Abstract
The cell nucleus is responsible for the storage, expression, propagation, and maintenance of the genetic material it contains. Highly organized macromolecular complexes are required for these processes to occur faithfully in an extremely crowded nuclear environment. In addition to chromosome territories, the nucleus is characterized by the presence of nuclear substructures, such as the nuclear envelope, the nucleolus, and other nuclear bodies. Other smaller structural entities assemble on chromatin in response to required functions including RNA transcription, DNA replication, and DNA repair. Experiments in living cells over the last decade have revealed that many DNA binding proteins have very short residence times on chromatin. These observations have led to a model in which the assembly of nuclear macromolecular complexes is based on the transient binding of their components. While indeed most nuclear proteins are highly dynamic, we found after an extensive survey of the FRAP literature that an important subset of nuclear proteins shows either very slow turnover or complete immobility. These examples provide compelling evidence for the establishment of stable protein complexes in the nucleus over significant fractions of the cell cycle. Stable interactions in the nucleus may, therefore, contribute to the maintenance of genome integrity. Based on our compilation of FRAP data, we propose an extension of the existing model for nuclear organization which now incorporates stable interactions. Our new “induced stability” model suggests that self-organization, self-assembly, and assisted assembly contribute to nuclear architecture and function.
Electronic supplementary material
The online version of this article (doi:10.1007/s10577-010-9161-8) contains supplementary material, which is available to authorized users.
Keywords: Nucleus, Fluorescence microscopy, FRAP, Multi-protein complex, Chromatin binding, Dynamics, Residence time, Induced stability, Assisted assembly, Self-assembly, Self-organization
Structure and function in the nucleus
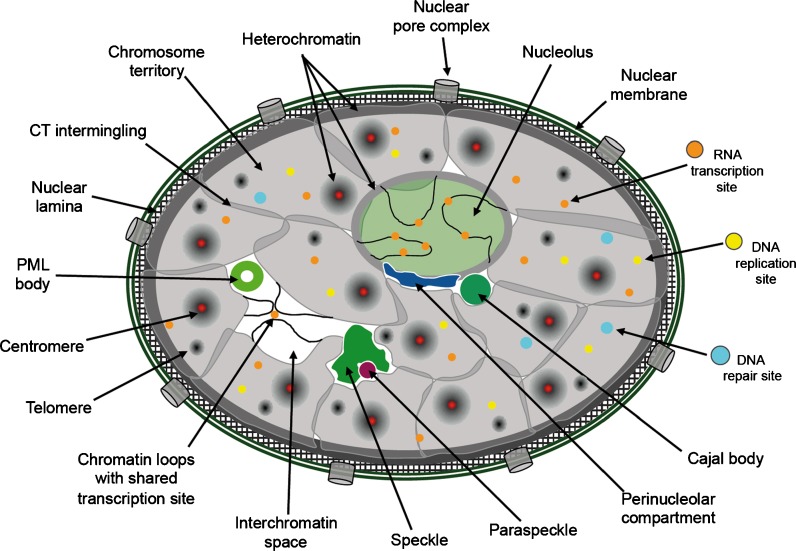
The storage, propagation, maintenance, and expression of the genetic material is executed by biochemical activities, namely DNA compaction/decompaction, DNA replication and segregation, DNA repair, and RNA transcription/processing, respectively (Diekmann and Hemmerich 2005). The corresponding machineries are highly structured, yet dynamic macromolecular assemblies (Misteli 2007) which must work on chromatin with high fidelity in a crowded nuclear environment (Richter et al. 2007). In addition, the mammalian cell nucleus contains a variety of subnuclear domains, nuclear bodies, or subnuclear compartments (Figs. 1 and 2). DNA is wrapped around nucleosomes and forms individual chromosomes which are compacted in mitosis (Fig. 2g). In interphase cells, chromosomes decondense into so-called chromosome territories (CTs), which occupy distinct volume regions (Figs. 1 and 2h) (Cremer et al. 2006; Heard and Bickmore 2007; Solovei et al. 2009; Finan et al. 2010). Staining of interphase chromatin using DNA dyes does not reveal CT structures but allows discrimination between transcriptionally active euchromatin and transcriptionally silent heterochromatin (Fig. 2e). Constitutive heterochromatin is mainly composed of pericentric DNA, and in this case, the chromosome’s centromere/kinetochore complex can be found embedded within or adjacent to this chromatin region (Fig. 2j) (Probst and Almouzni 2008). The nucleus obtains structural support through the nuclear lamina, which is attached to the nuclear double-membrane, together forming the nuclear envelope (Fig. 1). The nuclear envelope controls traffic of molecules between the cytoplasm and the nucleoplasm but has also emerged as a critical determinant in genome architecture (Starr 2009).
