Abstract
Cell signaling is a complex and highly regulated process. Post-translational modifications of proteins serve to sense and transduce cellular signals in a precisely coordinated manner. It is increasingly recognized that protein S-nitrosylation, the addition of a nitric oxide group to cysteine thiols, serves an important role in a wide range of signaling pathways. In spite of the large number of SNO-proteins now identified (~1000), the observed specificity of S-nitrosylation in terms of target proteins and specific cysteines within modified proteins is incompletely understood. Here we review the progress made in S-nitrosylation detection methods that have facilitated the study of the SNO-proteome under physiological and pathophysiological conditions, and some factors important in determining the SNO-proteome. Classification schemes for emergent denitrosylases and prospective ‘protein S-nitrosylases’ are provided.
Introduction
Of the many post-translational modifications of proteins, reversible redox modifications of critical cysteine residues have emerged as a key strategy used by cells for signaling. In particular, an increasing number of studies have identified important roles played by S-nitrosylation (the covalent modification of a reactive cysteine by nitric oxide, NO) in normal cellular function as well as in a wide range of pathophysiological conditions [1]. The influence of NO on cellular function was initially ascribed predominantly to the activation of guanylate cyclase (through formation of a heme-nitrosyl) and thus to enhanced production of cGMP [2]. However, S-nitrosylation is now well established as a major source of NO bioactivity [3], and proteins shown to be modified in situ by S-nitrosylation (SNO-proteins) participate in a wide range of biological processes including those involved in cellular trafficking [4], muscle contractility [5], apoptosis [6,7], and circulation [8]. Dysregulation of S-nitrosylation has also been implicated in numerous disease states [7,9,10]. The increasing prominence of S-nitrosylation has pointed to the need for the development of methods aimed at identifying the complement of SNO-proteins (the SNO-proteome) under various physiological and pathophysiological conditions. Most candidate methods are based on the biotin-switch technique (BST), as discussed below. In addition, it is now well established that targeting of S-nitrosylation between proteins and particularly between Cys residues within target proteins reflects the operation of a number of determinants of specificity, and we discuss the nature of those factors below.
Identification of SNO-proteins and SNO-sites
Early techniques for the detection and identification of SNO-proteins were limited: available methods such as Hg-coupled photolysis/chemiluminescence [11] could determine total SNO levels (or amounts of SNO in a isolated protein) but were unable to identify the individual SNO-proteins in situ or specific S-nitrosylated Cys residues. The development of the BST by Jaffrey et al. [12] not only made the identification of individual SNO-proteins more feasible, but also led to modified techniques that could identify SNO-sites in a high-throughput manner (Table 1). The three key steps involved in detecting SNO-proteins by BST involve: firstly, S-methylthiolation: blocking all free cysteine thiols using MMTS (S-methyl methanethiosulfonate); secondly, reduction: selective and specific reduction of SNOs to free thiols using ascorbate; thirdly, labeling: free thiols are labeled with biotin-HPDP (N-6-(Biotinamido)hexyl)-3’-(2’-pyridyldithio)-propionamide). The biotin-labeled proteins can be detected by immunoblotting for either biotin or for a specific protein(s) after pull-down with streptavidin beads. Although the specificity of SNO “reduction” (formerly denitrosation) by ascorbate has been questioned, artifactual SNO signals can be avoided in the BST by using stringent controls [13]. To avoid artifacts, it is crucial to perform the labeling step in the dark (to prevent reduction of biotin-HPDP) and to use metal chelators (to prevent ascorbate driven one-electron reductions of Cu2+ and Fe3+ ions, which may result in reactions that affect specificity) [13]. The specificity of SNO denitrosation by ascorbate is well-grounded on both chemical and thermodynamic grounds.
Table 1.
