial communities, and ecosystem processes, especially pertaining to the N cycle. For example, by comparing the relative contributions of nitrification and denitrification to total N2O emissions measured from a 15N incubation experiment with assessments of nitrifier and denitrifier community composition and abundance, Ma et al. (2008) found nitrification was the primary source of N2O emissions in wetland soils and that cultivation increased nitrifier but not denitrifier abundance. Avrahami et al. (2002) combined N2O flux measurements with community assays of the amoA and nirK genes and found, while greater ammonium additions increased nitrification and subsequent N2O release rates, that the community structure of the ammonia oxidizers was not changed. Relative abundances of terminal-restriction fragments (TRFs) of amplified nirK fragments increased, however, with increasing ammonium additions.
Agricultural soils receiving different C and N input, in terms of quality and quantity, are expected to differ in rates of microbial-mediated N transformations and C cycling. The aims of this study were to (i) examine the responses of AOB, denitrifier, and total bacterial abundances to differences in long-term N management in conventional (annual mineral fertilizer applications), low-input (mineral fertilizer and cover crop applied in alternating years), and organic (annual manure and cover crop additions) cropping systems across one growing season; and (ii) determine whether AOB and denitrifier densities are related to short-term N mineralization and nitrification rates across different soil microenvironments. We addressed these objectives in a long-term agricultural site where Kong et al. (2005) found significantly higher soil C stocks and greater soil aggregation in the organic compared to the conventional and low-input maize-tomato systems (Zea mays L.- Lycopersicum esculentum L.)]. We hypothesized that (i) the higher N inputs and soil N content of the organic system foster larger AOB and denitrifier communities than in the conventional and low-input systems, (ii) differences in potential gross N mineralization and nitrification rates across the systems correspond with AOB and denitrifier abundances, and (iii) amoA, nosZ, and 16S rRNA gene abundances are higher in the microaggregates than in the cPOM and silt-and-clay microenvironments. Real-time quantitative polymerase chain reactions (qPCR) were combined with 15N isotopic pool dilution assays to test our hypotheses.
2. Materials and Methods
2.1. Study site, experimental design, and field operations
The field study took place on the Long Term Research in Agricultural Sustainability (LTRAS) plots at the Russell Ranch experimental site (Davis, CA, USA; 38°32′24″ N 121°52′12″ W), which is located in a region characterized by wet winters and hot, dry summers. Two soil types dominate the site: i) Yolo silt loam (fine-silty, mixed, nonacid, thermic Typic Xerorthent) and ii) Rincon silty clay loam (fine, montmorillonitic, thermic Mollic Haploxeralf). The field study was conducted during the 2006 maize growing season in three maize-tomato (Zea mays L.- Lycopersicum esculentum L.) cropping systems (n=3): conventional (annual synthetic fertilizer applications), low-input (synthetic N fertilizer applied in alternate years with cover crop-N incorporated the years without synthetic N fertilization) and organic (annual composted manure and cover crop additions) (see description in Table 1). Since 1993, these maize-tomato cropping systems have been arranged in a completely randomized design with three 0.4 ha replicates under furrow irrigation and conventional tillage.
Table 1.
Distribution of soil microenvironments and total soil nitrogen (N) levels after cover crop incorporation (Ti sampling event) and descriptions of N inputs to the conventional, low-input, and organic maize-tomato systems. Lower-case letters indicate significant differences associated with a soil microenvironment effect on the proportions of soil microenvironments across the cropping systems (p<0.05). Total soil N assigned with different upper-case letters are significantly different at the p<0.05 level.
| Cropping System | Distribution of Soil Microenvironments |
Total Soil N | N inputs | |||
|---|---|---|---|---|---|---|
| cPOM (>250 μm) | Microaggregate (53–250 μm) | Silt-and-clay (<53 μm) | ||||
| (%) | (Mg ha−1) | Even Years of Cropping | Odd Years of Cropping | |||
| Conventional | 1.3c | 40.7b | 58.0a | 1.41B | NH4-NO3 fertilizer | NH4-NO3 fertilizer |
| Low-input | 2.0c | 33.0b | 64.7a | 1.33B | winter legume cover crop | NH4-NO3 fertilizer |
| Organic | 2.3c | 36.0b | 61.7a | 1.85A | winter legume cover crop and composted manure | winter legume cover crop and composted manure |
During the winter cover crop (hairy vetch: Vicia dasycarpa) growing season, microplots (1.0 × 1.0 m) were established in each cropping system replicate (3 treatments × 3 replicates = 9 microplots). In the final week of April 2006, the winter cover crop biomass in the microplots was mowed and roto-tilled into the microplots to a depth of 15 cm. Maize was direct-seeded into the conventional and then into the low-input and organic plots in the second and final weeks of May, respectively. At the start of May, composted manure was incorporated into the organic cropping system at a rate of 373 kg N ha−1, while in the first week of June, the conventional system received 51 kg N ha−1 as N-P-K starter and 170 kg N ha−1 as ammonium nitrate side-dressing. Subsequent simulations of field operations within the microplots (cultivation, weeding, etc.) were done by hand.
2.2. Soil sample collection and aggregate separation
On October 27, 2005 (T0 sampling event; before seeding of the winter cover crop), two soil cores (4 cm diameter; 0–15 cm) were collected from each microplot, composited, and sub-sampled for baseline soil measurements. During the 2006 maize growing season, soil core samples were collected from the microplots after cover crop incorporation (May 31st; Ti) and at harvest (September 13th; Th). Soil bulk density was determined on an individual soil core basis. Approximately 10 g of field-moist soil was sub-sampled for moisture content and the remainder was gently broken apart and large debris was removed. Of the latter, 10 g was air-dried, and the remainder was stored at −20°C until thawed at 4°C 20 min prior to fractionation or 2 mm sieving for 15N pool dilution assays. Air-dried soil samples were ground and analyzed for elemental N concentrations, using a PDZ Europa ANCA-GSL isotope ratio mass spectrometer (Sercon, Crewe, UK). To extrapolate elemental N values from g N kg−1 soil to a hectare-basis, we used the minimum equivalent soil mass to correct for potential temporal changes in soil bulk density (Ellert and Bettany, 1995; Lee et al., 2009).
For each thawed soil sample, 30 g subsamples were fractionated into three fractions: coarse particulate organic matter (cPOM: >250 μm), microaggregate (53–250 μm), and silt-and-clay (<53 μm), using a microaggregate isolator according to the methodology outlined in Six et al. (2000). Thawed soil samples were submerged in deionized water at room temperature for five minutes to slake the soil. The slaked soil was immersed in deionized water on top of a 250 μm mesh screen and shaken with 50 stainless steel beads (4 mm diameter) until only cPOM and sand were retained on the 250 μm mesh screen. During shaking, a continuous and steady stream of water flowed through the device to ensure that microaggregates were immediately flushed onto a 53 μm sieve and were not exposed to any further disruption by the beads. The material on the 53 μm sieve was manually sieved by moving the sieve in an up-and-down motion 50 times over a two minute period (Elliott, 1986), to separate water-stable microaggregates from silt-and-clay particles. All fractions were collected as separate soil suspensions, which were centrifuged at 5,000 rpm for 15 min at 4°C (Sorvall RC-5C Plus Superspeed centrifuge, Thermo Scientific). The supernatant was discarded and the remaining soil was lyophilized and stored at −20°C until further analysis. The different fractions served as the soil microenvironments of interest.
2.3. 15N isotopic pool dilution assays
The remainder of the thawed soil samples was 2 mm-sieved and air-dried for extractable mineral N and N transformation rate analyses. For extractable N measurements, 10 g subsamples of air-dried soil were extracted with 50 ml of 2 M KCl solution by shaking on a reciprocal shaker for 1 h and filtering through a Whatman No. 42 filter paper (ashless). The NH4+-N and NO3− -N concentrations of the soil extracts were analyzed colorimetrically by the Berthelot reaction for NH4+ (Forster, 1995), and by vanadium (III) chloride reduction for NO3− (Doane and Horwath, 2003).
