Abstract
Animals have evolved a bewildering diversity of mechanisms to determine the two sexes. Studies of sex determination genes – their history and function – in non-model insects and Drosophila have allowed us to begin to understand the generation of sex determination diversity. One common theme from these studies is that evolved mechanisms produce activities in either males or females to control a shared gene switch that regulates sexual development. Only a few small-scale changes in existing and duplicated genes are sufficient to generate large differences in sex determination systems. This review summarises recent findings in insects, surveys evidence of how and why sex determination mechanisms can change rapidly and suggests fruitful areas of future research.
Keywords: evolution, evolutionary forces, gene duplication, sex determination, sexual differentiation and selection
Introduction
The regulation and evolution of sexual development have long been of central interest in developmental and evolutionary biology. One key question has been how sexual fate is determined and regulated, giving rise to the sexually dimorphic traits that play such a dominant role in animal evolution and behaviour. Much has been learned about how sex determination is realised and is integrated into the developmental program from model systems; the insect Drosophila melanogaster, the nematode Caenorhabditis elegans and the mouse Mus musculus 1–4. These studies have not resolved the question of why the regulatory principles of sex determination are so bewilderingly different and how this diversity is generated molecularly.
Sex determination mechanisms can vary substantially between phylogenetically closely-related species 5–7 and even within a single species 8–10. The same mechanism can apparently be regulated by different genes 11, 12. This implies that these mechanisms evolve very rapidly, despite the antiquity of the two sexes.
Here, we review recent advances made in insects that have begun to uncover how diverse sex determination systems are generated and regulated 12–22. The evolutionary and molecular routes taken broaden our understanding of how and why novel regulatory controls of a developmental process evolve. The key questions we address in this paper are: (i) How are differences in sex determination molecularly realized; (ii) how did differences in sex determination evolve from a pre-existing genetic repertoire; (iii) what forces drive sex determination systems to diverge?
Since the first reports of a sex determination mechanism 23 and of the inherited basis of sex determination 24, 25 – both made in insects – an astonishing diversity of mechanisms has been uncovered in a variety of species and phylogenetic lineages 5, 7, 26, 27. Considering insects, classical genetic and cytological studies have identified a variety of genetic and environmental signals that determine the two sexes. In the fruit fly D. melanogaster, for instance, the double dose of X chromosomes 28 determines femaleness, a single dose maleness. Other dipteran insects (e.g. Musca domestica and Ceratitis capitata) employ a male-determining Y chromosome: females are XX and males are XY 13, 29. Butterflies present the opposite scenario, possessing a female-determining W chromosome: females are ZW and males are ZZ 7. In XY chromosomal systems the number of Y chromosomes can vary substantially 30; for instance in some species of the fruit fly Anastrepha, females are XX/XX and males XX/Y 26. Other species employ male and female determiners with no visible chromosomal differences (e.g. phorid fly Megaselia scalaris 10, Chironomus 9, 31). Male scale insects and white flies (both homopterans), thrips (thysanopterans) and hymenopteran insects (wasps, ants and bees) are haploid and females are diploid 7. The genetic basis of haplo/diploidy in several hymenopteran species is complementary sex determination; males are homo- or hemizygous and females heterozygous at a single locus 32, 33. In another hymenopteran species, the parasitic wasp Nasonia vitripennis, sex determination is consistent with a maternal imprinting mechanism 22. Differential elimination of sex chromosomes during the first stages of embryonic division is used as a sex determination mechanism in the fungus gnat Sciara 26. Females develop when one paternal X chromosome is lost from the 3X:2A zygote and males arise when two paternal X chromosomes are lost. Maternally-derived signals (e.g. the blowfly Chrysomya rufifacies) or environmentally-derived signals, such as the temperature of egg incubation (e.g. the gall midge Heteropeza and the fungus gnat Sciara), are also utilised as sex determination signals.
Here, we wish to limit our review to sex determination signals of the pathways that have been identified in insects. From the phylogenetic relationships of hymenopteran insects, at the base of the holometabolous insect branch (Fig. 1F), we can retrace the ancestral components that were shared by common ancestors and then explore which components evolved and established new sex determination systems.
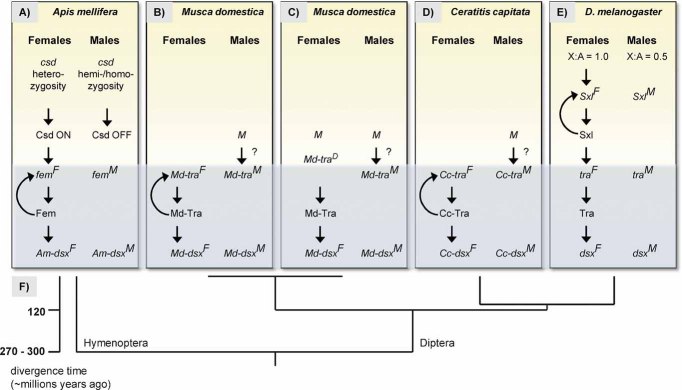
Figure 1.