Proteomic approaches for identification of S-nitrosylated proteins
| Technique | Description of method | Reference |
|---|---|---|
| Biotin switch | Conversion of SNO-cysteines to biotinylated cysteines, pull-down by avidin beads and detection by MALDI-TOF MS | [12] |
| SNOSID | Derived from BST, it enriches for biotinylated tryptic peptides and detection by LC-MS/MS | [14] |
| Detection by DAF | SNO-proteins separated on non-reducing diaminofluorescein gels are detected by release of NO by UV photolysis | [54] |
| His-Tag switch | Conversion of SNO-cysteines to His-Tag cysteines, pull-down by nickel beads, detection by MALDI-TOF MS | [55] |
| Quadrupole TOF-MS | Direct detection of SNO-peptides by LC-MS/MS on QTOF | [56] |
| Protein microarray | Modified BST to fluorescently label SNO-proteins on microarrays | [18] |
| SNO-RAC | Simplifies BST by the use of thiol-reactive resin to pull down specifically reduced SNO-proteins; identification by LC-MS/MS | [16] |
| SNO-DIGE | Labeling of specifically reduced SNO-proteins by thiol-reactive fluorescent tags and identification by 2D-gel electrophoresis | [17] |
| Organic mercurial-based | Direct capture of SNO-cysteines with phenylmercury compounds and detection by LC-MS/MS | [15] |
These basic steps have been modified to perform high–throughput identification of SNO-proteins along with the identification of specific sites of nitrosylation. A number of studies have used liquid chromatography-tandem mass spectrometry (LC-MS/MS) subsequent to trypsin digestion of proteins pulled down by streptavidin agarose [14,15]. SNO-RAC is a recent methodology that was derived from the BST and provides significant advantages. It combines the steps of reduction, labeling, and pull-down by the use of a thiol-reactive resin to bind free thiols reduced by ascorbate [16]. The method both simplifies the BST and allows for the “on-resin” trypsinization of proteins disulfide-bound to the resin. Thus, after washing the resin, the only peptides that remain bound contain cysteines that were denitrosated by ascorbate (and hence were S-nitrosylated in the sample). In addition, differential iTRAQ labeling “on-resin” can be combined with mass spectrometry for direct comparison of SNO-protein levels between samples.
Another advantage provided by SNO-RAC vis-à-vis the BST is better sensitivity for the detection of many proteins, particularly high-molecular-weight SNO-proteins such as the 565 kDa ryanodine receptor/Ca2+ channel (RyR) [16]. Finally, SNO-RAC can also be adapted for the analysis of other cysteine modifications by simply varying the agent used to generate free thiols, for example, by using hydroxylamine instead of ascorbate for the detection of S-acylated proteins [16].
Techniques based on 2D-gel electrophoresis have also been used to identify differences in the SNO-proteome between two conditions [17]. Following blocking of free thiols and specific reduction of S-nitrosylated cysteines as in the BST, labeling with different thiol-reactive fluorescent tags is used to differentiate between samples, followed by 2D-gel electrophoresis to separate and identify SNO-proteins. This approach is laborious and the number of SNO-proteins identified is low, emphasizing the advantage of SNO-RAC with regards to sample preparation and identification of SNO-proteins.
All techniques based on the BST are influenced by the abundance of the SNO-protein, and low abundance SNO-proteins may be overlooked. Protein microarrays were demonstrated to be an alternative to mass spectrometry-based assays for the identification of SNO-proteins [18]. The method uses a modified BST to biotin-label expressed, purified, and immobilized SNO-proteins on microarrays, followed by detection using anti-biotin antibody and fluorescently labeled secondary antibody. This method has certain advantages. The identification of SNO-proteins does not require mass spectrometry, and differing relative abundance of cellular SNO-proteins does not introduce bias. However, although the microarray approach is a useful tool to assess the effects of different NO donors and cofactors, it cannot be used to determine the SNO-proteome under physiological conditions in vivo. Most recently, Ischiropoulos and co-workers have developed an organic mercurial-based method to directly identify SNO-sites. The method provides a new alternative to the BST with great potential [15].
With the use of these sophisticated high-throughput approaches, about a thousand verified and potential S-nitrosylation sites have been identified [19]. The information from this growing list of SNO-proteins has been utilized to develop computational methods to identify SNO-sites [19]. A list of SNO-proteins that have been verified in human cells was compiled by Xue et al. [19] and selected proteins are presented in Table 2.
Table 2.