Potential gross mineralization and nitrification rates were measured using 15N pool dilution techniques (Barraclough, 1991). Four 10 g replicates of air-dried soil were brought to 20% water holding capacity (WHC) in 120 ml specimen cups. After equilibrating for 12 hours to avoid the period of stimulated microbial activity upon re-wetting air-dried soil (Birch, 1958), solutions of (15NH4)2SO4 (99 atom%) for potential gross mineralization incubations and K15NO3 (99 atom%) for potential gross nitrification incubations were added to two replicate samples, respectively. The addition of the 15N solutions resulted in an approximate soil moisture level of 60% WHC. The quantity of 15N added to each replicate was adjusted to achieve approximately 10 atom% 15N-enrichment of the NH4+ and NO3− pool sizes estimated from the extractable N measurements described above. All replicates were incubated at room temperature (25°C). For the potential gross mineralization assay, two replicates were destructively sampled after 3 h (t0) and 1 d (t1); whilst for the potential gross nitrification assay, the two remaining replicates were destructively sampled after 3 h (t0) and 3 d (t3). The different destructive sampling times for the two assays reflect differences in optimal incubation times that avoided re-mineralization of added N, as determined by preliminary experiments. After destructive sampling, samples were extracted with 2 M KCl, and the quantities of NH4+-N and NO3− -N were measured as described above. The 15N isotopic signatures of NH4+-N and NO3− -N were determined by diffusion onto acidified disks (Stark and Hart, 1996) and analyzed with a PDZ Europa 20–20 isotope ratio mass spectrometer (Crewe, United Kingdom; Stable Isotope Facility of the University of California, Davis).
Rates for potential gross mineralization were calculated using the following equation (Barraclough, 1991):
| (Equation 1) |
where m = potential gross mineralization rate, θ = the rate of change in the size of the NH4+ pool, = the 15N atom% excess of the NH4+ pool at t0, = the 15N atom% excess of the NH4+ pool at t1, and C0 = the size of the NH4+ pool at t0. Potential gross nitrification rates were also calculated using Equation 1, but with 15N enrichments, incubation times, and pool size changes associated with the NO3− pool.
2.4. Soil DNA extraction procedures
Genomic DNA was extracted from soil microenvironment samples using the Bio 101 Fast DNA SPIN kit for soil (BIO 101, Vista, CA). First, humic acid, a PCR inhibitor, was removed from the soil microenvironment sample as follows: 1 g of soil was incubated in 2 ml of 0.1% Na4P2O7 in 10 mM Tris-HCl buffer (pH 8.0)-1 mM ethylenediaminetetraacetic acid (EDTA), at room temperature for 30 min, centrifuged at 8,500 × g for 10 min at room temperature, and the supernatant was discarded. Total DNA was extracted from 0.5 g humic acid-washed sample according to the manufacturer’s instructions (BIO 101, Vista, CA). Extracted DNA was visualized by agarose (1.5% [wt/vol]) gel electrophoresis. DNA yield was quantified spectrofluorometrically using the Quant-iT fluorescent dye (Invitrogen, Carlsbad, CA).
2.5. Determination of 16S rRNA, amoA, and nosZ gene abundances
Abundances of total bacteria were determined using the real-time quantitative TaqMan PCR method as described in Suzuki et al. (2001). Real-time qPCR was performed in a 20 μl reaction mixture that consisted of 4 μl of template DNA (SOM fractions: ~2.5 ng μl−1; Whole soil: ~1.1 ng μl−1), 10 μl of TaqMan Universal PCR Master Mix (Applied Biosystems, NJ, USA), 4 μl of H2O, 0.8 μl of each primer, and 0.4 μl of the probe. The primers BACT1369F (5′-CGG TGA ATA CGT TCY CGG-3′), PROK1492R (5′-GGW TAC CTT GTT ACG ACTT-3′) and probe TM1389 (5′-CTT GTA CAC ACC GCC CGTC-3′) (Suzuki et al., 2001) were used at concentrations of 800 nM, 800 nM, and 200 nM, respectively. The PCR protocol for bacterial 16S rRNA quantification was as follows: 2 min at 50°C, 10 min at 95°C, and then 40 cycles consisting of 15 s at 95°C, and 1 min at 56°C. We acknowledge that novel bacterial sequences that were not targeted by the TaqMan probe that was used in this study to determine 16S rRNA gene abundances might have resulted in an underestimation of the total bacterial community size in our soil, which might subsequently have led to the calculation of higher relative abundances of the functional genes (e.g., amoA and nosZ).
Ammonia oxidizing and denitrifying communities were investigated by quantifying the abundances of the functional genes, amoA, which encodes for the active subunit of ammonia monooxygenase (Rotthauwe et al., 1997), and nosZ, which encodes for nitrous oxide reductase (Henry et al., 2006). Abundances of AOB and denitrifiers were both determined using the SYBR green real-time qPCR method. As described by Cavagnaro et al. (2007) for the amoA qPCR reaction, 1.2 μl of the A189 forward primer (5′-GNG ACT GGG ACT TCT GG-3′; 0.3 μM; Holmes et al., 1995) and 3.6 μl of the amoA-2R′ reverse primer (5′-CCC CTC KGS AAA GCC TTC TTC-3′; 0.9 μM; Rotthauwe et al., 1997) were used to amplify the amoA gene in a 20 μl reaction mixture with 5 μl of template DNA (SOM fractions: ~ 1.6 ng μl−1; Whole soil: ~1.0 ng μl−1). The PCR conditions were as follows: 15 s at 95°C, and then 40 cycles consisting of 15 s at 95°C, 30 s at 55°C, and 31 s at 72°C, followed by a dissociation stage of 15 s at 95°C, 30 s at 60°C, and 15 s at 95°C. The real-time qPCR nosZ assay was carried out according to modifications by Henry et al. (2006) in a reaction volume of 20 μl, and the assay mixture contained 10 μl of 2× ABI Power SYBR I Green PCR Master Mix, 0.8 μl of each nosZ primer (nosZ2F: 5′-CGC RAC GGC AAS AAG GTS MSS GT-3′, 0.3 μM and nosZ2R: 5′-CAK RTG CAK SGC RTG GCA GAA-3′, 0.3μM), and 5 μl of template DNA (SOM fractions: ~ 2.1 ng μl−1; Whole soil: ~1.0 ng μl−1). Thermal cycling conditions for the nosZ primers were as follows: an initial cycle of 95°C for 10 min, then six cycles of 95°C for 15 s, 65°C for 30 s, 72°C for 30 s, followed by 40 cycles of 95°C for 15 s, 60°C for 15 s, 72°C for 30 s, and 83°C for 30 s (acquisition data step), and, finally, one cycle at 95°C for 15 s and 60°C for 30 s, to 95°C for 15 s.
Fluorescence signals were used to calculate CT (threshold cycle) values for the amoA, nosZ, and 16S rRNA gene copies using the internal software of the thermocycler (7300 System SDS software, V1.2.2, Applied Biosystems, CA, USA). The parameter, CT (threshold cycle), is the cycle number at which the fluorescence emission crosses a threshold within the logarithmic increase phase.
We acknowledge that the high clay and high humic acid content associated with the silt-and-clay and the cPOM, respectively, may have affected DNA extraction efficiency and may have inhibited the qPCR reactions. We also acknowledge that an increase in a specific subpopulation of nitrifiers or denitrifiers in the whole soil or SOM fractions may not have been detected against a larger amoA- or nosZ-containing population that remained inactive. However, because 16S rRNA gene abundances were detected in all SOM fractions and in the whole soil, we are confident that our methods did not bias the gene abundance results.
