Abstract
Tau undergoes numerous post-translational modifications during the progression of Alzheimer’s disease (AD). Some of these changes accelerate tau aggregation, while others are inhibitory. AD-associated inflammation is thought to create oxygen and nitrogen radicals such as peroxynitrite (PN). In vitro, PN can nitrate many proteins, including tau. We have previously demonstrated that tau’s ability to form filaments is profoundly affected by treatment with PN and have attributed this inhibition to tyrosine nitration. However, PN is highly reactive and unstable leading to oxidative amino acid modifications through its free radical byproducts. To test whether PN can modify other amino acids in tau via oxidative modifications, a mutant form of the tau protein lacking all tyrosines (5XY→F) was constructed. 5XY→F tau readily forms filaments; however, like wild-type tau the extent of polymerization was greatly reduced following PN treatment. Since 5XY→F tau cannot be nitrated, it was clear that non-nitrative modifications are generated by PN treatment and that these modifications change tau filament formation. Mass spectrometry was used to identify these oxidative alterations in wild-type tau and 5XY→F tau. PN-treated wild-type tau and 5XY→F tau consistently displayed lysine formylation throughout tau in a non-sequence specific distribution. Lysine formylation likely results from reactive free radical exposure caused by PN treatment. Therefore, our results indicate that PN treatment of proteins in vitro cannot be used to study protein nitration as it likely induces numerous other random oxidative modifications clouding the interpretations of any functional consequences of tyrosine nitration.
Tau is a microtubule-associated protein likely involved in normal cytoskeleton function (1, 2). Tau transitions from its relatively soluble state into filamentous aggregates called neurofibrillary tangles (NFTs) (3), constituting one of the pathological hallmarks of Alzheimer’s disease (AD) (4–6). The anatomical distribution of filamentous tau inclusions correlates with the cognitive deficits diagnostic of AD, appearing first in the transentorhinal and entorhinal cortices prior to emerging in the hippocampus (7, 8).
Post-translational modifications such as phosphorylation (4, 9, 10) and truncation (11) are thought to contribute to tau conformational changes (12–14) that accelerate filament formation. By contrast, nitration of tyrosine residues in tau has been reported to inhibit tau filament formation in vitro (15–17).
Unlike phosphorylation, which is catalyzed by naturally occurring protein kinases, nitration is thought to be the result of a non-catalytic event that involves peroxynitrite (PN). PN is a nitrogen radical posited to form in situ as the result of inflammatory processes, such as those associated with AD (18). This compound is capable of nitrating tyrosine residues in vitro (19) through its reaction with carbon dioxide leading to secondary nitrating species (20) that generate 3-nitrotyrosine (3-NT) by the addition of a nitro group. This alteration is thought to modify a protein’s hydrogen bonding, electrostatic properties, and hydrophobicity. Several groups have shown that nitration has biological selectivity and can alter both function and folding properties of the affected protein (21–25). In addition to tau, a number of proteins implicated in neurodegenerative diseases have been shown to be nitrated (21–25).
In addition to nitration of tyrosines, PN can cause oxidative modification of other amino acids including cysteine, tryptophan, histidine, and methionine among others (20). In fact, Norris and colleagues (2003) provided evidence that PN treatment could change the behavior of proteins independent of tyrosine nitration (26). The treatment of an α-synuclein mutant lacking tyrosine residues with PN impaired fibril formation, suggesting involvement of other oxidative amino acid modifications (26). However, the nature of these other alterations was not identified.
Previously, we reported that nitration of individual tyrosine residues in tau appeared to result in inhibition of filament formation (16); however, we did not determine whether other oxidative modifications occurred. To address this issue, a tau mutant lacking all tyrosine residues (5XY→F tau) was used in assembly reactions both with and without PN treatment to determine whether PN treatment caused other oxidative modifications in addition to nitration in tau. Here we report that PN treatment of 5XY→F tau prevents filament formation, indicating that PN can cause modifications in tau other than tyrosine nitration. Additionally, using mass spectrometry we identified an apparently non-sequence specific oxidative modification, formylation of lysine residues, generated by PN treatment. Our results indicate that formylation of lysines is sufficient to alter the polymerization capacity of tau in vitro, suggesting that PN should not be used to study tyrosine nitration, without taking into consideration other oxidative modifications caused by PN exposure.
Materials and Methods
Recombinant Tau Proteins
Wild-type and mutant tau proteins were expressed using a pT7C-tau plasmid that drives the expression of full-length human tau (441 amino acids) fused to an amino terminal 6X His affinity tag (27). This isoform contains two N-terminal insertions (exons 2 and 3) and exon 10 which encompasses MTBR 2 (28–30). Genetic modifications were used to create different tau constructs by point mutations with a QuickChange Site-Directed Mutagenesis kit (Stratagene). Four tau mutants, each containing a single tyrosine (Y) residue at position Y18, Y29, Y197, or Y394 were generated by mutating all of the other native tyrosine residues to phenylalanine (F) (Table 1) as previously described (15). A fifth tau mutant, with only Y310, was not used in this study since that residue is seldom nitrated by PN in vitro (16). A tau mutant containing no tyrosine residues, termed 5XY→F, was made as previously described (15) and used as a negative control. The identity of all the tau mutants was confirmed using DNA sequencing (Genomics Core Facility, Northwestern University). These tau mutants and wild-type tau were then expressed in E. coli strain BL21 (DE3) and purified using a Talon metal affinity column (Clontech), followed by size-exclusion chromatography to further separate full length tau from incompletely translated tau proteins and proteolytic fragments (31). The purification was assayed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie Brilliant Blue staining.
Table 1.
Description of all tau constructs used in this studya
| Nomenclature | Description |
|---|---|
| WT | All 5 native tyrosines at sites: Y18, Y29, Y197, Y310, Y394 |
| Y18 | Single tyrosine at Y18; (Y29F, Y197F, Y310F, Y394F) |
| Y29 | Single tyrosine at Y29; (Y18F, Y197F, Y310F, Y394F) |
| Y197 | Single tyrosine at Y197; (Y18F, Y29F, Y310F, Y394F) |
| Y394 | Single tyrosine at Y394; (Y18F, Y29F, Y197F, Y310F) |
| 5XY→F | No tyrosine residues; (Y18F, Y29F, Y197F, Y310F, Y394F) |
Y – tyrosine; F – phenylalanine
Protein Nitration
PN (Cayman Chemicals) was used to nitrate tau (15). The PN concentration was measured spectrophotometrically at 302 nm in 0.3 M NaOH (ε302=1670 M−1 cm−1) prior to each experiment (22). Proteins were dialyzed into nitration buffer [100 mM potassium phosphate, 25 mM sodium bicarbonate (pH 7.4), and 0.1 mM diethylenetriaminepentaactetic acid] for 16 hours at 4°C (15). After dialysis, the Lowry method was used to determine protein concentrations with bovine serum albumin as a standard (32). The pH of the solution was kept constant at 7.4 throughout the nitration reaction (33). The mutant and control tau proteins were exposed to 100-fold molar excess PN twice in separate reactions with vigorous stirring for 30 seconds each time at room temperature (15). After PN treatment, the samples were passed through a size-exclusion column to remove any 3’,3’-dityrosine cross-linked proteins created during the nitration process. The final concentration of PN-treated proteins was measured as outlined above.