Representative S-nitrosylated proteins and their cellular functions
| Protein name | Gene name | Pathway |
|---|---|---|
| Type-1 angiotensin II receptor | AGTR1 | Receptor signaling |
| Dynamin-1/2 | DYN1, DYN2 | |
| Insulin receptor | INSR | |
| NMDA receptor | NMDAR2A, NMDAR1 | |
| Beta-arrestin 2 | ARRB2 | |
| MAP kinase 8 | MAPK8 | Signal transduction |
| Protein-tyrosine phosphatases, PTP1B, SHP1 | PTP1B, PTP1C | |
| Dexamethasone-induced Ras-related protein 1 | DEXRAS1 | |
| GTPase HRas, NRas | HRAS, NRAS | |
| Sarcoplasmic/endoplasmic reticulum Ca2+ ATPase | ATP2A1 | Muscle contraction |
| Myosin-9 | MYH9 | |
| Ryanodine receptor 1/2 | RYR1, RYR2 | |
| Apoptosis regulator Bcl-2 | BCL2 | Apoptosis |
| Caspase-3, -9 | CASP3, CASP9 | |
| E3 ubiquitin-protein ligase XIAP | XIAP | |
| Arginase-1 | ARG1 | NO synthesis |
| Argininosuccinate synthase | ASS1 | |
| Nitric oxide synthase, endothelial | NOS3 | |
| Glutaredoxin-1 | GRX | Redox regulation |
| Thioredoxin-1 | TRX1 | |
| Stress-70 protein, mitochondrial | HSPA9 | Heat shock response |
| Heat shock protein HSP 90-alpha | HSP90AA1 | |
| Elongation factor 1-alpha | EF1A | Gene regulation |
| Histone deacetylase 2 | HDAC2 | |
| Hypoxia-inducible factor 1-alpha | HIF1A | |
| Inhibitor of nuclear factor kappa-B kinase | IKKB | |
| Calcium/calmodulin-dependent protein kinase type II | CAMK2A | |
| E3 ubiquitin-protein ligase Mdm2 | MDM2 | |
| Nuclear factor NF-kappa-B p105 subunit | NFKB1 | |
| Tumor necrosis factor receptor superfamily | TNFRSF10A | |
| Hemoglobin | HBA1; HBA2 | Circulation |
| Tissue-type plasminogen activator | PLAT | |
| Voltage-gated potassium channel | KCNA8, KCNA5 | Membrane channels |
| Cytochrome c oxidase subunit 2 | MT-CO2 | Respiratory chain |
| Protein disulfide-isomerase | PDI | Protein homeostasis |
| Ubiquitin-conjugating enzyme E2 L3 | UBE2L3 | |
| Cytosolic phospholipase A2 | PLA2G4A | Others |
| Glucokinase | GCK | |
| Iron-responsive element-binding protein 2 | IREB2 | |
| Matrix metalloproteinase-9 | MMP9 | |
| Microsomal glutathione S-transferase 1 | MGST1 | |
| Syntaxin-1A | STX1A | |
| Vesicle-fusing ATPase | NSF |
Specificity of S-nitrosylation
Of the multiple cysteine residues available for S-nitrosylation within a protein, only specific residues are modified and are responsible for altering protein function [20,21]. Many contributing factors for specificity of nitrosylation have been proposed and these are discussed here. In addition, another important determinant of the cellular SNO-proteome is the presence and activation levels of denitrosylases.
Motifs
Based on the transnitrosylative transfer of NO groups from low-mass S-nitrosothiols to Cys93 in the β-subunit of hemoglobin, the propensity of a particular cysteine to be S-nitrosylated was proposed to be dependent in part on acidic and basic residues [22]. This motif could be present either in the flanking primary sequence or emerge in tertiary structure [15,23]. Greco et al. (2006) [23] suggested extending the motif in primary sequence to include acids and bases in the −6 through +6 positions around Cys residues that are sites of S-nitrosylation [23]. In a more recent study, Marino and Gladyshev [24] used 70 known SNO-Cys sites for identification of general features associated with S-nitrosylation and proposed that the acid-base motif, defined as close apposition of target Cys and charged residue(s), was not sufficient to account for a majority of cases of S-nitrosylation. However, acidic and basic side chains were more likely in an 8Å surround. The authors hypothesized that this relatively distant acid-base motif did not activate the target cysteine for reaction with NO or stabilize the SNO formed, but rather played a role in protein-protein interactions that could facilitate transnitrosylation. The specificity of the interaction would thereby subserve selective nitrosylation. In this regard, it is interesting to note that residues comprising the acid-base motif in hemoglobin, as originally conceived [22,25], were also implicated in regulating the interaction between protein subunits that determine hemoglobin conformation, and have subsequently been shown to participate in selective transnitrosylation of the red blood cell membrane protein AE1 (anion exchange protein 1, also known as Band 3 protein) [26] (see below and Table 3). Finally, Liu et al. reportedly verify the centrality of an acid-base motif in a subset of proteins nitrosylated by an atypical class of NO donor [27]. This result fits with the emerging understanding that stereochemistry and structure of the NO donor may be an important determinant of the efficiency and selectivity of S-nitrosylation [18].