2.6. Standard curve and quantification of amoA, nosZ, and 16S rRNA genes
External standard curves illustrating the relationship between amoA, nosZ, and 16S rRNA copy numbers and respective CT values were generated with 10-fold serial dilutions of known copy numbers of the amoA, nosZ, and 16S rRNA genes, respectively. DNA from three plasmids containing amoA, nosZ, and 16S rRNA gene fragments amplified from Nitrosomonas europaea ATCC 19718, Bradyrhizobium japonicum USDA 110, and Escherichia coli K-12, respectively, with the forward and reverse primers for amoA, nosZ, and 16S rRNA (listed above) were extracted with a Plasmid Mini Kit (Qiagen). Plasmid concentrations (ng μl−1) were measured spectrofluorometrically using the Quant-iT fluorescent dye method. Copy numbers of amoA, nosZ, and 16S rRNA were directly calculated from the respective concentrations of extracted plasmid DNA. The following equations were used to convert CT values to amoA, nosZ, and 16S rRNA copy numbers, respectively:
| (Equation 2) |
| (Equation 3) |
| (Equation 4) |
Standard curves were linear over six orders of magnitude and the detection limit was approximately 4.0×105 and 2.0×105 copies g dry soil−1 for the amoA and nosZ real-time qPCRs, respectively, and ~1.0×105 copies g dry soil−1 for the 16S rRNA real-time qPCR.
2.7. Data analyses
Whole soil N measurements and the distribution of aggregate size classes within Ti were analyzed using analyses of variance (ANOVA) by the PROC MIXED procedure of the Statistical Analysis System (SAS; SAS Institute, 2002). To compare differences in amoA, nosZ, and 16S rRNA gene abundances among the three sampling events, repeated measures analyses were performed using the PROC MIXED procedure. The data were analyzed as a completely randomized design, with cropping system as the main plot and plot as a random factor. A standard variance components covariance structure was specified with the TYPE=VC option in the model statement. Differences between means were calculated based on least significant difference tests, with the PDIFF option of the LSMEANS statement. Letters for mean separation in PROC MIXED were assigned using the macro PDMIX 800 (Saxton, 1998). All differences discussed were significant at the p<0.05 probability level, unless otherwise stated.
To assess the potential effects of the fractionation/wet-sieving procedure on recoveries of microbial populations in the SOM fractions, the gene copy numbers for amoA, nosZ, and 16S rRNA in the three soil microenvironments corresponding to the same soil sample, respectively, were summed together (i.e., copies g−1 soil for cPOM + microaggregates + silt-and-clay = summed copies for soil sample) and then compared to their corresponding gene abundances in the whole soil (for all soil samples). Although we did not expect the sum of the gene abundances in the SOM microenvironments to be identical to the abundances in the whole soil, the recoveries in the SOM fractions were not drastically different from abundances in the whole soil. Therefore, the wet-sieving procedure did not bias the gene abundances in the SOM fractions.
Gene abundances for amoA, nosZ, and 16S rRNA genes and correlations with environmental variables (NH4+ and NO3− concentrations and potential gross mineralization and nitrification rates) were analyzed with canonical correspondence analysis (CCA) (Lepš and Šmilauer, 2003), using the CANOCO software (Microcomputer Power, Ithaca, NY, USA). The Monte Carlo permutation test was used to test the significance of the correlations between microbial community gene abundances and measured environmental variables.
Initially, we intended to measure denitrification rates to compare with nosZ gene abundances. However, in the process of re-analyzing soils due to unforeseen set-backs, the amount of each soil sample dwindled and, as a result, we did not have enough soil to accurately conduct the denitrification rate measurements following aggregate separation, baseline soil measurements, three gene assays, and mineralization plus nitrification rate assays for each soil sample. Nevertheless, we feel that the molecular assays and N transformation rate experiments that were conducted encompass a wide breadth of N cycling processes.
3. Results
3.1. Physical and chemical soil properties
Across the three sampling events, total soil N levels were highest in the organic (1.85 Mg N ha−1) and similar between the conventional and low-input systems (1.41 and 1.33 Mg N ha−1, respectively). Total soil N levels were greater at T0 than at Ti and Th; however, because trends in N levels were similar across the sampling events within each of the soil microenvironments and N level distribution among the soil microenvironments at Th were similar to Ti, only total soil N concentrations at Ti are shown in Table 1. The distributions of soil microenvironments were similar between the cropping systems and did not change from T0 to Th. At Ti, the silt-and-clay fraction (<53 μm) was the greatest by weight proportion across the systems, representing 58.0% to 64.7% of the soil mass, while the microaggregate (53–250 μm) and cPOM (>250 μm) fractions comprised 36.6% and 1.9%, respectively, across the systems (Table 1).
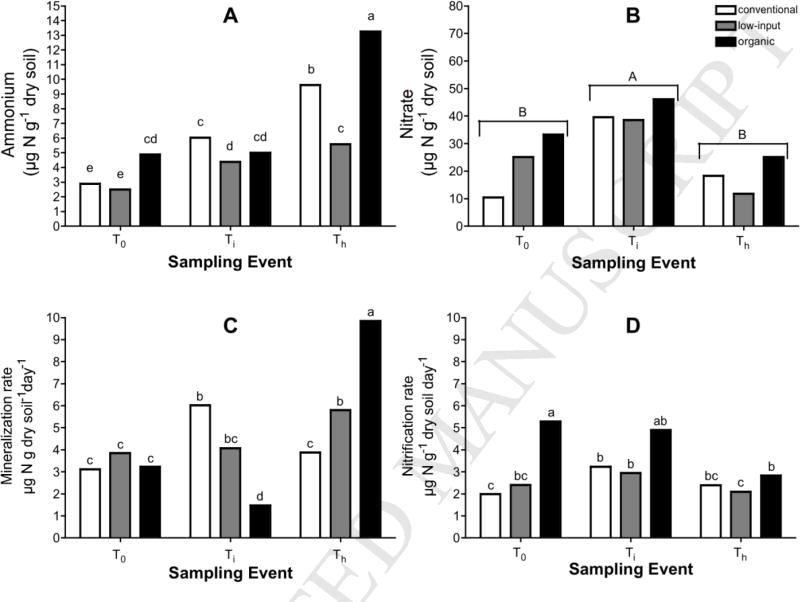
Ammonium concentrations were significantly affected by cropping system and a -cropping system*sampling event interaction, while only sampling event influenced NO3− concentrations (Fig. 1). At T0, NH4+ concentrations were higher in the organic (5.0 μg N g−1 dry soil) than in the conventional and low-input systems (2.9 and 2.2 μg N g−1 dry soil). After cover crop incorporation into each cropping system and immediately before the application of either mineral- or composted manure-N, NH4+ levels at Ti were greater than at T0 (5.2 versus 3.4 μg N g−1 dry soil). Concentrations of NH4+ at Ti decreased in the following order: conventional ≥ organic ≥ low-input (6.2, 5.0, and 4.4 μg N g−1 dry soil, respectively). At Th, NH4+ concentrations were the highest of the season and were greatest in the organic (13.3 μg N g−1 dry soil), intermediate in the conventional (10.0 μg N g−1 dry soil), and lowest in the low-input (5.6 μg N g−1 dry soil). Nitrate pools were at least twice as large as ammonium pools in the three cropping systems. Unlike NH4+ levels, NO3− concentrations peaked at Ti (41.2 μg N g−1 dry soil) and were lower at T0 and Th (23.1 and 20.1 μg N g−1 dry soil, respectively).
Figure 1.
Ammonium (A) and nitrate concentrations (B) (μg N g−1 dry soil) and potential gross mineralization (C) and nitrification rates (D) (μg N g−1 dry soil day−1) for the conventional, low-input, and organic cropping systems, before the start of the cover crop growing season (T0), after cover crop incorporation (Ti), and during maize harvest (Th). Bars represent means (n=3). Different letters within the respective figures for ammonium concentrations, gross mineralization rates, and gross nitrification rates indicate differences associated with a significant sampling event*cropping system effect (p<0.05). For nitrate concentrations, different letters symbolize differences among sampling events (p<0.05).