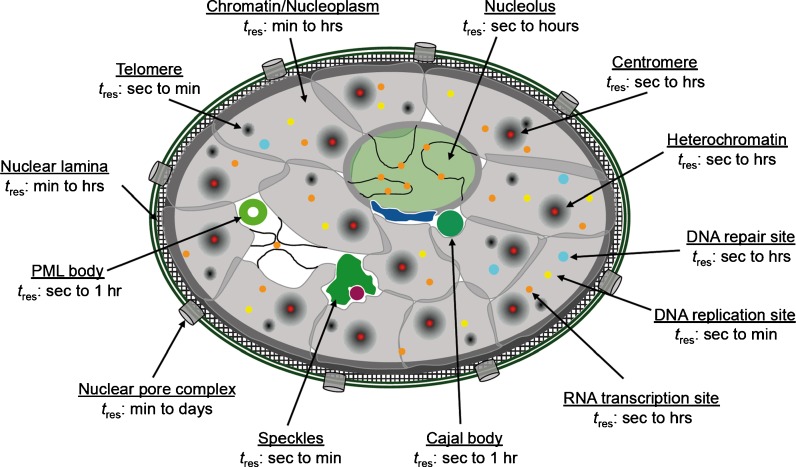
Fig. 1.
Compartmentalization of the mammalian cell nucleus. The mammalian cell nucleus contains chromatin in the form of chromosome territories (CTs). CTs may overlap at their touching borders (intermingling) or create the so-called interchromatin space (white). Constitutive heterochromatin is mainly found as pericentromeric chromatin in patches throughout the nuclear volume, at the nuclear periphery, as well as around nucleoli. Nuclear pore complexes, the double-layered nuclear membrane (dark green) and the meshwork-like nuclear lamina are structural hallmarks in the periphery of the nucleus. Chromatin loops with associated transcription factories may extrude out of chromosome territories within the nucleolus as well as throughout the nucleoplasm. Transcription (orange), replication (yellow), and DNA repair processes (light blue) usually occur in small domains with a diameter below 100 nm. A diverse set of nuclear bodies, such as speckles, paraspeckles, the perinucleolar compartment, Cajal bodies, or PML bodies are found in the interchromatin space
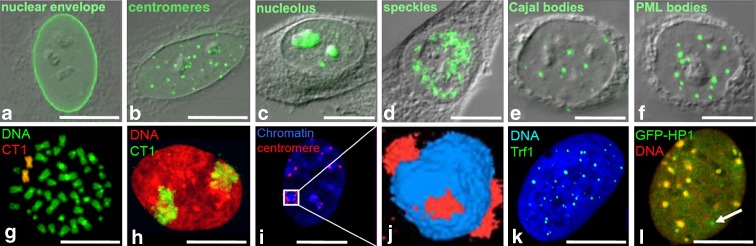
Fig. 2.