Table 3.
Substrate-based classification of protein S-nitrosylases and denitrosylases
| S-nitrosylases | Reference | Denitrosylases | Reference |
|---|---|---|---|
| Class I: Cys transnitrosylases | Class I: GSNO reductases | ||
| Hemoglobin (AE1 nitrosylase) | [26] | Alcohol dehydrogenase III | [39] |
| Caspase-3 (XIAP nitrosylase) | [36] | Carbonyl reductase | [57] |
| Thioredoxin-1 (Procaspase-3 nitrosylase) | [35] | Class II: Protein-SNO reductases | |
| Class II: Metal-thiol transnitrosylasesa | Thioredoxin-1 | [6] | |
| Hemoglobin | [37] | Thioredoxin-2 | [6] |
| Nitrophorin | [58] | Class III: Microbial denitrosylaseb | |
| Flavohemoglobin | [59] |
Note that in these proteins the auto-transfer of NO occurs from transition metal to thiol within and not between proteins.
Although not true denitrosylases, this class of NO-consuming enzymes has been shown to influence global SNO-protein levels [48].
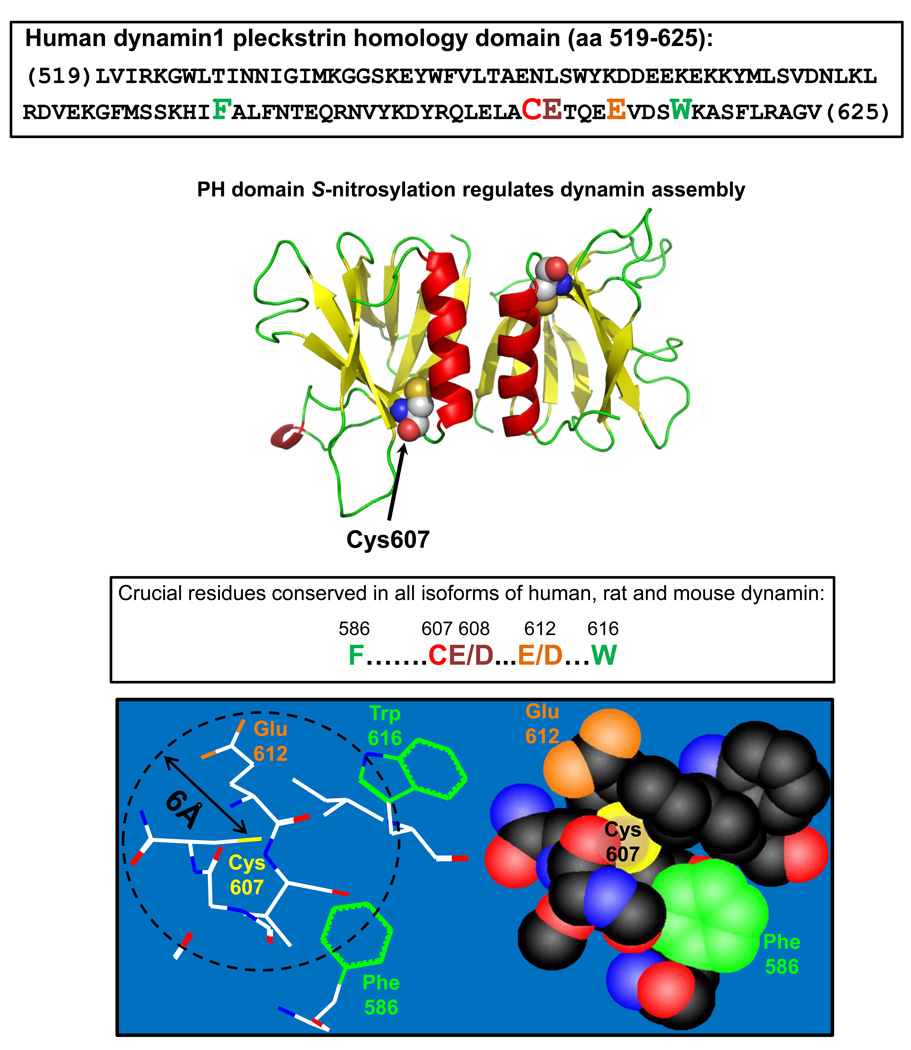
The interaction or sub-cellular co-localization of proteins with nitric oxide synthases (NOSs) would provide high local concentrations of NO. NO can react directly with thiols to form a radical anion intermediate that is oxidized to an S-nitrosothiol in the presence of an electron acceptor [28] and may also dismutate (to form NO+, which will nitrosate thiol) following dimerization upon aromatic residues [29]. Alternatively, NO may react with O2 to generate nitrosylating equivalents. Accordingly, several features of proteins may facilitate the effective reaction of target cysteine with NO. S-nitrosylation may be promoted by hydrophobic regions around the target cysteine (“hydrophobic motif”), including adjacent aromatic residues that serve to both increase thiol nucleophilicity (reactivity) and concentrate nitrosylating equivalents (to enable nitrosylation reactions) [3,29,30]. Strategic positioning of aromatic/Cys residues at protein/protein interfaces may also regulate protein-protein interactions, as illustrated in the case of β-catenin, where either Cys nitrosylation or Tyr phosphorylation will dissociate VE cadherin [31]. The importance of local hydrophobicity is emphasized in the ryanodine receptor, where the only cysteine residue (out of about 50) that is S-nitrosylated is located in a hydrophobic calmodulin-binding domain [32,33]. A modified motif that includes both acid/base and hydrophobic features has been proposed [3], and is exemplified in the protein dynamin (Figure 1), which undergoes S-nitrosylation downstream of the activation of G protein-coupled receptors [34]. The Cys-bearing motif in dynamin resides within a pleckstrin homology domain that subserves protein-protein and protein-lipid interactions, and is critical for the multimerization of dynamin.
Figure 1. S-nitrosylation of a crucial cysteine in the pleckstrin homology domain regulates dynamin multimerization.