3.2. Potential gross N transformation rates
Interactions between cropping systems and sampling events were observed for potential gross N transformation rates in the whole soil. Potential gross mineralization and nitrification rates were similar in magnitude despite the larger pool size of NO3− than NH4+ (Fig. 1). At T0, gross mineralization rates were not different among the conventional, low-input, and organic cropping systems (~3.42 μg N g−1 dry soil day−1). At Ti, shortly after incorporation of the winter cover crop biomass, gross mineralization rates were fastest in the conventional system (6.05 μg N g−1 dry soil day−1) and slowest in the organic system (1.53 μg N g−1 dry soil day−1). By the end of the season, the gross mineralization rates were 9.57, 5.27, and 3.89 μg N g−1 dry soil day−1, in the organic, low-input, and conventional systems, respectively. Potential gross nitrification rates were similar between the low-input and conventional systems (~2.56 μg N g−1 dry soil day−1) at T0, Ti, and Th, while rates for the organic systems (~4.34 μg N g−1 dry soil day−1) were generally the highest (but only significantly higher at T0; Fig. 1).
The ratio of gross nitrification rates over gross mineralization rates (Nit:Min) were calculated to assess competition between heterotrophic microorganisms and autotrophic nitrifiers for NH4+ (Table 2). In these soils, Nit:Min ratios were generally low, ranging from 0.29 to 4.0, with the Nit:Min ratio for the organic system rising from T0 (1.78) to Ti (4.0) and falling from Ti to Th (0.29). Cropping systems did not clearly influence the ratio of Nit:Min across the different sampling events.
Table 2.
Ratios of potential gross nitrification rates to gross mineralization rates across the conventional, low-input, and organic cropping systems, before the start of the cover crop growing season (T0), after cover crop incorporation (Ti), and during maize harvest (Th) (n=3). Values with different letters are significantly different at the p<0.05 level.
| Sampling Event | Cropping System | ||
|---|---|---|---|
| Conventional | Low-input | Organic | |
| T0 | 1.0 (bc) | 0.75 (c) | 1.78 (b) |
| Ti | 0.46 (c) | 0.69 (c) | 4.0 (a) |
| Th | 0.72 (c) | 0.48 (c) | 0.29 (c) |
3.3. Total bacterial, AOB, and denitrifier community abundances
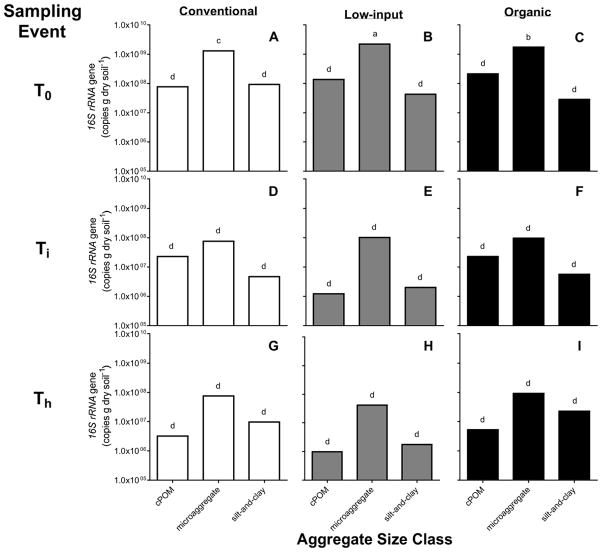
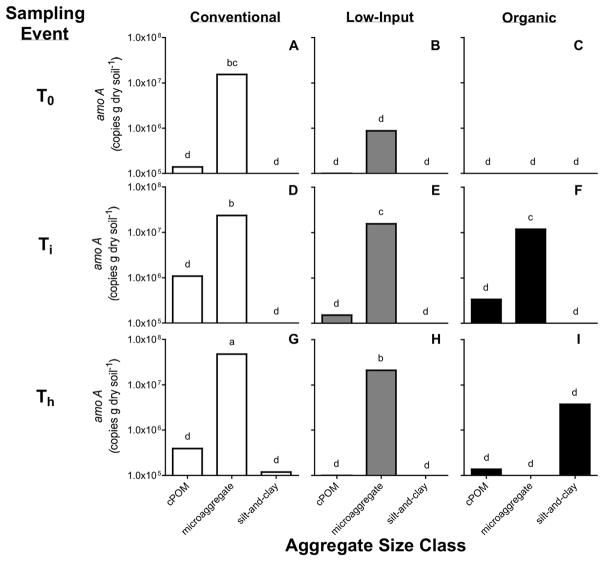
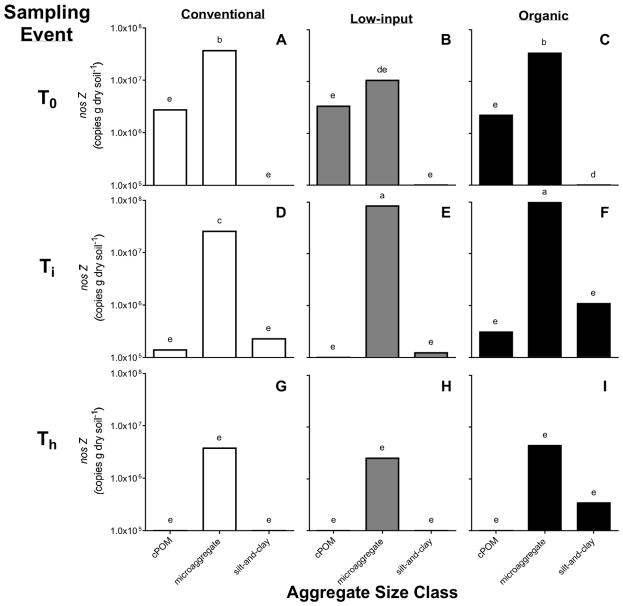
Total copy numbers for 16S rRNA genes averaged across the cropping systems increased from 1.06×108 copies g−1 soil at T0 to 3.34×108 and 7.70×108 copies g−1 soil at Ti and Th, respectively. The size of ammonia-oxidizing and denitrifying bacterial communities changed significantly over the course of the season, but not congruently. Total amoA gene abundances among cropping systems at T0 and Ti were ~6 and ~2 times higher in the organic (1.63×108 and 1.75×108 copies g−1 soil, respectively) than the conventional and low-input systems, respectively; however at Th, densities of amoA genes did not differ among the cropping systems (Table 3). Total nosZ copy numbers were similar among all cropping systems at Ti, but were also the lowest across the season (4.39×106 copies g−1 soil). Copies of nosZ were greater in the conventional and low-input systems than in the organic at Th. Copy numbers of nosZ at the end of the season in the conventional and low-input systems (Th) were also higher than at T0, and Ti, whereas the denitrifier abundances in the organic system were not significantly different throughout the season (Table 3).
Table 3.