Sex determination in insect model species, with their phylogenetic relationships representing ∼300 million years of evolution. The fruit fly D. melanogaster, the housefly M. domestica, the medfly Ceratis capitata and the honeybee Apis mellifera share a common pathway (indicated by a grey box) composed of the transformer (tra) gene and its downstream target doublesex (dsx). Female spliced tra transcripts (traF) give rise to Tra proteins that direct splicing of dsxF mRNAs, production of DsxF proteins and female development. When tra is spliced into the male variant, no Tra proteins are produced. This results in splicing of male dsxM mRNAs, DsxM proteins and male development. A: Sex in the honeybee (A. mellifera) is determined by the heterozygosity or homo-/hemizygosity of the csd gene. In females, different Csd proteins, derived from a heterozygous csd gene, direct the processing of female fem mRNAs (femF) 14. The fem gene is apparently an orthologue of the tra gene 12). Fem protein regulates female splicing of dsx, but also self-sustains the female splicing of fem. In males, inactive Csd proteins that are derived from the same alleles (homo- or hemizygous csd genes) result in a default splicing of fem (femM). (B and C) Models of two alternative sex determination systems that co-exist in M. domestica populations 15, 29. B: Sex is determined by the absence/presence of an unidentified male-determiner M. In the absence of M, maternally-derived Md-tra gene products establish an auto-regulative loop in females in which Md-Tra protein mediates the production of more female Md-tra mRNA. Presence of M impairs this tra auto-regulatory loop and also mediates the splicing of male Md-tra mRNAs. C: Sex in M. domestica can also be determined by a female determiner. Presence/absence of a tra allele, Md-traD(=FD), determines sexual fate. In females, the presence of Md-traD leads to female splice products and Md-Tra protein, even in the presence of the male-determiner M 15. In males, the male-determiner M mediates male Md-traM mRNAs in the absence of the Md-traD allele. D: Sex in C. capitata 13, 17 is determined by presence/absence of a, thus far, unidentified male-determiner M. In the absence of M, maternally-derived Cc-tra gene products appear to establish a Cc-tra auto-regulative loop 13. Presence of M mediates the splicing of male Cc-tra transcripts (traM). E: Sex in D. melanogaster is determined by the dose of X chromosomes 1, 28. Double doses of X in females activate the Sxl gene and expression of Sxl proteins. Sxl proteins direct splicing of female traF mRNAs that give rise to functional proteins. Sxl proteins also establish an auto-regulatory feedback loop by directing splicing of productive female SxlF mRNAs, which maintain the female state throughout development. In addition, there is an additional feedback activity in which Tra proteins stimulate Sxl positive auto-regulation 98. In males, the single dose of X chromosomes does not direct early Sxl protein expression. As a consequence the downstream regulatory decisions do not occur and male dsxM is produced. F: The evolutionary relationship of the species used in the comparison with their approximate time scale of divergence 40, 99.
In the next section, we summarise the different ways in which sex determination is achieved molecularly to regulate the proportions of males and females. We then turn to the question of how novel sex determination systems can evolve from the ancestral genetic repertoire, before discussing the forces that drive divergence in sex determination. Finally, we highlight potentially fruitful directions for future research.
Regulatory diversity and common principles of sex determination
Sex determination systems use different genes and regulatory mechanisms to establish activities in either males or females (Fig. 1). These activities regulate tra genes, which are key, upstream components of an ancestral sex-determining pathway 12–15, 17, 20–22.
In D. melanogaster, the double dose of X chromosomes 28 establishes feminising activity (Fig. 1E). The X chromosome encodes several transcription factors (e.g. runt, sisA, sisB) that, through the double dose in females, activate the Sxl gene. Sxl proteins in females are splicing factors that splice tra mRNAs in females to produce Tra protein. A single X chromosome results in the absence of the Sxl protein and, as a consequence, male tra mRNAs with a premature stop codon are produced.
In the honeybee Apis mellifera, heterozygosity of the complementary sex determiner gene (csd) establishes feminising activity 33, 34 (Fig. 1A). Csd allelic proteins derived from heterozygous csd activate the feminizer gene (fem) by directing splicing to form the female fem mRNAs that encode the Fem protein. Proteins derived from hemizygous (haploid, unfertilised eggs) or homozygous csd genes are non-active. As a result, the fem mRNAs are spliced into the male configuration that contains a premature translation stop codon. fem is apparently an orthologue of the tra genes 12.
The housefly, M. domestica, exhibits a number of different sex determination systems 8, 29, 35, 36 that co-exist in this species. There is a classical system (Fig. 1B) in which a dominant male-determiner on the Y chromosome provides a masculinising activity (males are X/M-Y and females X/X; the molecular nature of M has yet to be identified). Other systems can have the dominant male-determiners M on any of the five autosomal chromosomes, and even on the X chromosome. Furthermore, a dominant female-determiner (FD) exists in this species that establishes feminising activity. Females are FDM/FM and males are FM/FM (Fig. 1C). FD, the dominant female-determiner, is an allelic variant of the Md-tra gene 15 that produces female tra mRNAs and active Tra protein, even in the presence of the male-determiner M. In the absence of the FD allele, the male-determiner M ensures male-specific splicing of other Md-tra alleles and, consequently, the absence of active Md-Tra protein in males.
The medfly, C. capitata, also possesses a dominant male-determiner M on the Y chromosome that is responsible for masculinising activity 13, 17, 37 (Fig. 1D). The molecular nature of M is yet to be identified. In the presence of M, male Cc-tra mRNAs, but not Tra proteins, are produced. In the absence of M, maternally-derived Tra proteins direct splicing into productive female Cc-tra mRNAs.
These results imply that sex determination mechanisms in insects are used in two ways (Fig. 2); sex determination mechanisms in the zygote produce either feminising activities (D. melanogaster/A. mellifera) that switch tra genes ON, or they generate masculinising activities (C. capitata and M. domestica) that switch tra genes OFF. In the absence of these signals the pre-zygotic state of tra (‘default’ OFF or ON) is executed, resulting in male (D. melanogaster/A. mellifera), or female (C. capitata and M. domestica) development. tra gene regulation in N. vitripennis apparently follows this latter rule 22.
Figure 2.
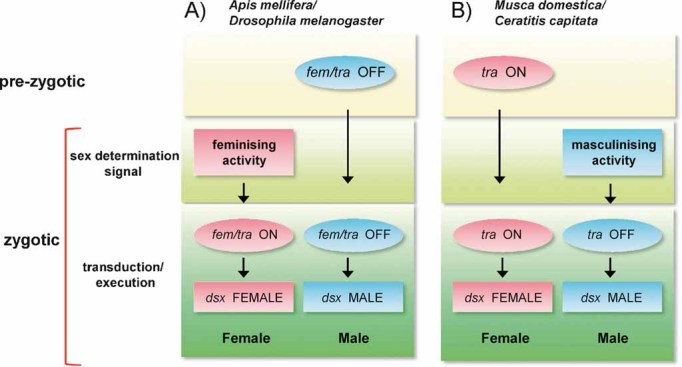
The regulatory principle underlying insect sex determination mechanisms. Species-specific, sex determination systems produce either a feminising or a masculinsing activity in the zygote to determine the two sexes in the proper proportions. In the absence of this activity, the pre-determined tra activity in the pre-zygote produces the alternative sex. A: Feminising activity in D. melanogaster and A. mellifera switches the tra gene from the pre-zygotic OFF (non-active) into the ON (active) state. B: Masculinising activity in C. capitata and M. domestica switches the tra gene from the pre-zygotic ON to the OFF state.