The S-nitrosylated Cys607 lies in a hydrophobic pocket (so-called “hydrophobic core” of the pleckstrin homology domain), which contains aromatic side-chains and an acid-base motif within a 6Å radius (“acid-base/hydrophobic” motif). The residues that form the proposed S-nitrosylation motif are conserved in mammalian dynamins and are highlighted in the pleckstrin homology domain sequence.
Transnitrosylation (‘Cys transnitrosylases’)
In addition to direct modification by NO or nitrosylating equivalents, transnitrosylation can lead to the nitrosylation of proteins, which may be of particular importance where target cysteines do not appear to be candidates for modification based on their reactivity. Transnitrosylation is the direct thiol-to-thiol transfer of an NO group from a small-molecular-weight S-nitrosothiol or a SNO-protein to a target protein (see Table 3), as in the case of transfer of an NO group from the β-chain Cys93 of SNO–hemoglobin to AE1 (see above) [26]. Similarly, SNO-thioredoxin-1 can nitrosylate procaspase-3 under some conditions [35], and SNO-caspase-3 can transnitrosylate the X-linked inhibitor of apoptosis (XIAP) and thereby inhibit its E3 ubiquitin ligase activity [36]. Interestingly, hemoglobin also provides a paradigmatic example of auto-S-nitrosylation in which NO bound to heme is transferred intramolecularly to thiol [37]. Proteins that directly introduce NO groups into target proteins may be considered “protein nitrosylases”, by direct analogy to protein kinases, which bind target proteins and introduce phosphate groups [25,38]. Thus, hemoglobin is an AE-1 nitrosylase, caspase 3 is a XIAP nitrosylase, and thioredoxin-1 is potentially a caspase-3 nitrosylase [35] (Table 3).
For many SNO-proteins, S-nitroso-glutathione (GSNO) can serve as an NO group donor. Genetic knockout of GSNO reductase (GSNOR), an enzyme that catalyses the denitrosylation of GSNO, results in high levels of SNO-proteins, which demonstrates that GSNO is in equilibrium with a pool of SNO-proteins [39,40].
Interaction with nitric oxide synthase (NOS)
The major sources of NO in cells are the NOSs. There are three major NOS isoforms: neuronal NOS (nNOS/NOS1), inducible NOS (iNOS/NOS2) and endothelial NOS (eNOS/NOS3), which use NADPH, tetrahydrobiopterin, and oxygen to convert L-arginine to L-citrulline with the release of NO [41]. Direct interaction and/or subcellular compartmentalization of NOS with the target protein provide a basis for specificity when NOS is the source of NO for S-nitrosylation. NOS can bind its target protein either directly, as in the case of the interaction of iNOS with cyclooxygenase-2 [42], or via an adaptor or scaffolding protein such as carboxy-terminal PDZ ligand of nNOS (CAPON), which mediates the interaction between Dexras1 and nNOS [43]. It was demonstrated recently that β–arrestin-2 interacts with and is S-nitrosylated by eNOS, which regulates β-adrenergic receptor trafficking [4]. In fact, eNOS binds to multiple proteins that regulate β-adrenergic receptor trafficking, emphasizing that eNOS is placed where it is needed for local action, rather than serving as a source of NO that diffuses to react indiscriminately with multiple effectors.