Right-Angle Laser Light Scattering (LLS)
Polymerization of both untreated and PN-treated wild-type tau and tau mutants was induced using the fatty acid inducer arachidonic acid (Cayman Chemicals) as previously described (34). This in vitro assembly paradigm accurately models tau polymerization at near physiological conditions (e.g. tau concentrations, temperature, pH and ionic strength) and the filaments are morphologically similar to tau filaments isolated from postmortem AD brains (35, 36). Wild-type tau and PN-treated tau mutants were individually diluted in polymerization buffer (10 mM N-(2-hydroxyethyl) piperazine-N-2-ethanesulfonic acid, 100 mM sodium chloride, 0.1 mM ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid, and 5 mM dithiothreitol, pH 7.6).
Two distinct experimental approaches were employed. First, the effects of PN treatment on tau polymerization were assessed by measuring polymerization without (baseline) and with PN treatment of wild-type tau and single tyrosine tau mutants. Each protein, at a concentration of 4 µM, was polymerized in the presence of 75 µM arachidonic acid. Second, the effects of PN-treated tau species on polymerization of untreated tau were assayed by measuring polymerization in a reaction containing a mixture of untreated wild-type tau (4 µM) and PN-treated tau proteins (wild-type or single Y mutants) at three concentrations (0.4 µM, 0.8 µM and 4 µM). In these experiments, arachidonic acid was added to each reaction mixture separately to achieve a final concentration of 75 µM. After arachidonic acid was added, untreated tau and PN-treated tau solutions were mixed (250 µL total volume) and transferred into fluorometer cells (5 mm path length; PGC Scientific). All samples measured were illuminated with vertically polarized laser light at a wavelength of 475 nm generated by a Lexel model 65 ion laser set at 5 mW. Images were captured at a 90° angle to the incident light beam and perpendicular to the direction of polarization using a digital camera (Electrim Corp., model EDC1000HR) operated by HiCam ’95 software (written by G. Albrecht-Buehler, Northwestern University (available at http://.basic.northwestern.edu/g-buehler/hicam.htm)). Images were collected at 0, 5, 10, 15, 30, 45, 60, 90, 120, 180, 240, and 300 minutes. Background levels were obtained by determining the scattering intensity before the addition of arachidonic acid (time 0). The levels of scattered light were measured as pixel intensity using Photoshop 7.0 and background values were subtracted from each time point to obtain an accurate assessment of mass. The IC50 values and maximal level of inhibition were calculated using GraphPad Prism 3.0 software and compared using a one-way analysis of variance (ANOVA) test with all multiple comparison procedures done using the Student-Newman-Keuls method.
Quantitative Transmission Electron Microscopy (EM)
After the final LLS reading, the samples were fixed in 2% (w/v) glutaraldehyde for EM analysis. Samples were absorbed onto 300 mesh, carbon-Formvar coated copper grids and then negatively stained using 2% (w/v) uranyl acetate (37). The grids were analyzed using a JEOL JEM-1220 EM at 60 kV at a magnification of 12,000X. Five random pictures were taken from each grid for analysis. Images were processed in Adobe Photoshop 7.0, and average filament number, mass, and length were quantified using Optimus 6.5 software (Media Cybernetics) as described previously (38). Average filament mass was determined through the summation of all the filaments, and average filament length was calculated by dividing filament mass by filament number. The level of statistical significance was set at p≤0.05, and the means were compared through a two-way ANOVA with all pairwise multiple comparison procedures done using the Holm-Sidak method.
Mass Spectrometry
Proteins were separated by 1-D SDS-PAGE and stained with Coomassie Blue. Bands of interest were excised and the proteins digested in situ with trypsin (Promega Corporation). The digests were analyzed by capillary high performance liquid chromatography-electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) on a Thermo Fisher LTQ fitted with a New Objective PicoView 550 nanospray interface. On-line HPLC separation of the digests was accomplished with an Eksigent NanoLC micro HPLC: column, PicoFrit™ (New Objective; 75 µm i.d.) packed to 10 cm with C18 adsorbent (Vydac; 218MS 5 µm, 300 Å). A mass spectral scan strategy was used in which a survey scan was acquired followed by data-dependent collision-induced dissociation (CID) spectra of the seven most intense ions in the survey scan above a set threshold. Mascot (Matrix Science) was used to search the uninterpreted CID spectra against a locally-generated protein database (containing the sequences of wild-type tau and the Y→F tau mutants) concatenated with the SwissProt database. For initial searches, the variable modifications that were considered were: hydroxylation (Pro); nitration (Tyr); oxidation (His, Met, Trp); and S-nitrosylation (Cys). In subsequent searches, lysine formylation was included in place of cysteine S-nitrosylation. Cross correlation of the Mascot results with X! Tandem and determination of protein and peptide identity probabilities were accomplished by Scaffold (Proteome Software).
Results
Peroxynitrite treatment causes both nitration and oxidative modifications
Previously, we demonstrated that PN treatment efficiently nitrates tyrosines in tau in vitro (15). In the absence of PN treatment, wild-type tau and the single tyrosine tau mutants readily formed filaments; however, polymerization of wild-type tau and the single tyrosine tau mutants is markedly decreased by PN treatment (15, 16). The more likely scenario in situ is the presence of a mixture of non-nitrated and nitrated tau proteins. Thus, it was of considerable interest to determine whether PN-treated (nitrated) tau can influence the assembly of untreated wild-type tau. The addition of PN-treated wild-type tau to untreated tau in varying ratios (1:10, 1:5, and 1:1) was tested to establish the effects of tau nitration on polymerization of unmodified tau. The resulting measurements were normalized to polymerization of untreated wild-type tau controls that were run with each experiment. Addition of PN-treated wild-type tau to untreated wild-type tau resulted in a reduction in the total filament mass formed, but the rates of filament assembly were unaffected (Figure 1A). Since there was no change in polymerization rate, we chose to evaluate filament mass as a steady-state endpoint, and defined inhibition as a reduction in filaments longer than 20 nm after 5 hours incubation. The addition of PN-treated wild-type tau to untreated wild-type tau resulted in a dose-dependent decrease in the amount of filaments formed proportional to the amount of PN-treated tau added to the reaction (Figure 1A).
Figure 1.
Effects of PN treatment on wild-type and 5XY→F tau polymerization as assayed by LLS. (A) PN treatment reduces polymerization of wild-type tau (▲) compared to untreated wild-type tau (■). The addition of PN-treated wild-type tau to untreated wild-type tau at ratios of 1:10 (▽), 1:5 (♦) and 1:1 (○) causes a dose-dependent reduction in the polymerization of wild-type tau. (B) PN treatment reduces polymerization of 5XY→F tau (▲) compared to untreated wild-type tau (■). The addition of PN-treated 5XY→F tau to untreated tau at ratios of 1:10 (▽), 1:5 (♦) and 1:1 (○) causes a dose-dependent reduction in polymerization. (C) Quantitative EM measurements for filament mass for both the PN-treated wild-type tau (black) and 5XY→F (gray) additions to untreated tau. In A–C, the error bars are ±SEM and n=3 for each condition (*=p<0.05).
This inverse relationship between the amount of PN-treated wild-type tau added and the mass of filaments formed indicated that the modifications caused by PN treatment prevented untreated wild-type tau from forming filaments. To confirm that the effects on polymerization were not a result of changing the ratio between total tau and arachidonic acid (36), a polymerization reaction was performed in which different concentrations of untreated wild-type tau (0.4 µM, 0.8 µM and 4 µM) were added to a second sample of 4 µM untreated wild-type tau. No change of the polymerization rate was noted but, as expected, there was an increase in the total number of filaments formed with an increase in the amount of untreated wild-type tau in the reaction (data not shown). These results indicated that the tau to arachidonic acid ratios used in this experimental paradigm did not inhibit polymerization.