Total copy numbers of 16S rRNA, amoA, and nosZ genes measured from whole soil, before the start of the cover crop growing season (T0), after cover crop incorporation (Ti), and during maize harvest (Th) (n=3). Values for amoA and nosZ with different upper-case and lower-case letters are significantly different within respective gene abundances (p<0.05). For 16S rRNA gene copy numbers, values followed by different lower-case letters are significantly greater than values for other sampling events, within a cropping system (p<0.05).
| 16S rRNA | amoA | nosZ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cropping System | Conventional | Low-input | Organic | Conventional | Low-input | Organic | Conventional | Low-input | Organic |
| Sampling Event | (No. gene copies g−1 soil) | ||||||||
| T0 | 1.22×108 (xy) | 2.48×108 (xy) | 2.25×108 (xy) | 2.48×107 (C) | 2.72×107 (C) | 1.63×108 (A) | 3.93×107 (bc) | 4.94×106 (d) | 7.71×106 (d) |
| Ti | 8.54×108 (y) | 6.01×107 (y) | 8.81×107 (y) | 9.34×107 (B) | 6.40×107 (BC) | 1.75×108 (A) | 4.43×106 (d) | 5.55×106 (d) | 3.20×106 (d) |
| Th | 1.25×109 (x) | 8.80×108 (x) | 1.83×108 (x) | 5.57×107 (BC) | 2.98×107 (C) | 3.44×107 (C) | 7.65×107 (a) | 5.58×107 (ab) | 8.83×106 (d) |
A significant interaction between cropping system, soil microenvironment, and sampling event effects was found for the amoA, nosZ, and 16S rRNA gene copy numbers of the soil microenvironments. Across the season, a cropping system effect was only observed for the copy numbers of amoA, nosZ, and 16S rRNA genes in the microaggregate fraction (Figs. 2, 3, and 4). Also, copy numbers associated with the microaggregate fraction were higher than gene abundances in the cPOM and silt-and-clay. Total bacterial abundance in the silt-and-clay was on average 2.79×107 copies g−1 soil, while copy numbers of amoA and nosZ in silt-and-clay ranged from 0 or below the detection limit to 3.88×106 copies g−1 soil and 0 or below-detection-limit to 4.38×107 copies g−1 soil, respectively. In the cPOM, copy numbers of amoA, nosZ, and 16S rRNA were on average 2.77×105, 1.09×106, and 5.32 ×107 copies g−1 soil, respectively.
Figure 2.
Mean copy numbers of 16S rRNA genes within coarse particulate organic matter (cPOM: >250 μm), microaggregate (53–250 μm), and silt-and-clay (<53 μm) soil microenvironments of the conventional (A, D, G), low-input (B, E, H), and organic (C, F, I) cropping systems, before the start of the cover crop growing season (T0), after cover crop incorporation (Ti), and during maize harvest (Th) (n=3). Bars shown with different letters are significantly different across soil microenvironments, cropping systems, and sampling events (p<0.05).
Figure 3.
Mean copy numbers of the amoA gene within coarse particulate organic matter (cPOM: >250 μm), microaggregate (53–250 μm), and silt-and-clay (<53 μm) soil microenvironments of the conventional (A, D, G), low-input (B, E, H), and organic (C, F, I) cropping systems, before the start of the cover crop growing season (T0), after cover crop incorporation (Ti), and during maize harvest (Th) (n=3). Bars shown with different letters are significantly different across soil microenvironments, cropping systems, and sampling events (p<0.05).
Figure 4.
Mean copy numbers of the nosZ gene within coarse particulate organic matter (cPOM: >250 μm), microaggregate (53–250 μm), and silt-and-clay (<53 μm) soil microenvironments of the conventional (A, D, G), low-input (B, E, H), and organic (C, F, I) cropping systems, before the start of the cover crop growing season (T0), after cover crop incorporation (Ti), and during maize harvest (Th) (n=3). Bars shown with different letters are significantly different across soil microenvironments, cropping systems, and sampling events (p<0.05).
3.4. Canonical correspondence analysis
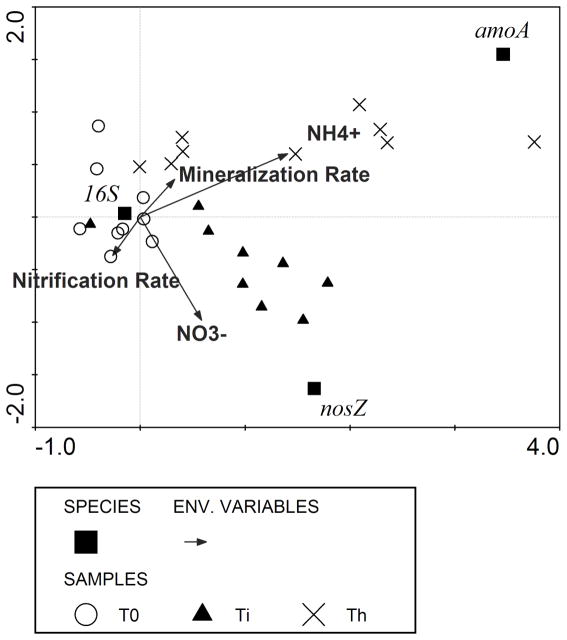
The first two canonical axes of the CCA of amoA, nosZ, and 16S rRNA gene abundances, NH4+ and NO3− concentrations, and potential gross mineralization and nitrification rates at T0, Ti, and Th explained 35.6% of the variance in the gene abundance data and 100% of variance in the total gene abundance-environment parameter relationship (Fig. 5). The first canonical axis, which explained 85.5% of the total gene abundance-environmental parameter variation (p=0.006), was dominated mainly by NH4+ concentrations (0.48) and, to a smaller degree, by NO3− concentrations (0.20). The second canonical axis explained an additional 14.5% of the total gene abundance-environmental parameter variation (p=0.006) and was also dominated by the effects of NO3− (0.27) and NH4+ concentrations (0.17), indicating that NO3− and NH4+ concentrations are the most influential environmental gradients, of those measured, on the distribution of nitrifier and denitrifier genes among samples. However, when sampling event was included as a covariable in the CCA, canonical axes 1 (x axis) and 2 (y axis) explained 98.2% and 1.8% of the gene abundance-environmental variable variance in the data, respectively, but the inclusion of seasonality in the CCA rendered the axes insignificant (data not shown).
Figure 5.
Canonical correspondence analysis (CCA) of measures of the 16S rRNA, amoA, and nosZ gene abundances (plotted as squares) in relation to soil environmental variables (plotted as vectors) for samples collected before the start of the cover crop growing season (T0: empty circles), after cover crop incorporation (Ti: filled triangles), and during maize harvest (Th: X symbols). Canonical axes 1 (x axis) and 2 (y axis) explain 85.5% and 14.5% of the gene abundance-environmental variable variance in the data, respectively (p=0.006).
4. Discussion
4.1. Long-term management effects on N transformation rates and AOB, denitrifier, and total bacterial community dynamics
For the past 13 years, cover crop- and composted manure-N have been incorporated into the organic system at the Russell Ranch at rates nearly two and five times greater than the rates of mineral-N and mineral- plus cover crop-N input to the conventional and low-input systems, respectively. It was expected that inorganic N availability and the results from the isotope dilution experiments would mirror these input differences between the cropping systems. However, short-term NH4+ and NO3− concentrations and potential gross N transformation rates did not demonstrate a consistently greater soil N supplying capacity for the organic compared to the conventional and low-input systems. At T0, which took place after the 2005 growing season, NH4+ levels were at their lowest, likely due to the removal of plant residues and the lack of irrigation at that time of the year. In addition, NH4+ levels were lower in the low-input and conventional systems than in the organic system, which has been suggested to release NH4+ more gradually than conventional systems (Burger and Jackson, 2003). Several months after mineral-and/or composted manure-N incorporation into the systems, NH4+ concentrations were the greatest for the season and reflected the relative rates of N input to the systems (organic > conventional > low-input). In contrast, NO3− levels peaked at Ti and were not different among the systems. Under incubation conditions, Burger and Jackson (2003) found that NO3− immobilization by heterotrophic microbes was greater in organic (~2.5 μg N g−1 d−1) than conventionally managed soil (~0.5 μg N g−1 d−1), as greater NO3− immobilization would be expected for systems with high soil C content. The reasoning for the lack of differences in NO3− concentrations among the cropping systems, as well as the discrepancies between dynamics observed in the NH4+- and NO3−-N levels in this study are unclear.