Subcellular co-localization of target proteins with NOS is well illustrated in the heart. NO has opposing effects on cardiac contractility and the basis for this is the differential subcellular localization of eNOS (sarcolemmal caveolae) and nNOS (sarcoplasmic reticulum) [44]. eNOS inhibits contractility via S-nitrosylation of the L-type Ca2+ channel, whereas nNOS facilitates contractility by S-nitrosylating and activating the ryanodine receptor (RyR2). In heart failure, relocation of nNOS from the sarcoplasmic reticulum to the plasma membrane has been implicated in pathophysiology [45]. By the same token, relocation of eNOS from cytosol to endoplasmic reticulum (ER) is necessary to effect the nitrosylation of ER-resident proteins that regulate ER function [46].
Emerging role of denitrosylation
Although the mechanisms of S-nitrosylation are of obvious importance in determining the endogenous SNO-proteome, accumulating evidence indicates a significant influence of denitrosylation. A number of enzymes with primary functions unrelated to nitrosylation have been reported to act as denitrosylases, including protein disulfide isomerase, xanthine oxidase, carbonyl reductase, glutathione peroxidase, and superoxide dismutase (reviewed in [47]). However, the physiological relevance of these activities is not clear.
The physiological relevance of two denitrosylase systems has been well established; GSNOR (here designated Class 1) and thioredoxin/thioredoxin reductase (Trx/TR) (Class 2). GSNOR functions to reduce GSNO, the main low-molecular-weight SNO, and is highly conserved from bacteria to man [39]. As noted above, GSNOR does not act directly on protein SNOs, but GSNO serves as an important source of NO groups that generate SNO-proteins, and GSNO and SNO-protein levels co-vary [40,48]. Breakdown of GSNO (by GSNOR) thus results in glutathione-dependent denitrosylation of a subset of SNO-proteins. The thioredoxin/thioredoxin reductase system, which has been well characterized as a disulfide reductase and a critical regulator of cellular redox status, has recently been shown to be a major SNO-protein denitrosylase in mammalian cells [6,49,50]. Earlier studies demonstrated a potential role for the thioredoxin system (Class 2) in promoting the denitrosylation of both protein SNOs and low-molecular-weight SNOs including GSNO (reviewed in [47]. Its function as a physiological denitrosylase was established by the demonstration that SNO-caspase 3 (which plays a central role in apoptosis) is denitrosylated and thereby activated by thioredoxin under pro-apoptotic conditions [6,51]. The emerging picture suggests that SNO-proteins are differentially susceptible to denitrosylation by enzymatic mechanisms or through interaction with GSH (which is regulated by GSNOR), and that the SNO-proteome will reflect the action of different denitrosylating mechanisms on different sets of SNO-proteins [16,50,52]. In microbes and plants, (flavo)hemoglobins represent a separate class of “denitrosylase” (Class 3). By eliminating NO enzymatically, they lower the steady state levels of nitrosylated proteins [48] (Table 3).
Conclusions
The SNO-proteome is expanding exponentially because of the introduction of more facile methods for the identification of SNO-proteins. The expanding range of SNO-proteins helps increase the understanding of the mechanisms of S-nitrosylation that confer specificity of targeting between and within proteins. Further, evidence of dysregulated S-nitrosylation in a wide spectrum of human diseases is increasing rapidly (reviewed in [1, 53]). The potential of proteomic analysis of S-nitrosylation in disease to yield therapeutic targets remains largely untapped, but advances in SNO-protein detection will facilitate research directed at the identification of SNO-protein candidates in specific diseases.