How do tra genes implement female and male development? Tra proteins are members of SR-type splice regulators that control female splicing of the dsx gene (Fig. 1). In the absence of Tra proteins, male dsx mRNAs are produced. This regulatory principle is shared among dipteran and hymenopteran insects 12–22, 38, 39 implying that this is an ancestral principle of sexual regulation in holometabolous insects (hymenopteran insects are at the base of holometabolous insects 40). tra gene regulation involves a positive feedback loop in females that generates even more Tra proteins and, thereby, a stable female state throughout development 13–15 (Fig. 1). The feedback activity of tra, and its' role in germ cell differentiation, have been co-opted in D. melanogaster by the Sxl gene, the next upstream component 1, 41, 42 (Fig. 1), suggesting that the Drosophila model system is derived in these respects.
Sex-specific splicing of the dsx gene has been identified in other phylogenetic lineages, including the lepidopteran insects 16, 17, 43–51. The sex-specific, spliced dsx transcripts encode transcription factors of the DM type that have an atypical zinc-finger domain. The proteins differ in females and males at their C-terminal ends 1, which control transcription of target genes differently 52. The role of dsx in sexual differentiation has been demonstrated in D. melanogaster 52–54, M. domestica 16 and the lepidopteran Bombyx mori 55, indicative of an ancestral role in integrating sexual differentiation within the general developmental program 56–63.
tra genes in insects may also regulate the fruitless (fru) genes that encode a BTP zinc-finger transcription factor sex-specifically 64. fru specifies sexual orientation and courtship behaviour in Drosophila by regulating differentiation processes in the nervous system 64, 65, together with Dsx protein 66. Consistent with an ancestral role in insect sex development, fru sex-specific splice products have been detected in the mosquito Anopheles gambiae 67, C. capitata 17 and the hymenopteran wasp N. vitripennis 68.
Evolutionary origin of novel sex determination mechanisms
Functional and evolutionary analyses of sex determination genes (Fig. 1A,C,E) has revealed that small-scale mutational changes in the nucleotide sequence from existing and from duplicated copies of genes can generate novel sex determination mechanisms (Fig. 3).
Figure 3.
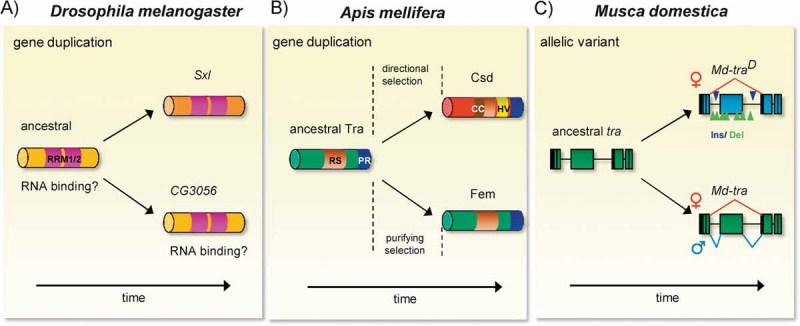
Mutational routes for the evolutionary origin of novel sex determination functions. A: The origin of the Sxl gene by gene duplication of CG3056 in dipteran insects 69. The encoding proteins are presented schematically. RRM 1 and RRM 2 denote the two RNA-binding domains. B: The origin of the csd gene by tandem gene duplication of the ancestral tra gene in the Apis lineage. The encoding proteins are presented schematically. The evolutionary rise of the csd gene was accompanied by the evolution of an asparagine/tyrosine-enriched repeat that varies in number in the different allelic specificities (denoted as HV: hyper-variable region) and with the origin of a putative coiled-coil domain (denoted as CC) possibly involved in protein binding 70, 71. Adaptive evolution (directional selection) was involved in shaping the evolutionary rise of the csd gene 12. The paralogous sister gene fem evolved under purifying selection, consistent with its ancestral function 100. RS denotes the arginine/serine-enriched (RS) domain and PR denotes the proline-rich domain. C: The evolutionary origin of the Md-traD allele from the ancestral Md-tra gene in M. domestica populations 15. The Md-tra genome structure is presented schematically. Blue triangles denote nucleotide insertions and the green triangles represent nucleotide deletions in the Md-traD allele that lost the ability to produce the male splice variant traM in the presence of M.
The evolutionary origin of the X dosage mechanism in D. melanogaster is associated with the evolutionary rise of the Sxl gene. The Sxl gene encodes an RNA-binding protein that originated in the dipteran lineage by gene duplication from a copy of the ancestral CG3056 gene 69 (Fig. 3A). The regulatory relationship of Sxl to the sex determination cascade arose through the evolution of Sxl protein-binding sites (poly-Uridine tracts between five and eight nucleotides in length) in the tra gene. Signalling of the double dose of X chromosomes by transcription factors (e.g. transcription factors runt, sisA, sisB) has apparently evolved through changes in the cis regulatory control of Sxl transcription.
The complementary sex determination gene in honeybees (Apis) evolved through gene duplication of the ancestral copy of the fem/tra gene within the last 60 million years 12 (Fig. 3B). The evolutionary rise of csd was accompanied by the insertion of a novel hyper-variable region that consists of asparagine/tyrosine-enriched repeats 70, 71 and by single nucleotide replacement changes that produced a novel coiled-coil motif 12. The hyper-variable region is thought to play a role in the recognition process of allelic differences (csd alleles are only active in the heterozygous state), whereas the putative coiled-coil domain appears to encode allelic protein-binding properties (Otte and Beye, unpublished results).
The novel, dominant female-determiner FD(= MdtraD) evolved in the housefly, M. domestica, from multiple small deletions and insertions in intron sequences of the tra gene 15 (Fig. 3C). The Md-traD allele is constitutively spliced in the productive female mode in the presence of a male-determiner M (Fig. 1C). Md-traD is a natural variant of the tra gene in housefly populations 29, suggesting that this mechanism originated recently.