Potential gross N mineralization and nitrification rates in the organic system, which received more organic matter-N inputs than the other systems, were not consistently higher than N transformation rates in the conventional and low-input systems; only at Th were potential gross mineralization rates higher in the organic system than in the low-input and conventional systems. The magnitude of the gross mineralization rates (Fig. 1), which were nearly two times faster than the <3 μg N g−1 soil day−1 reported in Burger and Jackson (2003) for the same conventional and organic cropping systems, implied that the NH4+ pool turned over daily. The discrepancies between our gross mineralization rates with those from Burger and Jackson (2003) may be related to our shorter incubation times (1 day versus 5 days). On the other hand, gross nitrification rates measured in this study were similar (~3 μg N g−1 soil day−1) to the rates reported in Burger and Jackson (2003). Gross nitrification rates for the systems reflected the overall trends in soil N content among the systems at T0 (e.g., organic > low-input ≥ conventional). However, like Burger and Jackson (2003), who found that gross nitrification rates were not statistically different between the conventional and organic soils, gross nitrification rates in our study became more similar among the three cropping systems over the course of the season. The lack of dramatic differences in potential gross nitrification rates among the systems at Ti and Th, suggested that, despite different quantities and qualities of N input, NH4+ levels were adequate in all three systems. In addition, the lack of differences suggested that the nitrification activity of the AOB populations was not limited by the supply of NH4+ or the soil conditions created by the additions of the different N inputs.
We postulated that AOB, denitrifier, and total bacterial community sizes would vary according to differences in the magnitudes of the measured inorganic N and soil C levels of the cropping systems (i.e., organic > low-input = conventional) and would correlate to the potential gross N transformation rates. In a long-term study conducted on soil types similar to those at Russell Ranch, Gunapala and Scow (1998) found that cover-crop- and manure-C inputs led to higher microbial biomass in organic compared to conventional cropping systems. Shi and Norton (2001) showed that available NH4+ limits both nitrification rates and the nitrifier population size. In our study, gross nitrification rates only weakly reflected NH4+ concentrations, while the abundances of amoA gene copies in the whole soil reflected soil C stock trends at T0 and Ti, but did not mirror measured NH4+ levels or potential gross nitrification rates. For example, copies of amoA genes at Ti were intermediate in the conventional system (Table 3), but NH4+ levels were similar to the organic and higher than the low-input system, respectively (Fig. 1). The lower AOB abundances in the conventional compared to the organic system at Ti and the similar AOB abundances between these two systems at Th do not concur with results from Okano et al. (2004). There, they found AOB communities to be significantly larger in annually fertilized than in unfertilized soil and suggested that ammonium fertilization has a long-term effect on the population sizes of AOB. Nevertheless, amoA gene abundances in this study are on the same order of magnitude as their study on the same maize-tomato systems (0–15 cm). The higher ratios of Nit:Min that we calculated for the organic than conventional system suggested that autotrophic nitrifiers were more competitive for NH4+ than the heterotrophic microorganisms (Hart et al., 1994). This result suggested that AOB gene copies should have been higher in the organic than the conventional and low-input systems, which was indeed found to be the case at T0 and Ti.
It was expected that the higher organic C inputs from the cover crop and composted manure additions into the organic system would lead to a greater abundance of nosZ gene copies, because organic fertilizers contain readily available C compounds, which can stimulate denitrification (Paul and Beauchamp, 1989; Rochette et al., 2000). Dambreville et al. (2006) reported that the denitrifying community, determined using the narG and nosZ genes as molecular markers in restriction fragment length polymorphism (RFLP) assays, was more diverse in the plot amended with organic (composted pig manure) fertilizer than the plot amended with mineral (ammonium nitrate) fertilizer. Using qPCR to quantify the nosZ gene, Hallin et al. (2009) found nearly one order of magnitude more nosZ gene copies in systems receiving organic fertilizer (solid cattle manure and sewage sludge) compared to the system receiving only ammonium sulfate. Hence, the lower abundance of nosZ gene copies across the season in the organic system was unexpected. Furthermore, the lower abundance of nosZ gene copies in the organic cropping system did not reflect NO3− concentrations, which were not different across the cropping systems. Interestingly, the denitrifier abundances increased in the conventional and low-input systems across the season, but remained lower and did not change in the organic system. We found that the ratios of nosZ:16S rRNA gene abundances within the systems did not significantly change across the season (data not shown), thereby implying that the size of the denitrifier communities increased along with the total bacterial community. Complex interactions between soil water content, O2 content, availability of C, and NO3−-N levels are understood to be environmental factors that affect microbiological denitrification (Firestone, 1982; Tiedje, 1988 Soil moisture and NO3−-N levels, were similar across the three cropping systems. Hence, in our study, the increase in denitrifier abundances in the conventional and low-input systems compared to the lack of change in the organic systems, may be linked more to the effects of C availability on the denitrifier community, as C availability has been recognized as one of the most important regulatory factors controlling denitrification in soil (Burford and Bremner, 1975; Weier et al., 1993).
4.2. Soil microenvironments select for N cycling communities
Our amoA, nosZ, and 16S rRNA gene abundance data agree with other reports that suggest the location of soil microorganisms within the soil matrix can affect their metabolism and activity (Gupta and Germida, 1988; Lensi et al., 1995; Phillippot et al., 1996; Ranjard et al., 2000). Ammonia oxidizing, denitrifier, and total bacterial communities were generally larger in the microaggregate (Figs. 2, 3, and 4) than in the cPOM (>250 μm) and silt-and-clay. This was not surprising as microaggregates are important in the protection and stabilization of C (Jastrow, 1996; Six et al., 1998; Gale et al., 2000) and might provide better physical protection for microorganisms than the cPOM and silt-and-clay (Ranjard and Richaume, 2001). Moreover, C availability has been shown to increase rates of denitrification (Beauchamp et al., 1989; McCarty and Bremmer, 1992; Luo et al., 1999), and especially within the more anoxic environment of aggregate structures (Sexstone et al., 1985; Parkin, 1987; Drury et al., 2004).
Our results suggest that the microaggregate is a microenvironment that is more conducive to microbial proliferation and nitrifying and denitrifying activity than the cPOM and silt-and-clay. Consequently, our hypothesis that amoA, nosZ, and 16S rRNA gene abundances are higher in the microaggregate than in the cPOM and silt-and-clay fractions was corroborated. Furthermore, significant cropping system*soil microenvironment effects found for the gene abundances indicate that crop management interacts with the aggregates in selecting for nitrifiers, denitrifiers, and total bacterial communities. Larger denitrifier populations in the microaggregates of the organic and low-input compared to the conventional system at Ti may indicate that microaggregates of the organic and low-input systems have greater C availability and NO3− concentrations than the conventional system. In contrast, the higher abundance of AOB in the microaggregates of the conventional system suggested that this microenvironment was characterized by NH4+ levels and conditions relatively more favorable to AOB than the conditions associated with microaggregates of the low-input and organic systems. Additionally, the consistent growth of nitrifiers across the season, particularly in the microaggregates of the conventional system, concurrent with a decrease in denitrifiers, suggested a steady increase in availability of O2 over the season as the soil became drier.
4.3. Relationship between microbial community abundance and N processing
The combined significance of the first and second canonical axes in the first CCA of this study (p=0.006) suggested that AOB and denitrifier community abundances correlated to potential gross N mineralization and nitrification rates and to NH4+ and NO3− concentrations; however, the first and second canonical axes were no longer significant when sampling event was included as a covariable in the CCA. Therefore, our data suggested that seasonality substantially influences the relationship between gene abundances and inorganic N concentrations and transformation rates, and that this relationship is a weak one. The lack of correlation between NO3− concentrations and nosZ gene abundance is in agreement with Kandeler et al. (2006), who found that the soil nitrate content could not explain the abundance of 16S rRNA, narG, nirK, nirS, and nosZ genes in the early stage of primary succession in a glacier foreland. Moreover, other studies have also found that gene abundance and microbial community composition cannot predict corresponding ecosystem processes. For example, Ma et al. (2008) found that, although different nitrifier and denitrifier communities were specific to landforms within cultivated versus uncultivated wetlands, these differences were not related to land-use or landform differences in N2O emissions. Similarly, Dandie et al. (2008) found that differences in denitrifier community compositions and abundances in a potato (Solanum tuberosum L.) cropping system were not related to denitrification rates and N2O emissions.