Acknowledgements
We thank Douglas Hess for helpful suggestions during the preparation of this manuscript. The authors acknowledge support from the National Institutes of Health (RO1 HL096673, RO1 HL095463, RO1 HL059130) and DARPA (N66001-10-C-2015).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest published within the period of review have been highlighted as
• of special interest
- 1.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murad F. Cyclic guanosine monophosphate as a mediator of vasodilation. J Clin Invest. 1986;78:1–5. doi: 10.1172/JCI112536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess DT, Matsumoto A, Kim S-O, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 4. Ozawa K, Whalen EJ, Nelson CD, Mu Y, Hess DT, Lefkowitz RJ, Stamler JS. S-nitrosylation of beta-arrestin regulates beta-adrenergic receptor trafficking. Mol Cell. 2008;31:395–405. doi: 10.1016/j.molcel.2008.05.024. • The authors show that a crucial component of GPCR signaling, b-arrestin 2, interacts with and is S-nitrosylated at a single cysteine by endothelial NO synthase (eNOS).
- 5.Xu L, Eu JP, Meissner G, Stamler JS. Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science. 1998;279:234–237. doi: 10.1126/science.279.5348.234. [DOI] [PubMed] [Google Scholar]
- 6. Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. • Using caspase-3 as a target, this paper identifies a physiological denitrosylase role for thioredoxin.
- 7.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singel DJ, Stamler JS. Chemical physiology of blood flow regulation by red blood cells: the role of nitric oxide and S-nitrosohemoglobin. Annu Rev Physiol. 2005;67:99–145. doi: 10.1146/annurev.physiol.67.060603.090918. [DOI] [PubMed] [Google Scholar]
- 9. Wei W, Li B, Hanes MA, Kakar S, Chen X, Liu L. S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci Transl Med. 2:9ra13. doi: 10.1126/scitranslmed.3000328. • Dysregulated S-nitrosylation is implicated in hepatocarcinogenesis via impairment of a DNA repair system.
- 10.Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci U S A. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Science STKE. 2001;2001:1–9. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 13.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radical Biology and Medicine. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. • A description of a resin-assisted capture method (SNO-RAC) for improved detection and identification of SNO-proteins.
- 17.Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, Smith RAJ, Murphy MP. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem. J. 2010;430:49–59. doi: 10.1042/BJ20100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster MW, Forrester MT, Stamler JS. A protein microarray-based analysis of S-nitrosylation. Proc Natl Acad Sci U S A. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, Ren J. GPS-SNO: Computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS ONE. 2010;5:e11290. doi: 10.1371/journal.pone.0011290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selvakumar B, Huganir RL, Snyder SH. S-nitrosylation of stargazin regulates surface expression of AMPA-glutamate neurotransmitter receptors. Proc Natl Acad Sci U S A. 2009;106:16440–16445. doi: 10.1073/pnas.0908949106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman MA, Senga T, Ito S, Hyodo T, Hasegawa H, Hamaguchi M. S-nitrosylation at cysteine 498 of c-Src tyrosine kinase regulates nitric oxide-mediated cell invasion. J Biol Chem. 2009;285:3806–3814. doi: 10.1074/jbc.M109.059782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO Signals: translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 23.Greco TM, Hodara R, Parastatidis I, Heijnen HFG, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: A modified acid-based motif and the emerging role of trans-nitrosylation. Journal of Molecular Biology. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamler JS, Lamas S, Fang FC. Nitrosylation: the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 26.Pawloski JR, Hess DT, Stamler JS. Export by red blood cells of nitric oxide bioactivity. Nature. 2001;409:622–626. doi: 10.1038/35054560. [DOI] [PubMed] [Google Scholar]
- 27.Liu M, Hou J, Huang L, Huang X, Heibeck TH, Zhao R, Pasa-Tolic L, Smith RD, Li Y, Fu K, et al. Site-specific proteomics approach for study protein S-nitrosylation. Anal Chem. 2010;82:7160–7168. doi: 10.1021/ac100569d. [DOI] [PubMed] [Google Scholar]
- 28.Gow AJ, Buerk DG, Ischiropoulos H. A novel reaction mechanism for the formation of S-nitrosothiol in vivo. J Biol Chem. 1997;272:2841–2845. doi: 10.1074/jbc.272.5.2841. [DOI] [PubMed] [Google Scholar]
- 29.Zhao YL, Bartberger MD, Goto K, Shimada K, Kawashima T, Houk KN. Theoretical evidence for enhanced NO dimerization in aromatic hosts: implications for the role of the electrophile (NO)(2) in nitric oxide chemistry. J Am Chem Soc. 2005;127:7964–7965. doi: 10.1021/ja042247s. [DOI] [PubMed] [Google Scholar]
- 30.Rafikova O, Rafikov R, Nudler E. Catalysis of S-nitrosothiols formation by serum albumin: The mechanism and implication in vascular control. Proc Natl Acad Sci U S A. 2002;99:5913–5918. doi: 10.1073/pnas.092048999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thibeault S, Rautureau Y, Oubaha M, Faubert D, Wilkes BC, Delisle C, Gratton JP. S-Nitrosylation of beta-catenin by eNOS-derived NO promotes VEGF-induced endothelial cell permeability. Mol Cell. 2010;39:468–476. doi: 10.1016/j.molcel.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Eu JP, Sun J, Xu L, Stamler JS, Meissner G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
- 33.Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci U S A. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y. Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc Natl Acad Sci U S A. 2006;103:1295–1300. doi: 10.1073/pnas.0508354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell DA, Morton SU, Fernhoff NB, Marletta MA. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc Natl Acad Sci U S A. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, et al. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. • The authors demonstrate direct transnitrosylation from SNO-caspase-3 to XIAP, which inhibits its E3 ligase activity.