To determine whether nitration was causing inhibition of tau polymerization as opposed to other oxidative changes induced by PN treatment, we tested the effects of PN treatment on 5XY→F tau protein. In the absence of PN treatment, the 5XY→F tau mutant, in which all five of the tyrosine residues have been replaced with phenylalanines, assembled essentially the same as wild-type tau under the same experimental conditions in vitro (16). However, PN treatment caused a decrease in the ability of 5XY→F tau to polymerize into filaments, as assayed with LLS (Figure 1B). Furthermore, PN-treated 5XY→F tau caused a dose-dependent inhibition of untreated wild-type tau assembly (Figure 1B), much like the PN-treated wild-type tau. Quantitative EM assays indicated little significant difference between the mass of filaments formed when either PN-treated wild-type tau or PN-treated 5XY→F tau was incubated with untreated wild-type tau (Figure 1C). Representative EM images used for filament mass quantification in the PN-treated 5XY→F additions are shown in Figure 2. Taken together, these data suggest that PN treatment caused modifications other than tyrosine nitration that are capable of inhibiting tau filament assembly to a similar extent in both wild-type tau and 5XY→F tau.
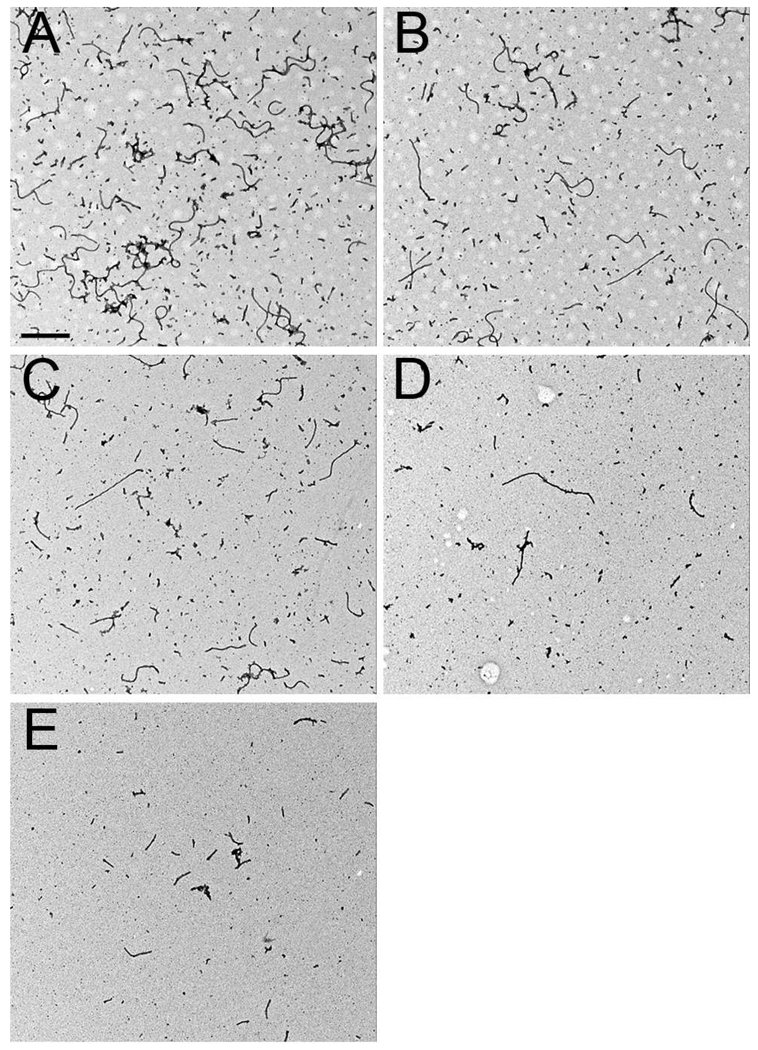
Figure 2.
PN treatment reduces the amount of filaments formed as assessed by EM. The EM images depict negatively stained tau filaments that are representative of those used for quantitative EM measurements. PN-treated 5XY→F tau (E) formed less filaments compared to untreated wild-type tau (A). A dose-dependent reduction in filament formation was observed when PN-treated 5XY→F tau was added to untreated wild-type tau at ratios of 1:10 (B), 1:5 (C) and 1:1 (D). Scale bars are 0.5 µm.
Mass spectrometry revealed nitration and oxidative modifications caused by PN
HPLC-electrospray ionization tandem mass spectrometry analysis of tryptic digests of wild-type tau and 5XY→F tau was used to indentify PN-derived tau modifications. In the initial Mascot database searches of tandem-MS data, nitration, S-nitrosylation and oxidation of methionine were included as possible modifications. Tyrosine nitration was detected only in PN-treated wild-type tau samples (Figure 3), and S-nitrosylation was not found in any of the samples, confirming our previous results (15).
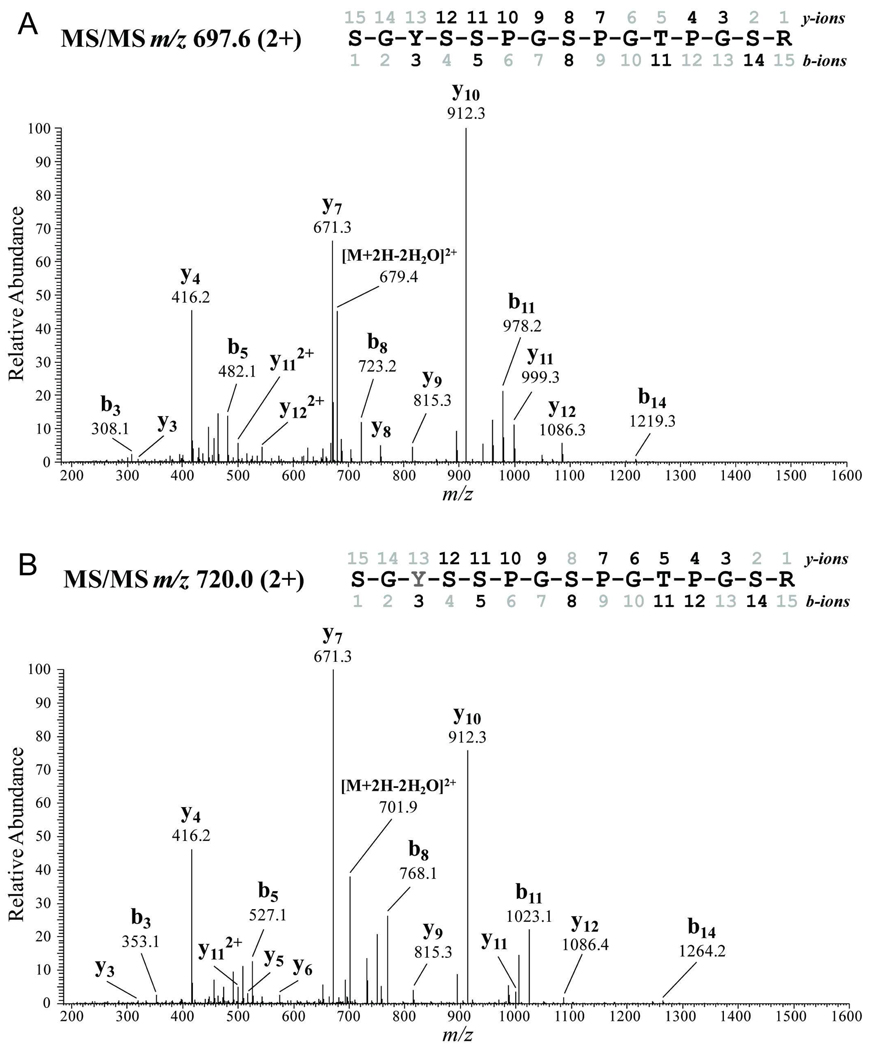
Figure 3.