These molecular analyses reveal how nucleotide changes in duplicated copies of genes and allelic variants can produce new sex determination genes and diverse mechanisms. Sex-determining variants, however, initially originated from rare mutational events in single individuals in a population. To understand how and why these initially rare variants became fixed in populations and species, we have to apply population genetic principles that account for the forces of evolutionary change. Multiple fixation processes through evolutionary time have generated the diversity of sex determination systems that we see today in the insects.
Forces driving the divergence of sex determination systems
Random genetic drift and directional (positive) selection are the evolutionary forces that can initially drive rare sex-determining variants throughout populations of any given species. Directional selection enhances the probability of fixation, given the random fluctuations of genetic variants in a population caused by genetic drift. We do not review here the body of theoretical work that has been done in the field of the evolution of sex determination, but rather focus on the empirical molecular evidence that has so far been documented.
Positive selection, driven by fitness gains in individual population members, is a plausible source for the fixation of new sex determination systems. For instance, phylogenetic surveys in hymenopteran species suggest that the complementary sex determination mechanism has been replaced in some highly inbreeding hymenopteran species (e.g. parasitic wasps) 22, 32, 72. Complementary sex determination under inbreeding results in large numbers of diploid males that cannot reproduce (only haploid males are fertile); suggestive of an evolutionary advantage for alternative sex determination mechanisms in this case.
Remarkably, once new sex determination mechanisms have evolved, nature does not stop generating new sex determination genes. An ongoing divergence process can be observed in several species in which alternative sex determination systems co-exist 9, 10, 15 and in lineages in which a sex determiner gene has been replaced, but not the underlying mechanism 11, 12. Recurrent, directional selection regimes that can continuously drive sex determination systems to diverge have been proposed through several hypotheses: adjustment of sex ratios, intra-locus sexual conflict, the degeneration of sex determiners, sex ratio drive 6, 73–78 and sexual selection 79–81.
The adjustment of the sex ratio hypothesis states that the rise of new sex determiners can be selectively advantageous in sub-divided or inbreeding populations or when male and female fitness are affected differently by environmental factors 73, 74, because this will allow adjustment for the optimal sex ratio. The intralocus sexual conflict hypothesis argues that a newly evolved dominant male- or female-determiner will increase in frequency when it is closely linked to a gene with beneficial effects in the sex this gene determines 75, 76. The degeneration hypothesis states that the cessation or reduction of meiotic recombination at primary, sex-determining loci results in a gradual loss of the efficiency of selection (for instance, the efficient removal of deleterious mutations in a population), resulting in an evolutionary degeneration of a sex determiner gene 6, 12, 82, 83. A malfunction in sex determination favours directional selection of an alternative, novel sex determiner. The degeneration hypothesis implies an evolutionary paradox: although meiotic recombination and the mixing of genetic material require two sexes, the genes that establish them may lack the evolutionary advantage of sexual reproduction. The sex ratio drive hypothesis suggests that X-linked, meiotic drive factors increase in frequency by inducing the loss of Y chromosome-containing sperm 77, 78. There is, thus far, no direct evidence that any one of these directional selection scenarios favoured the rise of novel, sex determination systems. These hypotheses are also difficult to test, as it is not known whether fitness gains or losses currently observable arise from novel sex determiner genes themselves, or are associated with other, secondary effects (for instance, suppression of recombination around the sex determination gene or accumulation of sex-specific beneficial alleles) 84.
The function of sex determination hierarchies (Fig. 1) is inconsistent with the view that sex determination systems evolve because of sexual selection. The sex determination signals of M. domestica, C. capitata and A. mellifera all regulate tra genes that control sexual development in its entirety (Fig. 1). They do not specify a particular set of sexually-dimorphic traits on which sexual selection can operate.
There are also examples in which the evolutionary rise of a new sex determination system is inconsistent with the pre-conditions of any of the directional selection hypotheses. Multiple male-determiners, for instance, that co-exist in the dipteran insects M. domestica and M. scalaris 10, 35, 85, cannot be used to adjust sex ratios, nor do different male-determiners promote sexual conflict. It is not plausible that evolutionary degeneration of a preceding sex determiner (i.e. through the accumulation of deleterious mutations) would directionally select for multiple male sex determiners. Fitness differences of M. domestica populations that either have an autosomal M or a Y-M sex determination mechanism 86 can also be explained by secondary effects and genetic differences associated with X/Y chromosomal backgrounds.
Another explanation for the evolutionary rise of alternative male-determiners is genetic drift. A higher evolutionary origin rate by mutation will increase the probability of fixation (see 87 for a theoretical analysis of how genetic drift can drive genetic pathway evolution). Multiple male-determiners (>4) in M. domestica and M. scalaris populations could reflect the increased mutation rate by which new repressing activities of tra genes can evolve from the entire gene repertoire of the genome. A repressing activity can be established at different levels of tra gene regulation, suggesting a high rate of origin. In contrast, female-determiners evolve more rarely (indeed, only one female-determiner has been identified in M. domestica, but none in M. scalaris) because it is less likely that a new function arises by mutation that properly controls tra pre-mRNA splicing and protein production.
The null-hypothesis of genetic drift has been rejected in the case of the csd gene. An excess of non-synonymous over synonymous, neutral nucleotide changes has been identified during the evolutionary origin of csd (Fig. 3B) 12. More amino acid-encoding nucleotides have been replaced directly after csd origin than neutral changes that solely became fixed by random genetic drift. Intriguingly, some of these new amino acids form a putative coiled-coil domain that appears to alter binding properties between Csd allelic proteins (Otte and Beye, unpublished results). The cause of directional selection during the evolutionary rise of csd is not understood and cannot be explained by an adjustment of sex ratios or an intra-locus sexual conflict. Complementary sex determination systems allow control over sex ratios (males derive from unfertilised eggs, females from fertilised eggs) and are not targets of intra-locus sexual conflict and sex ratio drive (as they are inherited in both sexes). Complementary sex determination systems can reside in genomic regions of substantially-reduced meiotic recombination 70, 88, implying that evolutionary degeneration may operate. Malfunction of the preceding complementary sex determiner could have selected for a new sex determiner; csd.