The current approaches used to study microbial communities with respect to soil processes have made substantial inroads in our understanding of the linkages between the two. Nevertheless, to determine whether a relationship exists between ecosystem N processes and soil microbial population densities, future studies must ensure that approaches to measuring these relationships are not confounded by temporal or spatial biases. Moreover, the methods used to quantify nitrifiers and denitrifiers must strive to encompass all microorganisms responsible for nitrification and denitrification. Recently, Leininger et al. (2006) found a high abundance and wide distribution of archaeal ammonia monooxygenase (amoA) genes in the soil environment. To comprehensively characterize the nitrifying populations in soil, the abundances of autotrophic ammonia-oxidizing bacteria of the β- and γ-subgroups of proteobacteria should be considered along with the abundances of archaea.
4.4. Conclusions
This work sought to relate the abundances of AOB, denitrifiers, and total bacterial communities within different soil microenvironments to their activity along a gradient of long-term N management regimes. We found that, although crop management influenced AOB and denitrifier community abundance, changes in AOB and denitrifier community sizes did not relate to changes in NH4+ or NO3− concentrations and that potential gross N mineralization and nitrification rates were decoupled from nitrifier and denitrifier community abundances. Consequently, our study did not show a strong link between nitrifier and denitrifier abundances and N cycling processes. This study did find that microaggregates were characterized by larger nitrifier, denitrifier, and total bacterial communities that fluctuated more than communities in the cPOM and silt-and-clay fractions, particularly in low-input and organic systems; hence, the microaggregate appears to be a microenvironment that is conducive for nitrifiers and denitrifiers (e.g., higher C accumulation and NO3− content), and is, consequently, a potential hotspot for N2O loss.
Acknowledgments
Many thanks to Marisa Alcorta and the entire staff at the Russell Ranch for their assistance in the field. We also thank Engil Isadora Pujol Pereira, Alice Yan, and Theresa Yim for their invaluable work in the laboratory. Thanks to Neil Willits and Dr. John Battles for assisting us with the statistical analyses in this study and to two anonymous reviewers for their comments to improve this manuscript. Funding for this project was provided by the Kearney Foundation of Soil Science, University of California, a grant from the Western Sustainable Agriculture Research and Education Program, and by grant number 5 P42 ES04699 from the National Institute of Environmental Health Sciences, NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avrahami S, Conrad R, Braker G. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Applied and Environmental Microbiology. 2002;68:5685–5692. doi: 10.1128/AEM.68.11.5685-5692.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough D. The use of mean pool abundances to interpret 15N tracer experiments. I. Theory. Plant and Soil. 1991;131:89–96. [Google Scholar]
- Beare MH, Parmelee RW, Hendrix PF, Cheng W, Coleman DD, Crossley DA., Jr Microbial and faunal interactions and effects on litter nitrogen and decomposition in agroecosystems. Ecological Monographs. 1992;62:569–591. [Google Scholar]
- Beauchamp EG, Trevors JT, Paul JW. Carbon sources for bacterial denitrification. Advances in Soil Science. 1989;10:113–142. [Google Scholar]
- Birch HF. The effect of soil drying on humus decomposition and nitrogen availability. Plant and Soil. 1958;10:9–31. [Google Scholar]
- Bossio DA, Scow KM, Gunapala N, Graham KJ. Determinants of soil microbial communities: Effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microbial Ecology. 1998;36:1–12. doi: 10.1007/s002489900087. [DOI] [PubMed] [Google Scholar]
- Bronick C. Soil structure and management: a review. Geoderma. 2005;124:3–22. [Google Scholar]
- Burford JR, Bremner JM. Relationships between the denitrification capacities of soils and total, water-soluble and readily decomposable soil organic matter. Soil Biology & Biochemistry. 1975;7:389–394. [Google Scholar]
- Burger M, Jackson LE. Microbial immobilization of ammonium and nitrate in relation to ammonification and nitrification rates in organic and conventional cropping systems. Soil Biology & Biochemistry. 2003;35:29–36. [Google Scholar]
- Cavagnaro TR, Jackson LE, Scow KM, Hristova KR. Effects of arbuscular mycorrhizas on ammonia oxidizing bacteria in an organic farm soil. Microbial Ecology. 2007;54:618–626. doi: 10.1007/s00248-007-9212-7. [DOI] [PubMed] [Google Scholar]
- Dambreville C, Hallet S, Nguyen C, Morvan T, Germon JC, Philippot L. Structure and activity of the denitrifying community in a maize-cropped field fertilized with composted pig manure or ammonium nitrate. FEMS Microbiology Ecology. 2006;56:119–131. doi: 10.1111/j.1574-6941.2006.00064.x. [DOI] [PubMed] [Google Scholar]
- Dandie C, Burton DL, Zebarth BJ, Henderson SL, Trevors JT, Goyer C. Changes in bacteria denitrifier community abundance over time in an agricultural field and their relationship with denitrification activity. Applied and Environmental Microbiology. 2008;74:5997–6005. doi: 10.1128/AEM.00441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doane TA, Horwath WR. Spectrophotometric determination of nitrate with a single reagent. Analytical Letters. 2003;36:2713–2722. [Google Scholar]
- Drury CF, Yang XM, Reynolds WD, Tan CS. Influence of crop rotation and aggregate size on carbon dioxide production and denitrification. Soil and Tillage Research. 2004;79:87–100. [Google Scholar]
- Ellert BH, Bettany JR. Calculation of organic matter and nutrients stored in soils under contrasting management regimes. Canadian Journal of Soil Science. 1995;75:529–538. [Google Scholar]
- Elliott ET. Aggregate structure and carbon, nitrogen, and phosphorus in native and cultivated soils. Soil Science Society of America Journal. 1986;50:627–633. [Google Scholar]
- Firestone MK. Biological denitrification. In: Stevenson FJ, editor. Nitrogen in Agricultural Soils. American Society of Agronomy; Madison: 1982. pp. 289–326. [Google Scholar]
- Forster JC. Soil nitrogen. In: Alef K, Nannipieri P, editors. Methods in Applied Soil Microbiology and Biochemistry. Academic Press; San Diego. CA: 1995. pp. 79–87. [Google Scholar]
- Frey SD, Elliott ET, Paustian K. Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biology & Biochemistry. 1999;31:573–585. [Google Scholar]
- Gale WJ, Cambardella CA, Bailey TB. Root-derived carbon and the formation and stabilization of aggregates. Soil Science Society of America Journal. 2000;64:201–207. [Google Scholar]
- Gunapala N, Scow KM. Dynamics of soil microbial biomass and activity in conventional and organic farming systems. Soil Biology & Biochemistry. 1998;30:805–816. [Google Scholar]
- Gupta VVSR, Germida JJ. Distribution of microbial biomass and its activity in different soil aggregate size classes as affected by cultivation. Soil Biology & Biochemistry. 1988;20:777–786. [Google Scholar]
- Hallin S, Jones CM, Schloter M, Philippot L. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME Journal. 2009;3:597–605. doi: 10.1038/ismej.2008.128. [DOI] [PubMed] [Google Scholar]
- Hart SC, Nason GE, Myrold DD, Perry DA. Dynamics of gross nitrogen transformations in an old-growth forest: the carbon connection. Ecology. 1994;75:880–891. [Google Scholar]