Tandem mass spectra of wild-type tau. (A) Untreated wild-type tau contained unmodified amino acids. (B) In contrast, PN-treated wild-type tau contained a shift of 45 mass units on Tyr197 (Y, gray), indicative of a nitrotyrosine. Detected N-terminal (b-series) and C-terminal (y-series) ions are indicated on the spectra and peptides comprising amino acids 195–209 are depicted.
Additional Mascot database searches were conducted using an “error tolerant” strategy in which all modifications listed in UniMod were considered to determine whether other modifications resulting from PN treatment were present. The results indentified an additional mass of 28 on numerous lysine residues, randomly distributed in both PN-treated wild-type tau and 5XY→F tau (Figure 4). The observed mass shift is consistent with lysine formylation (see Discussion). Since full-length tau contains 44 lysine residues, accounting for 9.98% of the 441 amino acids (Figure S1), the high frequency of formylation observed in both PN-treated wild-type and 5XY→F tau suggests that lysine formylation is responsible for inhibition of tau polymerization.
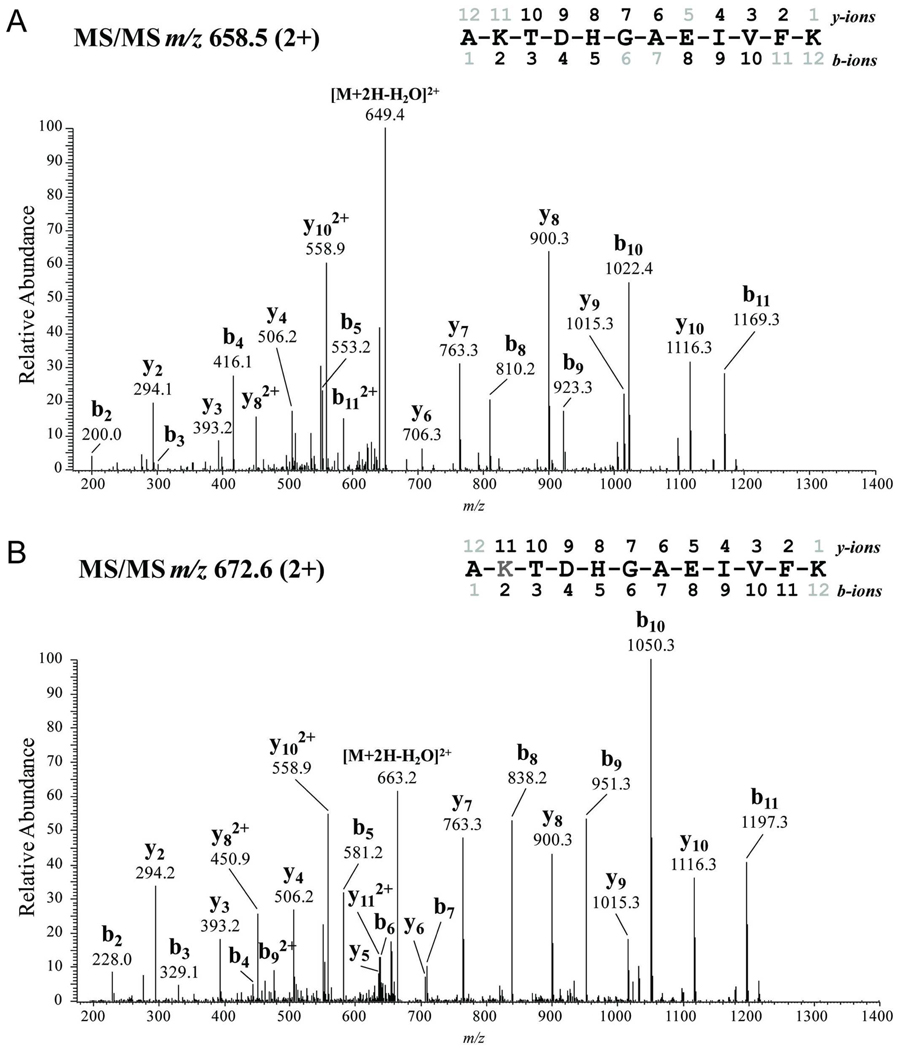
Figure 4.
Tandem mass spectra of 5XY→F tau. (A) Untreated 5XY→F tau contained unmodified amino acids. (B) In contrast, PN-treated 5XY→F tau has a shift of 28 mass units on Lys385 (K, gray), indicative of a formyl-lysine. Detected N-terminal (b-series) and C-terminal (y-series) ions are indicated on the spectra and peptides comprising amino acids 384–395 are depicted.
Tau nitrated at Y197 retains its polymerization capabilities
Attempts were made to nitrate tau without causing any effects on the polymerization profile of the 5XY→F protein by using alternative methods. First, both wild-type tau and 5XY→F were exposed to different concentrations of PN (0.01–100 molar excess of PN compared to tau) to determine if there was a concentration of PN that could cause nitration and not alter the polymerization capacity of the control protein. At all concentrations, PN caused a consistent inhibitory effect on polymerization of 5XY→F (data not shown). Another nitration method was used that involves a transition metal center and nitrative reagents that have the capacity to nitrate a protein. However, this method caused unexpected results in that it drastically inhibited polymerization in 5XY→F protein, and the filaments that formed from wild-type tau did not morphologically resemble filaments typically generated from tau polymerization (data not shown).
Since the lysine modification detected in our experiments appeared to have a substantial influence on tau polymerization, we reasoned that only the effects that were significantly different from PN-treated 5XY→F tau could be attributable to tyrosine nitration. The concentration of each tau mutant at which 50% inhibition (IC50) was achieved is reported in Table 2. The IC50 values of PN-treated wild-type tau, Y18 and Y197 are statistically significant when each is compared to PN-treated Y29, Y394 and 5XY→F. In addition to the differences in IC50 values of the various tau constructs, the amount of maximum inhibition of wild-type tau polymerization must also be taken into consideration to determine if any tau mutant is significantly different from PN-treated 5XY→F tau. In comparing the maximal inhibition of wild-type tau polymerization, PN-treated Y197 was the only tau construct statistically significant from all other PN-treated tau mutants, wild-type tau and 5XY→F (Table 2). Surprisingly, nitration at Y197 appears to reduce the inhibitory effects of lysine formylation as PN-treated Y197 tau formed significantly more filaments alone and when added to untreated wild-type tau compared to the PN-treated 5XY→F polymerization profiles (Figure 5A). Comparative analysis of the IC50 values and maximal inhibition for all tau species used in this study (Table 2) revealed that PN-treated Y394 tau was an appropriate representative of the similarity of the PN-treated single tyrosine mutants compared to 5XY→F (Figure 5B). In contrast, PN-treated Y197 tau was statistically significant when compared to both the IC50 and maximal inhibition of 5XY→F, and the maximal inhibition achievable with this mutant was lower than the other variants (56.6% inhibition). The observation that Y197 tau cannot inhibit normal tau assembly to the same extent as the other tau mutants could indicate that nitration at site 197 stabilizes the soluble conformation of the protein, making it more assembly competent.