Fruitful routes of future research
There is a need to characterise more sex determination genes. Discovering other regulatory mechanisms and their routes of evolutionary origin will broaden our understanding of how and why these key controls of developmental processes have evolved. Characterisation of sex determiners will be greatly facilitated by next-generation sequencing technologies, especially for species in which sex-determining factors can be linked to short genomic regions of otherwise freely-recombining chromosomes (e.g. neo Y chromosomes) 9, 10, 31, 35, 85, 89. Sequencing DNA from pools of male and female progenies of a single cross will identify sex-specific nucleotide differences. These sex-specific differences will identify candidate genes. The function of these genes can be further tested using RNAi-induced knock-down studies.
Nucleotide sequences of novel sex determiners will allow us to trace the evolutionary history 12, 15, 69 and the evolutionary forces that shaped their origin 12. By applying population genetic tests on nucleotide sequences (i.e. dN/dS ratio, MacDonald Kreitman test, Tajima's D, among others 90), we can directly identify evidence for the cause of evolutionary change 12 in the responsible genes. This powerful approach has been widely neglected in evolutionary developmental biology.
An open question is when did the key sex-determining functions of tra (Figs. 1, 3) evolve? tra genes apparently have no sex-determining function outside of insects (see the crustacean case Daphnia magna 91), but it is not known whether hemimetabolous insects use these tra key functions. The fast-evolving tra genes may be identified by a conserved 30 amino acid motif that has been identified in non-Drosophila species 12, 13, 15, 92, 93. tra has not been identified in the sequenced B. mori genome 94, 95, which may reflect a lack of sufficient conservation, evolutionary loss, or a lack of corresponding sequence information.
What is widely unknown is how sexual dimorphic traits can evolve so rapidly by changes in the underlying developmental program. Reproductive structures, behaviours and secondary sexual characteristics are some of the most variable and changeable features among insects; some of which evolve through sexual selection (Darwin 1871). Morphological changes may evolve by the regulatory control of dsx target genes [by either modification, loss or novel origin of cis regulatory elements (CREs) 52] or by recruiting other genes (i.e. transcription factors) to the sex-determining process. Investigation into what aspects of morphology dsx controls in different species could give insights into whether dsx is a conserved key determinant of sexual dimorphic differentiation. Transgenic studies have shown that at least some aspects of dsx sexual differentiation are conserved in the dipteran insect M. domestica 16 and the lepidopteran B. mori 55, 96. Expression of male and female Dsx proteins, with the help of transgenic tools in a dsx null mutant background, would facilitate study into which aspects of sexual differentiation are controlled by dsx (RNAi-induced knockdown of dsx by injection procedures have, thus far, failed in several insects) and whether other key components in non-Drosophila species have yet to be identified. The evolution of abdominal pigmentation and pheromone production in some Drosophila species have been shown to be caused by small-scale evolutionary changes of CREs of Dsx protein target genes 52, 63, 97. A way to identify putative shared and evolved dsx target genes is to characterise binding sites of Dsx proteins in informative insect species by chromatin immuno-precipitation (ChiP) and next-generation sequencing.
Conclusion
Recent studies in insects have shed some light on how and why sex determination systems evolve 1, 12, 15, 69. Gain-of-function alleles of tra, the double dose of X-linked transcription factors that activate Sxl, or Csd proteins derived from a heterozygous csd gene are the molecular signals of diverse sex determination mechanisms, such as a dominant female-determining system, an X:A ratio and a complementary sex determination system. These organism-specific signals all share transduction of their activities to common tra genes; the upstream component of an ancestral pathway tra->dsx in holometabolous insects. Diverse signals utilise two regulatory principles to determine the two sexes (Fig. 2): they either produce feminising activities that switch tra genes ON, or they produce masculinising activities that switch tra genes OFF. In the absence of these activities, the pre-zygotic activity of tra (‘default state’ either ON or OFF) executes male or female development. tra genes in most studied insects maintain the sexually-determined state epigenetically through a positive regulatory feedback loop of pre-mRNA splicing 13.
Diverse mechanisms can evolve through small-scale nucleotide changes in regulatory and coding regions from existing or from duplicated genes. From the ease and rate that novel sex determiners arise by such changes 12, even within a species 10, 15, 35, 85, we suggest that non-adaptive forces (mutation and genetic drift) are also possible sources of novel sex determination systems. We suggest that genetic drift should serve as a null hypothesis in future work.
An excess of non-synonymous changes over synonymous, neutral changes 12 in the csd gene of honeybees shows that directional selection can enhance the evolutionary rise of novel sex determiner genes 12. Evolutionary degeneration, due to lack of recombination of the preceding complementary sex determiner gene, may have caused such directional selection 6, 12, 82, 83.
By applying molecular evolutionary and genetic approaches, we are just beginning to understand both the evolutionary routes and the molecular mechanisms that have generated the enormous diversity of sex determination mechanisms. These insights will greatly broaden our knowledge of how novel control mechanisms and regulatory principles evolve in developmental processes.
Acknowledgments
We very much thank Peter Dearden for his helpful comments on the manuscript; Daniel Bopp, Guiseppe Saccone and Walter Traut for discussion; and Robert Page (ASU) for hosting MB during spring break in 2009. We apologise to those whose work could not be cited due to space restrictions. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG).