- Henry S, Bru D, Stres B, Hallet S, Philippot L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Applied and Environmental Microbiology. 2006;72:5181–5189. doi: 10.1128/AEM.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiology Letters. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- Jastrow JD. Soil aggregate formation and the accrual of particulate and mineral-associated organic matter. Soil Biology & Biochemistry. 1996;28:656–676. [Google Scholar]
- Kandeler E, Deiglmayr K, Tscherko D, Bru D, Philippot L. Abundance of narG, nirS, nirK, and nosZgenes of denitrifying bacteria during p rimary successions of a glacier foreland. Applied and Environmental Microbiology. 2006;72:5957–5962. doi: 10.1128/AEM.00439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalchuk GA, Stephen JR. Ammonia-oxidizing bacteria: a model formolecular microbial ecology. Annual Review of Microbiology. 2001;55:485–529. doi: 10.1146/annurev.micro.55.1.485. [DOI] [PubMed] [Google Scholar]
- Kramer AW, Doane TA, Horwath WR, van Kessel C. Combining fertilizer and organic inputs to synchronize N supply in alternative cropping systems in California. Agriculture, Ecosystems and Environment. 2002;34:43–50. [Google Scholar]
- Lee J, Hopmans JW, Rolston DE, Baer SG, Six J. Determining soil carbon stock changes: Simple bulk density corrections fail. Agriculture, Ecosystems, and Environment. 2009;134:251–256. [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. Arachae predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Lensi R, Clays-Josserand A, Jocteur Monrozier L. Denitrifiers and denitrifying activity in size fractions of a mollisol under permanent pasture and continuous cultivation. Soil Biology & Biochemistry. 1995;27:61–69. [Google Scholar]
- Lepš J, Šmilauer P. Multivariate Analysis of Ecological Data Using CANOCO. University Press; Cambridge, UK: 2003. [Google Scholar]
- Luo J, Tillman RW, Ball PR. Factors regulating denitrification in a soil under pasture. Soil Biology & Biochemistry. 1999;31:913–927. [Google Scholar]
- Ma WK, Bedard-Haughn A, Siciliano SD, Farrell RE. Relationship between nitrifier and denitrifier community composition and abundance in predicting nitrous oxide emissions from ephemeral wetland soils. Soil Biology & Biochemistry. 2008;40:1114–1123. [Google Scholar]
- McCarty GW, Bremner JM. Availability of organic carbon for denitrification of nitrate in subsoils. Biology and Fertility of Soils. 1992;14:219–222. [Google Scholar]
- Miller MN, Zebarth BJ, Dandie CE, Burton DL, Goyer C, Trevors JT. Denitrifier community dynamics in soil aggregates under permanent grassland and arable cropping systems. Soil Science Society of America Journal. 2009;73:1843–1851. [Google Scholar]
- Mummey DL, Holben WE, Six J, Stahl PD. Spatial stratification of soil microbial populations in diverse soils: Rubrobacteria and Gemmatimonads are abundant in water-stable microaggregate interiors while Acidobacteria are primarily associated with macroaggregates. Microbial Ecology. 2006;51:404–411. doi: 10.1007/s00248-006-9020-5. [DOI] [PubMed] [Google Scholar]
- Okano Y, Hristova KR, Leutenegger CM, Jackson LE, Denison RF, Gebreyesus B, Lebauer D, Scow KM. Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Applied and Environmental Microbiology. 2004;70:1008–1016. doi: 10.1128/AEM.70.2.1008-1016.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin TB. Soil microsites as a source of denitrification variability. Soil Science Society of America Journal. 1987;51:1194–1199. [Google Scholar]
- Paul JW, Beauchamp EG. Effect of carbon constituents in manure on denitrification in soil. Canadian Journal of Soil Science. 1989;69:49–61. [Google Scholar]
- Philippot L, Renault P, Sierra J, Henault C, Clays-Josserand A, Chenu C, Chaussod R, Lensi R. Dissimilatory nitrite-reductase provides a competitive advantage to Pseudomonas spp. RTC01 to colonise the centre of soil aggregates. FEMS Microbiology Ecology. 1996;21:175–185. [Google Scholar]
- Ranjard L, Richaume AS. Quantitative and qualitative microscale distribution of bacteria in soil. Research in Microbiology. 2001;152:707–716. doi: 10.1016/s0923-2508(01)01251-7. [DOI] [PubMed] [Google Scholar]
- Ranjard L, Poly F, Combrisson J, Richaume A, Gourbière F, Thioulouse J, Nazaret S. Heterogeneous cell density and genetic structure of bacterial Pools associated with various soil microenvironments as determined by enumeration and DNA fingerprinting approach (RISA) Microbial Ecology. 2000;39:263–272. [PubMed] [Google Scholar]
- Recous S, Mary B, Faurie G. Microbial immobilization of ammonium and nitrate in cultivated soils. Soil Biology & Biochemistry. 1990;22:913–922. [Google Scholar]
- Robertson GP. Nitrogen use efficiency in row-crop agriculture: crop nitrogen use and soil nitrogen loss. In: Jackson LE, editor. Ecology in Agriculture. San Diego: Academic Press; 1997. pp. 347–363. [Google Scholar]
- Rochette P, Bochove EV, Prévost D, Angers DA, Côté D, Bertrand N. Soil carbon and nitrogen dynamics following application of pig slurry for the 19th consecutive years: II. Nitrous oxide fluxes and mineral nitrogen. Soil Science Society of America Journal. 2000;64:1396–1403. [Google Scholar]
- Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Applied and Environmental Microbiology. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakala WD, Cadisch G, Giller KE. Interactions between residues of maize and pigeonpea and mineral N fertilizers during decomposition and N mineralization. Soil Biology & Biochemistry. 2000;32:679–688. [Google Scholar]
- Saxton AM. Proceedings of 23rd SAS Users Group Intl. SAS Institute; Cary, NC: 1998. A macro for converting mean separation output to letter groupings in Proc Mixed; pp. 1243–1246. [Google Scholar]
- Scow KM. Soil microbial communities and carbon flow in agroecosystems. In: Jackson LE, editor. Ecology in Agriculture. San Diego: Academic Press; 1997. pp. 367–413. [Google Scholar]
- Seech AG, Beauchamp E. Denitrification in soil aggregates of different sizes. Soil Science Society of America Journal. 1988;52:1616–1621. [Google Scholar]
- Sexstone AJ, Revsbech NP, Parkin TB, Tiedje JM. Direct measurement of oxygen profiles and denitrification rates in soil aggregates. Soil Science Society of America Journal. 1985;49:645–651. [Google Scholar]
- Sexstone AJ, Parkin TB, Tiedje JM. Denitrification response to soil wetting in aggregate and unaggregated soil. Soil Biology & Biochemistry. 1988;20:767–769. [Google Scholar]
- Shi W, Norton JM. Microbial control of nitrate concentrations in an agricultural soil treated with dairy waste compost or ammonium fertilizer. Soil Biology & Biochemistry. 2000;32:1453–1457. [Google Scholar]
- Six J, Elliott ET, Paustian K, Doran J. Aggregation and soil organic matter accumulation in cultivated and native grassland soils. Soil Science Society of America Journal. 1998;62:1367–1377. [Google Scholar]
- Six J, Elliott ET, Paustian K. Soil macroaggregate turnover and microaggregate formation: A mechanism for C sequestration under no-tillage agriculture. Soil Biology & Biochemistry. 2000;32:2099–2103. [Google Scholar]
- Suzuki MT, Taylor LT, Delong EF. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environmental Microbiology. 2001;3:323–331. doi: 10.1046/j.1462-2920.2001.00198.x. [DOI] [PubMed] [Google Scholar]
- Stark JM, Hart SC. Diffusion technique for preparing salt solutions, Kjeldahl digests, and persulfate digests for nitrogen-15 analysis. Soil Science Society of America Journal. 1996;60:1846–1855. [Google Scholar]
- Tiedje JM. Ecology of denitrification and dissimilatory nitrate reduction to ammonium. In: Zehnder AJB, editor. Biology of Anaerobic Microorganisms. John Wiley; New York: 1988. pp. 179–244. [Google Scholar]
- Weier KL, Doran JW, Power JF, Walters DT. Denitrification and the dinitrogen/nitrous oxide ratio as affected by soil water, available carbon, and nitrate. Soil Science Society of America Journal. 1993;57:66–72. [Google Scholar]