Table 2.
Polymerization inhibition kinetics for each PN-treated tau constructa
| Tau | IC50 (µM) | Max Inhibition (% of wild-type tau) |
|---|---|---|
| WT | 1.922 ± 0.608* | 90.3 ± 0.73 |
| Y18 | 1.329 ± 0.459* | 90.5 ± 1.95 |
| Y29 | 0.283 ± 0.107 | 89.1 ± 1.17 |
| Y197 | 0.913 ± 0.146* | 56.6 ± 5.79** |
| Y394 | 0.475 ± 0.029 | 88.0 ± 0.47 |
| 5XY→F | 0.512 ± 0.027 | 77.4 ± 1.35 |
IC50 – concentration at 50% inhibition
p≤0.05 compared to PN-treated Y29, Y394 and 5XY→F constructs
p≤0.05 compared to all other constructs
Figure 5.
PN-treated Y197 had a significantly different polymerization profile compared to 5XY→F tau and all other single tyrosine mutants. (A) PN treatment reduces polymerization in Y197 (▲) compared to untreated wild-type tau (■). The addition of PN-treated Y197 to untreated wild-type tau at ratios of 1:10 (▽), 1:5 (♦) and 1:1 (○) causes a dose-dependent reduction in polymerization. (B) PN treatment reduces polymerization in Y394 (▲) compared to untreated tau (■). The addition of PN-treated Y394 to untreated tau at ratios of 1:10 (▽), 1:5 (♦) and 1:1 (○) causes a dose-dependent reduction in polymerization. (C) Quantitative EM indicated that PN-treated Y197 tau (black) forms significantly more filaments than PN-treated 5XY→F tau (light gray) when polymerized alone, and more filaments are formed when PN-treated Y197 tau is co-incubated with untreated wild-type tau compared to when PN-treated 5XY→F tau is co-incubated. In contrast, PN-treated Y394 tau (dark gray) behaves the same as PN-treated 5XY→F tau when polymerized in isolation and when co-incubated with untreated wild-type tau. In A–C the error bars are ±SEM and n=3 for each condition (*=p<0.05).
Quantitative EM analyses confirmed that PN-treated wild-type tau, Y18, Y29 and Y394 single tyrosine tau mutants were not significantly different when compared to PN-treated 5XY→F tau in inhibiting the assembly of wild-type tau. In contrast, the total filament mass of PN-treated Y197 tau was significantly greater than PN-treated Y394 and 5XY→F tau when polymerized alone or at the 1:1 dilution with untreated tau (Figure 5C). In addition, when polymerized in isolation, PN-treated Y197 exhibited a statistically significant increased amount of filaments compared to all the other PN-treated single tyrosine tau mutants and wild-type tau (data not shown). However, at the 1:10 and 1:5 dilutions, Y197 was not different from PN-treated 5XY→F tau (Figure 5C). These findings indicate that, after PN treatment, the Y197 single tyrosine mutant can readily assemble despite the presence of lysine formylation.
Discussion
Evidence reported in the literature clearly demonstrates that tyrosine nitration occurs in several proteins in neurodegenerative diseases (39). Understanding the way nitration affects protein function is crucial to determining how this modification plays a role in disease pathogenesis. Indeed, previous studies have confirmed that tau is nitrated during the progression of AD (40–42). Currently, the prevailing hypothesis is that PN, created by inflammatory processes (e.g. reaction between superoxide and nitric oxide), is the primary nitrating agent (43). Paradoxically, only a relatively small number of proteins appear to be nitrated in situ, and addition of a nitrate group on a single tyrosine residue can be sufficient to cause protein dysfunction (21). The current study demonstrates that PN treatment can nitrate tau, but, under the conditions used, there were additional modifications (e.g. lysine formylation) that had a substantial effect on tau polymerization. This indicates that PN treatment under these common experimental conditions is not specific for tyrosine nitration in vitro.
PN is a potent one- or two-electron oxidant that can spontaneously form •OH and •NO2 radicals (44). PN does not directly react with tyrosine but oxidizes and nitrates through radical byproducts (19). The free radicals interact with tyrosine and can abstract a hydrogen atom from the amino acid creating a tyrosyl radical. This radical can recombine with •NO2 to form 3-nitrotyrosine. The protocol used to nitrate and oxidize tau in this report is a well-established and commonly used method, but in most cases, additional modifications by PN exposure are not considered. Previous studies have established that PN causes not only nitration of tyrosines, but can also oxidize and/or nitrate cysteine, methionine, tryptophan, histidine, and phenylalanine among other amino acid residues. (19, 20). In the absence of CO2 (in the form of bicarbonate), cysteine is the amino acid that reacts the fastest with PN (20). In the presence of CO2, cysteine oxidation is decreased by 50% because the direct reaction of PN with CO2 outcompetes the reaction with the thiol of the cysteine (20). Using mass spectrometry, we confirmed that the two cysteines within the wild-type tau were not modified by PN. The common use of PN as a seemingly convenient nitrating agent in vitro stems from the increase of nitrotyrosine formation in the presence of CO2, created through the use of a 25 mM bicarbonate buffer during PN treatment (45). Also, the nitration reaction can be promoted through exposure of the tyrosine, association with a negative charge, and absence of proximal cysteines (22). For these reasons, as well as confirmatory mass spectrometry data (15, 45–47), PN exposure became a widely used method for protein nitration.
Our data clearly demonstrate that non-nitrative modifications occur in vitro and emphasize the need for proper controls when working with PN. A particularly striking discovery of our studies was uncovered when we examined PN-treated 5XY→F tau. After PN treatment, 5XY→F tau was unable to self-assemble into filaments, and it decreased the polymerization capacity of the unmodified tau species when co-incubated in solution. This suggests that PN treatment is causing an oxidative change that, in some manner, affects the ability of the untreated wild-type tau in solution to polymerize. These results closely mirrored those found with an α-synuclein mutant lacking tyrosines that was treated with PN (26). Norris and coworkers (2003) showed that PN treatment significantly reduced the amount of fibrils formed by an α-synuclein mutant lacking all tyrosine residues in vitro (26). While the authors acknowledged that the PN treatment must have caused other oxidative modifications, the identities of these modifications were not determined. Hence, it is possible that lysine formylation may have played a role in functionally altering α-synuclein as well, but this cannot be ascertained without further analysis.
Lysine formylation was not only found in the 5XY→F tau, but also in PN-treated wild-type tau. Formylation is the addition of HCO to the primary amino group on the side chain of lysine, eliminating a basic site on the protein. Although this is the most likely modification that could occur in our experimental paradigm, other groups have demonstrated that PN can create formaldehyde in a reaction with dimethyl sulfoxide (DMSO) (48, 49). Even though DMSO is not present in our reaction, creation of formaldehyde through the reaction of PN and the bicarbonate buffer could cause formylation of the lysine residues in tau. Other possibilities that would create an additional mass of 28, such as a dimethyl modification of lysine, cannot be completely excluded; however, there seems to be little likelihood of lysine dimethylation under these experimental conditions. Interestingly, lysine formylation did not appear to be site-specific as this modification was found throughout the protein. The fact that this modification was found at numerous lysine residues within the protein and that it changes the charge of the amino acid certainly could explain its disruption of tau polymerization in vitro. However, since the modification appeared to be non-sequence specific, it might not be possible to elucidate a clear mechanism for the influence of the formyl-lysine residues on tau polymerization.