References
- 1.Cline TW, Meyer BJ. Vive la difference: males vs. females in flies vs. worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 2.Zarkower D. Somatic sex determination. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.84.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lovell-Badge R, Canning C, Sekido R. Sex-determining genes in mice: building pathways. Novartis Found Symp. 2002;244:4–18. [PubMed] [Google Scholar]
- 4.Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–4. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 5.Marshall Graves JA. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet. 2008;42:565–86. doi: 10.1146/annurev.genet.42.110807.091714. [DOI] [PubMed] [Google Scholar]
- 6.Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–14. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 7.Bull JJ. Evolution of Sex Determining Mechanisms. Menlo Park, California: Benjamin/Cummings Publishing Company; 1983. [Google Scholar]
- 8.Franco MG, Rubini PG, Vecchi M. Sex-determinants and their distribution in various populations of Musca domestica L. of Western Europe. Genet Res. 1982;40:279–93. doi: 10.1017/s0016672300019157. [DOI] [PubMed] [Google Scholar]
- 9.Thompson PE, Bowen JS. Interactions of differentiated primary sex factors in Chironomus tentans. Genetics. 1972;70:491–3. doi: 10.1093/genetics/70.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traut W. Sex determination in the fly Megaselia scalaris, a model system for primary steps of sex chromosome evolution. Genetics. 1994;136:1097–1104. doi: 10.1093/genetics/136.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuda M. Sex determination in the teleost medaka, Oryzias latipes. Annu Rev Genet. 2005;39:293–307. doi: 10.1146/annurev.genet.39.110304.095800. [DOI] [PubMed] [Google Scholar]
- 12.Hasselmann M, Gempe T, Schiott M, Nunes-Silva CG, et al. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature. 2008;454:519–22. doi: 10.1038/nature07052. [DOI] [PubMed] [Google Scholar]
- 13.Pane A, Salvemini M, Bovi PD, Polito C, et al. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development. 2002;129:3715–25. doi: 10.1242/dev.129.15.3715. [DOI] [PubMed] [Google Scholar]
- 14.Gempe T, Hasselmann M, Schiott M, Hause G, et al. Sex determination in honeybees: two separate mechanisms induce and maintain the female pathway. PLoS Biol. 2009;7:e1000222. doi: 10.1371/journal.pbio.1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hediger M, Henggeler C, Meier N, Perez R, et al. Molecular characterization of the key switch F provides a basis for understanding the rapid divergence of the sex-determining pathway in the housefly. Genetics. 2010;184:155–70. doi: 10.1534/genetics.109.109249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hediger M, Burghardt G, Siegenthaler C, Buser N, et al. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev Genes Evol. 2004;214:29–42. doi: 10.1007/s00427-003-0372-2. [DOI] [PubMed] [Google Scholar]
- 17.Salvemini M, Robertson M, Aronson B, Atkinson P, et al. Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene of female sex determination. Int J Dev Biol. 2009;53:109–20. doi: 10.1387/ijdb.082681ms. [DOI] [PubMed] [Google Scholar]
- 18.Alvarez M, Ruiz MF, Sanchez L. Effect of the gene doublesex of Anastrepha on the somatic sexual development of Drosophila. PLoS One. 2009;4:e5141. doi: 10.1371/journal.pone.0005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagos D, Ruiz MF, Sanchez L, Komitopoulou K. Isolation and characterization of the Bactrocera oleae genes orthologous to the sex determining Sex-lethal and doublesex genes of Drosophila melanogaster. Gene. 2005;348:111–21. doi: 10.1016/j.gene.2004.12.053. [DOI] [PubMed] [Google Scholar]
- 20.Concha C, Scott MJ. Sexual development in Lucilia cuprina (Diptera, Calliphoridae) is controlled by the transformer gene. Genetics. 2009;182:785–98. doi: 10.1534/genetics.109.100982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagos D, Koukidou M, Savakis C, Komitopoulou K. The transformer gene in Bactrocera oleae: the genetic switch that determines its sex fate. Insect Mol Biol. 2007;16:221–30. doi: 10.1111/j.1365-2583.2006.00717.x. [DOI] [PubMed] [Google Scholar]
- 22.Verhulst EC, Beukeboom LW, van de Zande L. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science. 2010;328:620–3. doi: 10.1126/science.1185805. [DOI] [PubMed] [Google Scholar]
- 23.Dzierzon J. Gutachten über die von Herrn Direktor Stöhr im ersten und zweiten Kapitel des General-Gutachtens aufgestellten Fragen. Eichstädter Bienenzeitung. 1845;1:109–113. 119–121. [Google Scholar]
- 24.McClung CE. The accessory chromosome – sex determinant? Biol Bull. 1902;3:43–84. [Google Scholar]
- 25.Wilson EB. The chromosomes in relation to determination of sex in insects. Science. 1905;22:500–2. doi: 10.1126/science.22.564.500. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez L. Sex-determining mechanisms in insects. Int J Dev Biol. 2008;52:837–56. doi: 10.1387/ijdb.072396ls. [DOI] [PubMed] [Google Scholar]
- 27.Schutt C, Nöthiger R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development. 2000;127:667–77. doi: 10.1242/dev.127.4.667. [DOI] [PubMed] [Google Scholar]
- 28.Erickson JW, Quintero JJ. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 2007;5:e332. doi: 10.1371/journal.pbio.0050332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubendorfer A, Hediger M, Burghardt G, Bopp D. Musca domestica, a window on the evolution of sex-determining mechanisms in insects. Int J Dev Biol. 2002;46:75–9. [PubMed] [Google Scholar]
- 30.Marin I, Baker BS. The evolutionary dynamics of sex determination. Science. 1998;281:1990–4. doi: 10.1126/science.281.5385.1990. [DOI] [PubMed] [Google Scholar]
- 31.Kraemer C, Schmidt ER. The sex determining region of Chironomus thummi is associated with highly repetitive DNA and transposable elements. Chromosoma. 1993;102:553–62. doi: 10.1007/BF00368348. [DOI] [PubMed] [Google Scholar]
- 32.Cook JM. Sex determination in the hymenoptera: a review of models and evidence. Heredity. 1993;71:421–35. [Google Scholar]
- 33.Beye M. The dice of fate: the csd gene and how its allelic composition regulates sexual development in the honey bee, Apis mellifera. BioEssays. 2004;26:1131–9. doi: 10.1002/bies.20098. [DOI] [PubMed] [Google Scholar]
- 34.Beye M, Hasselmann M, Fondrk MK, Page RE, et al. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 2003;114:419–29. doi: 10.1016/s0092-8674(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 35.Inoue H, Fukumori Y, Hiroyoshi T. Mapping of autosomal male-determining factors of the housefly, Musca domestica L., by means of sex-reversal. Jpn J Genet. 1983;58:451–61. [Google Scholar]