- 37.Luchsinger BP, Rich EN, Gow AJ, Williams EM, Stamler JS, Singel DJ. Routes to S-nitrosohemoglobin formation with heme redox and preferential reactivity in the beta subunits. Proc Natl Acad Sci U S A. 2003;100:461–466. doi: 10.1073/pnas.0233287100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benhar M, Stamler JS. A central role for S-nitrosylation in apoptosis. Nat Cell Biol. 2005;7:645–646. doi: 10.1038/ncb0705-645. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 40.Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, et al. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 41.Bredt DS, Snyder SH. Nitric oxide: a physiologic messenger molecule. Annu Rev Biochem. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- 42.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 43.Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: A G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 44.Barouch LA, Harrison RW, Skaf MW, Rosas GO, Cappola TP, Kobeissi ZA, Hobai IA, Lemmon CA, Burnett AL, O'Rourke B, et al. Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature. 2002;416:337–339. doi: 10.1038/416337a. [DOI] [PubMed] [Google Scholar]
- 45.Hare JM, Stamler JS. NO/redox disequilibrium in the failing heart and cardiovascular system. J Clin Invest. 2005;115:509–517. doi: 10.1172/JCI200524459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwakiri Y, Satoh A, Chatterjee S, Toomre DK, Chalouni CM, Fulton D, Groszmann RJ, Shah VH, Sessa WC. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proc Natl Acad Sci U S A. 2006;103:19777–19782. doi: 10.1073/pnas.0605907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 48.Foster MW, Liu L, Zeng M, Hess DT, Stamler JS. A genetic analysis of nitrosative stress. Biochemistry. 2009;48:792–799. doi: 10.1021/bi801813n. [DOI] [PubMed] [Google Scholar]
- 49.Holmgren A, Lu J. Thioredoxin and thioredoxin reductase: Current research with special reference to human disease. Biochemical and Biophysical Research Communications. 2010;396:120–124. doi: 10.1016/j.bbrc.2010.03.083. [DOI] [PubMed] [Google Scholar]
- 50.Benhar M, Thompson JW, Moseley MA, Stamler JS. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry. 2010;49:6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 52.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chemistry & Biology. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lima B, Forrester MT, Hess DT, Stamler JS. S-nitrosylation in cardiovascular signaling. Circ Res. 2010;106:633–646. doi: 10.1161/CIRCRESAHA.109.207381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King M, Gildemeister O, Gaston B, Mannick JB. Assessment of S-nitrosothiols on diaminofluorescein gels. Anal Biochem. 2005;346:69–76. doi: 10.1016/j.ab.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 55.Camerini S, Polci ML, Restuccia U, Usuelli V, Malgaroli A, Bachi A. A novel approach to identify proteins modified by nitric oxide: the HIS-TAG switch method. J Proteome Res. 2007;6:3224–3231. doi: 10.1021/pr0701456. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Liu T, Wu C, Li H. A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1353–1360. doi: 10.1016/j.jasms.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bateman RL, Rauh D, Tavshanjian B, Shokat KM. Human carbonyl reductase 1 Is an S-nitrosoglutathione reductase. J Biol Chem. 2008;283:35756–35762. doi: 10.1074/jbc.M807125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weichsel A, Maes EM, Andersen JF, Valenzuela JG, Shokhireva TK, Walker FA, Montfort WR. Heme-assisted S-nitrosation of a proximal thiolate in a nitric oxide transport protein. Proc Natl Acad Sci U S A. 2005;102:594–599. doi: 10.1073/pnas.0406549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hausladen A, Gow AJ, Stamler JS. Nitrosative stress: Metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]