Little is known about lysine formylation, and certainly, the effects of lysine formylation have not been studied in the context of tau function, its presence in AD and its role in disease pathogenesis. However, lysine formylation has been identified as a post-translational modification of histones and other nuclear proteins (50, 51). Multiple formylation sites in histones have been shown to be important in DNA binding and may interfere with chromatin function, leading to cellular dysfunction and diseases such as cancer (50). Additionally, lysine formylation can be stimulated by oxidative stress (51). Oxidation of histones and lipid peroxidation have been suggested to contribute to the vulnerability of DNA to oxidation in AD brain (52). Oxidative stress creates free carbonyls that arise from side-chain oxidations of lysine as well as other amino acids (53). It has been demonstrated that carbonyl formation is a major oxidative modification that can result from PN in addition to the nitration of tyrosine phenols (42). Alzheimer’s disease tissue has been recognized to have increased carbonyl reactivity associated with neurofibrillary changes; indicative of an increase in oxidative stress during neurodegeneration (42, 54). In addition, the distribution of nitrotyrosine reactivity in situ in AD has been reported to be very similar to the staining of free carbonyls (42, 54). Other studies have demonstrated that 4-hydroxy-2-nonenal (HNE) a product of oxidative stress, can modify the nucleophilic side chains of several amino acids including lysine (55, 56). Oxidation by HNE causes carbonyl enhancement of lysines and this modification stabilizes the Alz50 epitope (57). It has been reported that this increase in the Alz50 conformation mediated by HNE is also dependent upon the phosphorylation of tau (58). These studies of HNE mediated lysine modifications suggest that carbonyl-modified tau may be a crucial component of the change from soluble tau to NFTs (57, 58). Also, selective carbamoylation of lysines reduces the capacity of tau to induce microtubule assembly and induces aggregation of tau into filaments (59) These studies strengthen the conclusion that lysine modifications may alter the normal polymerization profile of tau; however, further research is required to elucidate the effects of lysine formylation on tau folding and function.
A critically important finding in this work is the fact that PN cannot be used in vitro as a nitrating agent under these commonly used experimental conditions because it causes additional oxidative modifications that must be taken into consideration. These results bring into question the interpretation of our previous study conducted with the site-specific nitrated tau mutants (16). Importantly, we did confirm the polymerization results initially reported with PN-treated single tyrosine mutants (16), but it was necessary to reinterpret the data from this past publication in light of these new findings. The polymerization profile of 5XY→F tau after PN treatment must be taken into consideration prior to assaying the effect of nitration at specific tyrosine residues. Only one single tyrosine tau mutant had a significantly different effect on filament formation when compared to 5XY→F tau. The single tyrosine mutant Y197 appeared to be less affected by PN treatment in that it could still form filaments even though some of its lysines were likely formylated. Tyrosine 197 is located within the proline rich region of tau where there is also an abundance of lysine residues (Figure S1). This ability for PN-treated Y197 to form filaments could suggest that nitration at Y197 neutralizes the negative effect of formylation on the assembly process to some degree. Nitration at this site could also affect the ability of the surrounding lysines to be formylated. Further research is necessary to determine the method by which nitration at Y197 may affect filament formation in disease progression.
There are numerous important implications of the current study. First and foremost, PN cannot be used to study nitration in vitro under experimental conditions utilizing a bicarbonate buffer, as random formylation of lysines will likely mask any change due to nitrotyrosine formation. Second, if proteins known to contain 3-nitrotyrosine modifications are isolated from tissue, but do not contain formylated lysines, it is unlikely that PN is the effector responsible for tyrosine nitration. Third, accumulating data and results from our study supports the view that the likelihood of PN as a nitrating agent in situ is very low since nitrated proteins (including tau) have been found in normal cells and animals (60–62). Lastly, if PN is not responsible for nitration then one must posit other means for the nitrative modifications observed. Since nitration appears to be selective and specific in situ, it is tempting to speculate that nitration occurs through a more specific, enzymatic reaction; however, such a nitrotransferase has yet to be described. That said, a nitrotyrosine denitrase has been identified that can remove nitro groups from previously nitrated proteins (63).
In summary, our work indicates that PN treatment of tau causes nitration on tyrosines and non-sequence specific addition of formyl groups to lysine residues. This work likely invalidates the common experimental protocol that uses PN as a nitrating agent in the presence of a bicarbonate buffer on most proteins and suggests that alternative means must be discovered or developed to study the effect of nitration on protein function in vitro.
Supplementary Material
Acknowledgement
We gratefully acknowledge Dr. George Perry for sharing an unpublished protocol that was used as an alternate method to nitrate tau, Andres Silva for technical assistance, and Juan F. Reyes for inspiration and critical reading of the manuscript.
This work was supported by NIH Awards AG032091 (L.I.B.) and AG014449 (L.I.B.).
Abbreviations
- AD
Alzheimer’s disease
- ANOVA
analysis of variance
- CID
collision-induced dissociation
- DMSO
dimethyl sulfoxide
- EM
electron microscopy
- HNE
4-hydroxy-2-nonenal
- HPLC
high performance liquid chromatography
- IC50
concentration that causes 50% inhibition
- LLS
laser light scattering
- 3-NT
3-nitrotyrosine
- NFT
neurofibrillary tangle
- PN
peroxynitrite
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEM
standard error of the mean
- WT
wild-type tau
Footnotes
Supporting Information Available:
Even though the distribution of lysine formylation was non-sequence specific, it was of interest to examine the relationship between the lysine and tyrosine residues of tau. Lysine residues are found throughout tau with an abundance found within the proline rich region, which also contains tyrosine 197. There is a lysine located in close proximity of each of the tyrosine sites studied in this report. Figure S1 highlights the location of the tyrosine and lysine residues within the tau sequence. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Johnson GVW, Hartigan JA. Tau protein in normal and Alzheimer's disease brain: An update. Alzheimer's Disease Review. 1998;3:125–141. doi: 10.3233/jad-1999-14-512. [DOI] [PubMed] [Google Scholar]