- 36.McDonald IC, Evenson P, Nickel CA, Johnson OA. Housefly genetics: isolation of a female determining factor on chromosome 4. Ann Entomol Soc Am. 1978;71:692–4. [Google Scholar]
- 37.Willhoeft U, Franz G. Identification of the sex-determining region of the Ceratitis capitata Y chromosome by deletion mapping. Genetics. 1996;144:737–45. doi: 10.1093/genetics/144.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKeown M, Belote JM, Baker BS. A molecular analysis of transformer, a gene in Drosophila melanogaster that controls female sexual differentiation. Cell. 1987;48:489–99. doi: 10.1016/0092-8674(87)90199-1. [DOI] [PubMed] [Google Scholar]
- 39.Butler B, Pirrotta V, Irminger-Finger I, Nöthiger R. The sex-determining gene tra of Drosophila: molecular cloning and transformation studies. EMBO J. 1986;5:3607–13. doi: 10.1002/j.1460-2075.1986.tb04689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savard J, Tautz D, Richards S, Weinstock GM, et al. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res. 2006;16:1334–8. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsh JL, Wieschaus E. Is sex determination in germ line and soma controlled by separate genetic mechanisms? Nature. 1978;272:249–51. doi: 10.1038/272249a0. [DOI] [PubMed] [Google Scholar]
- 42.Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of Sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65:229–39. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- 43.Shearman DC, Frommer M. The Bactrocera tryoni homologue of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol Biol. 1998;7:355–66. doi: 10.1046/j.1365-2583.1998.740355.x. [DOI] [PubMed] [Google Scholar]
- 44.Sievert V, Kuhn S, Traut W. Expression of the sex determining cascade genes Sex-lethal and doublesex in the phorid fly Megaselia scalaris. Genome. 1997;40:211–4. doi: 10.1139/g97-030. [DOI] [PubMed] [Google Scholar]
- 45.Kuhn S, Sievert V, Traut W. The sex-determining gene doublesex in the fly Megaselia scalaris: conserved structure and sex-specific splicing. Genome. 2000;43:1011–20. doi: 10.1139/g00-078. [DOI] [PubMed] [Google Scholar]
- 46.Ohbayashi F, Suzuki MG, Mita K, Okano K, et al. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm, Bombyx mori. Mol Genet Metab. 2001;128:145–58. doi: 10.1016/s1096-4959(00)00304-3. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz MF, Eirin-Lopez JM, Stefani RN, Perondini AL, et al. The gene doublesex of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. Dev Genes Evol. 2007;217:725–31. doi: 10.1007/s00427-007-0178-8. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz MF, Stefani RN, Mascarenhas RO, Perondini AL, et al. The gene doublesex of the fruit fly Anastrepha obliqua (Diptera, Tephritidae) Genetics. 2005;171:849–54. doi: 10.1534/genetics.105.044925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho S, Huang ZY, Zhang J. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics. 2007;177:1733–41. doi: 10.1534/genetics.107.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dearden PK, Wilson MJ, Sablan L, Osborne PW, et al. Patterns of conservation and change in honey bee developmental genes. Genome Res. 2006;16:1376–84. doi: 10.1101/gr.5108606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cristino AS, Nascimento AM, Costa LF, Simoes ZL. A comparative analysis of highly conserved sex-determining genes between Apis mellifera and Drosophila melanogaster. Genet Mol Res. 2006;5:154–68. [PubMed] [Google Scholar]
- 52.Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat Rev Genet. 2009;10:797–804. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- 53.Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 54.Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki MG, Funaguma S, Kanda T, Tamura T, et al. Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev Genes Evol. 2003;213:345–54. doi: 10.1007/s00427-003-0334-8. [DOI] [PubMed] [Google Scholar]
- 56.Vincent S, Perkins LA, Perrimon N. Doublesex surprises. Cell. 2001;106:399–402. doi: 10.1016/s0092-8674(01)00468-8. [DOI] [PubMed] [Google Scholar]
- 57.Kopp A, Duncan I, Godt D, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–9. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- 58.Keisman EL, Baker BS. The Drosophila sex determination hierarchy modulates wingless and decapentaplegic signalling to deploy dachshund sex-specifically in the genital imaginal disc. Development. 2001;128:1643–56. doi: 10.1242/dev.128.9.1643. [DOI] [PubMed] [Google Scholar]
- 59.Christiansen AE, Keisman EL, Ahmad SM, Baker BS. Sex comes in from the cold: the integration of sex and pattern. Trends Genet. 2002;18:510–6. doi: 10.1016/s0168-9525(02)02769-5. [DOI] [PubMed] [Google Scholar]
- 60.Keisman EL, Christiansen AE, Baker BS. The sex determination gene doublesex regulates the A/P organizer to direct sex-specific patterns of growth in the Drosophila genital imaginal disc. Dev Cell. 2001;1:215–25. doi: 10.1016/s1534-5807(01)00027-2. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez L, Guerrero I. The development of the Drosophila genital disc. BioEssays. 2001;23:698–707. doi: 10.1002/bies.1099. [DOI] [PubMed] [Google Scholar]
- 62.Sanchez L, Gorfinkiel N, Guerrero I. Sex determination genes control the development of the Drosophila genital disc, modulating the response to Hedgehog, Wingless and Decapentaplegic signals. Development. 2001;128:1033–43. doi: 10.1242/dev.128.7.1033. [DOI] [PubMed] [Google Scholar]
- 63.Williams TM, Selegue JE, Werner T, Gompel N, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–23. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryner LC, Goodwin SF, Castrillon DH, Anand A, et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–89. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 65.Demir E, Dickson BJ. Fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–94. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 66.Rideout EJ, Dornan AJ, Neville MC, Eadie S, et al. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 2010;13:458–66. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gailey DA, Billeter JC, Liu JH, Bauzon F, et al. Functional conservation of the fruitless male sex-determination gene across 250 Myr of insect evolution. Mol Biol Evol. 2006;23:633–43. doi: 10.1093/molbev/msj070. [DOI] [PubMed] [Google Scholar]