- 2.Buee L, Bussiere T, Buee-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and the role in neurodegenerative disorders. Brain Res Brain Res Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Goedert M. Tau protein pathology in neurodegenerative diseases. Trends Neurosci. 1998;21:428–433. doi: 10.1016/s0166-2236(98)01337-x. [DOI] [PubMed] [Google Scholar]
- 4.Wood JG, Mirra SS, Pollock NJ, Binder LI. Neurofibrillary tangles of Alzheimer disease share antigenic determinants with the axonal microtubule-associated protein tau (tau) [published erratum appears in Proc Natl Acad Sci U S A 1986 Dec;83(24):9773] Proc Natl Acad Sci U S A. 1986;83:4040–4043. doi: 10.1073/pnas.83.11.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosik KS, Joachim CL, Selkoe DJ. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc Natl Acad Sci U S A. 1986;83:4044–4048. doi: 10.1073/pnas.83.11.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundke-Iqbal I, Iqbal K, Quinlan M, Tung YC, Zaidi MS, Wisniewski HM. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J Biol Chem. 1986;261:6084–6089. [PubMed] [Google Scholar]
- 7.Arriagada PV, Growdon JH, Hedley-Whyte ET, Hyman BT. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer's disease. Neurology. 1992;42:631–639. doi: 10.1212/wnl.42.3.631. [DOI] [PubMed] [Google Scholar]
- 8.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 9.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci U S A. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonso AC, Grundke-Iqbal I, Iqbal K. Alzheimer's disease hyperphosphorylated tau sequesters normal tau into tangles of filaments and disassembles microtubules. Nat Med. 1996;2:783–787. doi: 10.1038/nm0796-783. [DOI] [PubMed] [Google Scholar]
- 11.Gamblin TC, Chen F, Zambrano A, Abraha A, Lagalwar S, Guillozet AL, Lu M, Fu Y, Garcia-Sierra F, LaPointe N, Miller R, Berry RW, Binder LI, Cryns VL. Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer's disease. Proc Natl Acad Sci U S A. 2003;100:10032–10037. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghoshal N, Garcia-Sierra F, Fu Y, Beckett LA, Mufson EJ, Kuret J, Berry RW, Binder LI. Tau-66: Evidence for a novel tau conformation in Alzheimer's disease. J Neurochem. 2001;77:1372–1385. doi: 10.1046/j.1471-4159.2001.00346.x. [DOI] [PubMed] [Google Scholar]
- 13.Ghoshal N, Garcia-Sierra F, Wuu J, Leurgans S, Bennett DA, Berry RW, Binder LI. Tau Conformational Changes Correspond to Impairments of Episodic Memory in Mild Cognitive Impairment and Alzheimer's Disease. Exp Neurol. 2002;177:475–493. doi: 10.1006/exnr.2002.8014. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Sierra F, Ghoshal N, Quinn B, Berry RW, Binder LI. Conformational changes and truncation of tau protein during tangle evolution in Alzheimer's disease. J Alzheimer's Dis. 2003;5:65–77. doi: 10.3233/jad-2003-5201. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds MR, Berry RW, Binder LI. Site-specific nitration and oxidative dityrosine bridging of the tau protein by peroxynitrite: implications for Alzheimer's disease. Biochemistry. 2005;44:1690–1700. doi: 10.1021/bi047982v. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds MR, Berry RW, Binder LI. Site-specific nitration differentially influences tau assembly in vitro. Biochemistry. 2005;44:13997–14009. doi: 10.1021/bi051028w. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds MR, Lukas TJ, Berry RW, Binder LI. Peroxynitrite-mediated tau modifications stabilize preformed filaments and destabilize microtubules through distinct mechanisms. Biochemistry. 2006;45:4314–4326. doi: 10.1021/bi052142h. [DOI] [PubMed] [Google Scholar]
- 18.Beckman JS, Chen J, Crow JP, Ye YZ. Reactions of nitric oxide, superoxide and peroxynitrite with superoxide dismutase in neurodegeneration. Prog Brain Res. 1994;103:371–380. doi: 10.1016/s0079-6123(08)61151-6. [DOI] [PubMed] [Google Scholar]
- 19.Beckman JS. Oxidative damage and tyrosine nitration from peroxynitrite. Chem. Res. Toxicol. 1996;9:836–844. doi: 10.1021/tx9501445. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez B, Radi R. Peroxynitrite reactivity with amino acids and proteins. Amino Acids. 2003;25:295–311. doi: 10.1007/s00726-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 21.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem Biophys Res Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 22.Souza JM, Daikhin E, Yudkoff M, Raman CS, Ischiropoulos H. Factors determining the selectivity of protein tyrosine nitration. Arch Biochem Biophys. 1999;371:169–178. doi: 10.1006/abbi.1999.1480. [DOI] [PubMed] [Google Scholar]
- 23.Greenacre SA, Ischiropoulos H. Tyrosine nitration: localisation, quantification, consequences for protein function and signal transduction. Free Radical Res. 2001;34:541–581. doi: 10.1080/10715760100300471. [DOI] [PubMed] [Google Scholar]
- 24.MacMillan-Crow LA, Crow JP, Thompson JA. Peroxynitrite-mediated inactivation of manganese superoxide dismutase involves nitration and oxidation of critical tyrosine residues. Biochemistry. 1998;37:1613–1622. doi: 10.1021/bi971894b. [DOI] [PubMed] [Google Scholar]
- 25.Guittet O, Decottignies P, Serani L, Henry Y, Le Marechal P, Laprevote O, Lepoivre M. Peroxynitrite-mediated nitration of the stable free radical tyrosine residue of the ribonucleotide reductase small subunit. Biochemistry. 2000;39:4640–4648. doi: 10.1021/bi992206m. [DOI] [PubMed] [Google Scholar]
- 26.Norris EH, Giasson BI, Ischiropoulos H, Lee VM. Effects of oxidative and nitrative challenges on alpha-synuclein fibrillogenesis involve distinct mechanisms of protein modifications. J Biol Chem. 2003;278:27230–27240. doi: 10.1074/jbc.M212436200. [DOI] [PubMed] [Google Scholar]
- 27.Carmel G, Mager EM, Binder LI, Kuret J. The structural basis of monoclonal antibody Alz50's selectivity for Alzheimer's disease pathology. J Biol Chem. 1996;271:32789–32795. doi: 10.1074/jbc.271.51.32789. [DOI] [PubMed] [Google Scholar]
- 28.Lee G, Cowan N, Kirschner M. The primary structure and heterogeneity of tau protein from mouse brain. Science. 1988;239:285–288. doi: 10.1126/science.3122323. [DOI] [PubMed] [Google Scholar]
- 29.Kosik KS, Orecchio LD, Binder L, Trojanowski JQ, Lee VM, Lee G. Epitopes that span the tau molecule are shared with paired helical filaments. Neuron. 1988;1:817–825. doi: 10.1016/0896-6273(88)90129-8. [DOI] [PubMed] [Google Scholar]
- 30.Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 31.Abraha A, Ghoshal N, Gamblin TC, Cryns V, Berry RW, Kuret J, Binder LI. C-terminal inhibition of tau assembly in vitro and in Alzheimer's disease. J Cell Sci. 2000;113:3737–3745. doi: 10.1242/jcs.113.21.3737. [DOI] [PubMed] [Google Scholar]
- 32.Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 33.Ischiropoulos H, al-Mehdi AB. Peroxynitrite-mediated oxidative protein modifications. FEBS Lett. 1995;364:279–282. doi: 10.1016/0014-5793(95)00307-u. [DOI] [PubMed] [Google Scholar]