- 68.Bertossa RC, van de Zande L, Beukeboom LW. The fruitless gene in Nasonia displays complex sex-specific splicing and contains new zinc finger domains. Mol Biol Evol. 2009;26:1557–69. doi: 10.1093/molbev/msp067. [DOI] [PubMed] [Google Scholar]
- 69.Traut W, Niimi T, Ikeo K, Sahara K. Phylogeny of the sex-determining gene Sex-lethal in insects. Genome. 2006;49:254–62. doi: 10.1139/g05-107. [DOI] [PubMed] [Google Scholar]
- 70.Hasselmann M, Vekemans X, Pflugfelder J, Koeniger N, et al. Evidence for convergent nucleotide evolution and high allelic turnover rates at the complementary sex determiner gene of Western and Asian honeybees. Mol Biol Evol. 2008;25:696–708. doi: 10.1093/molbev/msn011. [DOI] [PubMed] [Google Scholar]
- 71.Hasselmann M, Beye M. Signatures of selection among sex-determining alleles of the honey bee. Proc Natl Acad Sci USA. 2004;101:4888–93. doi: 10.1073/pnas.0307147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heimpel GE, de Boer JG. Sex determination in the hymenoptera. Annu Rev Entomol. 2008;53:209–30. doi: 10.1146/annurev.ento.53.103106.093441. [DOI] [PubMed] [Google Scholar]
- 73.Hamilton WD. Extraordinary sex ratios. A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science. 1967;156:477–88. doi: 10.1126/science.156.3774.477. [DOI] [PubMed] [Google Scholar]
- 74.Trivers RL, Willard DE. Natural selection of parental ability to vary the sex ratio of offspring. Science. 1973;179:90–2. doi: 10.1126/science.179.4068.90. [DOI] [PubMed] [Google Scholar]
- 75.Rice WR. On the instability of polygenic sex determination: the effect of sex-specific selection. Evolution. 1986;40:633–9. doi: 10.1111/j.1558-5646.1986.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 76.Van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007;449:909–12. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- 77.Sandler L, Novitski E. Meiotic drive as an evolutionary force. Am Nat. 1957;41:105–10. [Google Scholar]
- 78.Temin RG. The independent distorting ability of the Enhancer of Segregation Distortion, E(SD), in Drosophila melanogaster. Genetics. 1991;128:339–56. doi: 10.1093/genetics/128.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pomiankowski A, Nothiger R, Wilkins A. The evolution of the Drosophila sex-determination pathway. Genetics. 2004;166:1761–73. doi: 10.1534/genetics.166.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh RS, Artieri CG. Male sex drive and the maintenance of sex: evidence from Drosophila. J Hered. 2010;101(Suppl 1):S100–6. doi: 10.1093/jhered/esq006. [DOI] [PubMed] [Google Scholar]
- 81.Singh RS, Kulathinal RJ. Male sex drive and the masculinization of the genome. BioEssays. 2005;27:518–25. doi: 10.1002/bies.20212. [DOI] [PubMed] [Google Scholar]
- 82.Charlesworth B, Charlesworth D. A model for the evolution of dioecy and gynodioecy. Am Nat. 1978;112:975–97. [Google Scholar]
- 83.Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–28. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 84.Charlesworth D, Charlesworth B. Sex chromosomes: evolution of the weird and wonderful. Curr Biol. 2005;15:R129–31. doi: 10.1016/j.cub.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 85.Denholm I, Franco MG, Rubini PG, Vecchi M. Identification of a male determinant on the X chromosome of housefly (Musca domestica L.) populations in south east England. Genet Res. 1983;42:311–22. [Google Scholar]
- 86.Hamm RL, Gao JR, Lin GG, Scott JG. Selective advantage for IIIM males over YM males in cage competition, mating competition, and pupal emergence in Musca domestica L. (Diptera: Muscidae) Environ Entomol. 2009;38:499–504. doi: 10.1603/022.038.0225. [DOI] [PubMed] [Google Scholar]
- 87.Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007;8:803–13. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- 88.Hasselmann M, Beye M. Pronounced differences of recombination activity at the sex determination locus (SDL) of the honey bee, a locus under strong balancing selection. Genetics. 2006;174:1469–80. doi: 10.1534/genetics.106.062018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gadau J, Gerloff CU, Kruger N, Chan H, et al. A linkage analysis of sex determination in Bombus terrestris (L.) (Hymenoptera: Apidae) Heredity. 2001;87:234–42. doi: 10.1046/j.1365-2540.2001.00919.x. [DOI] [PubMed] [Google Scholar]
- 90.Excoffier L, Heckel G. Computer programs for population genetics data analysis: a survival guide. Nat Rev Genet. 2006;7:745–58. doi: 10.1038/nrg1904. [DOI] [PubMed] [Google Scholar]
- 91.Kato Y, Kobayashi K, Oda S, Tatarazako N, et al. Sequence divergence and expression of a transformer gene in the branchiopod crustacean, Daphnia magna. Genomics. 2010;95:160–5. doi: 10.1016/j.ygeno.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 92.Kulathinal RJ, Skwarek L, Morton RA, Singh RS. Rapid evolution of the sex-determining gene, transformer: structural diversity and rate heterogeneity among sibling species of Drosophila. Mol Biol Evol. 2003;20:441–52. doi: 10.1093/molbev/msg053. [DOI] [PubMed] [Google Scholar]
- 93.McAllister BF, McVean GA. Neutral evolution of the sex-determining gene transformer in Drosophila. Genetics. 2000;154:1711–20. doi: 10.1093/genetics/154.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mita K, Kasahara M, Sasaki S, Nagayasu Y, et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- 95.Xia Q, Zhou Z, Lu C, Cheng D, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–40. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 96.Suzuki MG, Funaguma S, Kanda T, Tamura T, et al. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol Dev. 2005;7:58–68. doi: 10.1111/j.1525-142X.2005.05007.x. [DOI] [PubMed] [Google Scholar]
- 97.Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7:e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Siera SG, Cline TW. Sexual backtalk with evolutionary implications: stimulation of the Drosophila sex-determination gene Sex-lethal by its target transformer. Genetics. 2008;180:1963–81. doi: 10.1534/genetics.108.093898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yeates DK, Wiegmann BM, Courtney GW, Meier M, et al. Phylogeny and systematics of Diptera: two decades of progress and prospects. Zootaxa. 2007;1668:565–90. [Google Scholar]
- 100.Hasselmann M, Lechner S, Schulte C, Beye M. Origin of a function by tandem gene duplication limits the evolutionary capability of its sister copy. Proc Natl Acad Sci USA. 2010;107:13378–83. doi: 10.1073/pnas.1005617107. [DOI] [PMC free article] [PubMed] [Google Scholar]