- 34.Gamblin TC, King ME, Dawson H, Vitek MP, Kuret J, Berry RW, Binder LI. In vitro polymerization of tau protein monitored by laser light scattering: Method and application to the study of FTDP-17 mutants. Biochemistry. 2000;39:6136–6144. doi: 10.1021/bi000201f. [DOI] [PubMed] [Google Scholar]
- 35.Wilson DM, Binder LI. Free fatty acids stimulate the polymerization of tau and amyloid beta peptides. In vitro evidence for a common effector of pathogenesis in Alzheimer's disease. Am J Pathol. 1997;150:2181–2195. [PMC free article] [PubMed] [Google Scholar]
- 36.King ME, Ahuja V, Binder LI, Kuret J. Ligand-dependent tau filament formation: Implications for Alzheimer's disease progression. Biochemistry. 1999;38:14851–14859. doi: 10.1021/bi9911839. [DOI] [PubMed] [Google Scholar]
- 37.Gamblin TC, King ME, Kuret J, Berry RW, Binder LI. Oxidative regulation of fatty acid-induced tau polymerization. Biochemistry. 2000;39:14203–14210. doi: 10.1021/bi001876l. [DOI] [PubMed] [Google Scholar]
- 38.King ME, Gamblin TC, Kuret J, Binder LI. Differential assembly of human tau isoforms in the presence of arachidonic acid. J Neurochem. 2000;74:1749–1757. doi: 10.1046/j.1471-4159.2000.0741749.x. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds MR, Berry RW, Binder LI. Nitration in neurodegeneration: deciphering the "Hows" "nYs". Biochemistry. 2007;46:7325–7336. doi: 10.1021/bi700430y. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds MR, Reyes JF, Fu Y, Bigio EH, Guillozet-Bongaarts AL, Berry RW, Binder LI. Tau nitration occurs at tyrosine 29 in the fibrillar lesions of Alzheimer's disease and other tauopathies. Journal of Neuroscience. 2006;26:10636–10645. doi: 10.1523/JNEUROSCI.2143-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyes JF, Reynolds MR, Horowitz PM, Fu Y, Guillozet-Bongaarts AL, Berry R, Binder LI. A possible link between astrocyte activation and tau nitration in Alzheimer's disease. Neurobiol Dis. 2008;31:198–208. doi: 10.1016/j.nbd.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith MA, Richey Harris PL, Sayre LM, Beckman JS, Perry G. Widespread peroxynitrite-mediated damage in Alzheimer's disease. J Neurosci. 1997;17:2653–2657. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Vliet A, Eiserich JP, Halliwell B, Cross CE. Formation of reactive nitrogen species during peroxidase-catalyzed oxidation of nitrite. A potential additional mechanism of nitric oxide-dependent toxicity. J Biol Chem. 1997;272:7617–7625. doi: 10.1074/jbc.272.12.7617. [DOI] [PubMed] [Google Scholar]
- 44.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci U S A. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greis KD, Zhu S, Matalon S. Identification of nitration sites on surfactant protein A by tandem electrospray mass spectrometry. Arch Biochem Biophys. 1996;335:396–402. doi: 10.1006/abbi.1996.0522. [DOI] [PubMed] [Google Scholar]
- 46.Crow JP, Ye YZ, Strong M, Kirk M, Barnes S, Beckman JS. Superoxide dismutase catalyzes nitration of tyrosines by peroxynitrite in the rod and head domains of neurofilament-L. J Neurochem. 1997;69:1945–1953. doi: 10.1046/j.1471-4159.1997.69051945.x. [DOI] [PubMed] [Google Scholar]
- 47.Yi D, Smythe GA, Blount BC, Duncan MW. Peroxynitrite-mediated nitration of peptides: characterization of the products by electrospray and combined gas chromatography-mass spectrometry. Arch Biochem Biophys. 1997;344:253–259. doi: 10.1006/abbi.1997.0218. [DOI] [PubMed] [Google Scholar]
- 48.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci U S A. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denicola A, Freeman BA, Trujillo M, Radi R. Peroxynitrite reaction with carbon dioxide/bicarbonate: kinetics and influence on peroxynitrite-mediated oxidations. Arch Biochem Biophys. 1996;333:49–58. doi: 10.1006/abbi.1996.0363. [DOI] [PubMed] [Google Scholar]
- 50.Wisniewski JR, Zougman A, Mann M. Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008;36:570–577. doi: 10.1093/nar/gkm1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci U S A. 2007;104:60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drake J, Petroze R, Castegna A, Ding Q, Keller JN, Markesbery WR, Lovell MA, Butterfield DA. 4-Hydroxynonenal oxidatively modifies histones: implications for Alzheimer's disease. Neurosci Lett. 2004;356:155–158. doi: 10.1016/j.neulet.2003.11.047. [DOI] [PubMed] [Google Scholar]
- 53.Levine RL, Williams JA, Stadtman ER, Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 54.Smith MA, Perry G, Richey PL, Sayre LM, Anderson VE, Beal MF, Kowall N. Oxidative damage in Alzheimer's. Nature. 1996;382:120–121. doi: 10.1038/382120b0. [DOI] [PubMed] [Google Scholar]
- 55.Xu G, Liu Y, Kansal MM, Sayre LM. Rapid cross-linking of proteins by 4-ketoaldehydes and 4-hydroxy-2-alkenals does not arise from the lysine-derived monoalkylpyrroles. Chem Res Toxicol. 1999;12:855–861. doi: 10.1021/tx990056a. [DOI] [PubMed] [Google Scholar]
- 56.Liu Q, Raina AK, Smith MA, Sayre LM, Perry G. Hydroxynonenal, toxic carbonyls, and Alzheimer disease. Mol Aspects Med. 2003;24:305–313. doi: 10.1016/s0098-2997(03)00025-6. [DOI] [PubMed] [Google Scholar]
- 57.Takeda A, Smith MA, Avila J, Nunomura A, Siedlak SL, Zhu X, Perry G, Sayre LM. In Alzheimer's disease, heme oxygenase is coincident with Alz50, an epitope of tau induced by 4-hydroxy-2-nonenal modification. J Neurochem. 2000;75:1234–1241. doi: 10.1046/j.1471-4159.2000.0751234.x. [DOI] [PubMed] [Google Scholar]
- 58.Liu Q, Smith MA, Avila J, DeBernardis J, Kansal M, Takeda A, Zhu X, Nunomura A, Honda K, Moreira PI, Oliveira CR, Santos MS, Shimohama S, Aliev G, de la Torre J, Ghanbari HA, Siedlak SL, Harris PL, Sayre LM, Perry G. Alzheimer-specific epitopes of tau represent lipid peroxidation-induced conformations. Free Radic Biol Med. 2005;38:746–754. doi: 10.1016/j.freeradbiomed.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Farias GA, Vial C, Maccioni RB. Functional domains on chemically modified tau protein. Cell Mol Neurobiol. 1993;13:173–182. doi: 10.1007/BF00735373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giannopoulou E, Katsoris P, Polytarchou C, Papadimitriou E. Nitration of cytoskeletal proteins in the chicken embryo chorioallantoic membrane. Arch Biochem Biophys. 2002;400:188–198. doi: 10.1016/S0003-9861(02)00023-1. [DOI] [PubMed] [Google Scholar]
- 61.Aulak KS, Koeck T, Crabb JW, Stuehr DJ. Dynamics of protein nitration in cells and mitochondria. Am J Physiol Heart Circ Physiol. 2004;286:H30–H38. doi: 10.1152/ajpheart.00743.2003. [DOI] [PubMed] [Google Scholar]
- 62.Rayala SK, Martin E, Sharina IG, Molli PR, Wang X, Jacobson R, Murad F, Kumar R. Dynamic interplay between nitration and phosphorylation of tubulin cofactor B in the control of microtubule dynamics. Proc Natl Acad Sci U S A. 2007;104:19470–19475. doi: 10.1073/pnas.0705149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamisaki Y, Wada K, Bian K, Balabanli B, Davis K, Martin E, Behbod F, Lee YC, Murad F. An activity in rat tissues that modifies nitrotyrosine-containing proteins. Proc Natl Acad Sci U S A. 1998;95:11584–11589. doi: 10.1073/pnas.95.20.11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.