Visualization of nuclear compartments. a–f A diverse set of nuclear substructures, including the nuclear envelope, centromeres, nucleoli, Cajal bodies, or PML bodies can be visualized by confocal immunofluorescence microscopy employing specific antibodies. g, h Visualization of Chromosome 1 (CT1) by fluorescence in situ hybridization (FISH) on a metaphase spread or in an interphase nucleus of MRC-5 cells. i, j Staining of chromocenters (blue) and associated centromeres in a mouse cell nucleus demonstrates the clustering of pericentromeric heterochromatin of several chromosomes. k Telomeres can be visualized by FISH against telomeric DNA or, as shown here, by immuno-detection of Trf1 (green). l Nuclear distribution of GFP-tagged heterochromatin protein 1 (green) in relation to chromatin (DNA, red) reveals its accumulation in chromocenters in the nucleus of a mouse fibroblast. In addition, GFP-HP1 is distributed diffusely throughout the euchromatic region as well as in small dots representing PML nuclear bodies (white arrow). Size bar, 10 μm
The most prominent subnuclear domains include the nucleolus, the perinucleolar compartment, speckles, paraspeckles, Cajal bodies, and promyelocytic leukemia (PML) bodies (Figs. 1 and 2). In addition, a variety of other nuclear bodies have been identified such as PcG bodies, Gemini bodies (Gems), the OPT domain, cleavage bodies, and the SAM68 nuclear body (Spector 2001; Handwerger and Gall 2006). Subnuclear structures are macromolecular complexes that consist of membrane-less accumulations of specific sets of functionally related molecules. For example, components of the ribosome biogenesis pathway are predominantly confined to the nucleolus. First thought to be exclusively devoted to the synthesis of ribosomal RNA and assembly of ribosomal subunits, it has become clear that the nucleolus serves a variety of additional functions, including regulation of mitosis, cell-cycle progression, proliferation, and various stress responses (Raska et al. 2006; Sirri et al. 2008).
The biochemical function(s) of the other subnuclear domains are less clear or unknown. PML bodies attract a selected set of nuclear proteins which are functionally quiet promiscuous. Therefore, PML bodies have been implicated in the regulation of diverse cellular functions, such as the induction of apoptosis and cellular senescence, inhibition of proliferation, maintenance of genomic stability, and antiviral responses (Bernardi and Pandolfi 2007). PML bodies are positionally stable structures at which controlled molecular traffic and posttranslational modifications may regulate the activity of specific proteins throughout the genome and the epigenome in response to various cellular stresses (Bernardi and Pandolfi 2007; Torok et al. 2009). Speckles, also referred to as interchromatin granule clusters, are enriched in pre-mRNA splicing factors. At the microscopic level, speckles appear as irregular, punctate domains varying in size and shape (Fig. 2). They are considered to be the main sites for storage, assembly, and/or recycling of the essential spliceosome components (Lamond and Spector 2003). Because highly transcribed genes are found in the periphery of speckles and also other subnuclear domains, they may also serve to efficiently integrate and regulate mRNA transcription and mRNA processing machineries (Zhao et al. 2009). Cajal bodies (CBs) are involved in the biogenesis of several classes of small ribonucleoprotein particles (snRNPs) as well as their modification (Gall 2000; Matera et al. 2009). Resembling the speckles/gene association mentioned above, CBs associate specifically with histone and snRNA genes. This colocalization is transcription-dependent, requires expression of snRNA coding regions, and is probably based on an energy-driven motor activity in the nucleus (Frey and Matera 2001; Dundr et al. 2007).
In mammalian cell nuclei, DNA replication, RNA transcription, and repair of damaged DNA occurs in dot-like structures with a mean diameter of ∼100 nm (Fig. 3). With respect to transcription and replication, these focal sites have been coined “factories” as each site contains all of the enzymatic activity required. A general model was recently suggested for the organization of all genomes in which the transcription factories play a central role (Cook 1999; Cook 2010). Notably, the model proposes that active RNA polymerases do not move along their templates during elongation but are bound to a factory acting both as motors that reel in their templates and as fixed structural entities that hold active chromatin loops in place (Cook 2010). DNA replication also occurs at similarly specialized subnuclear sites where the factors directly or indirectly involved in replication are concentrated (Fig. 3b) (Leonhardt et al. 2000). Finally, repair of damaged DNA at focal sites throughout the genome is also a dynamic process that requires careful orchestration of a multitude of enzymes, adaptor proteins, and chromatin constituents (Fig. 3c) (Lukas et al. 2005).
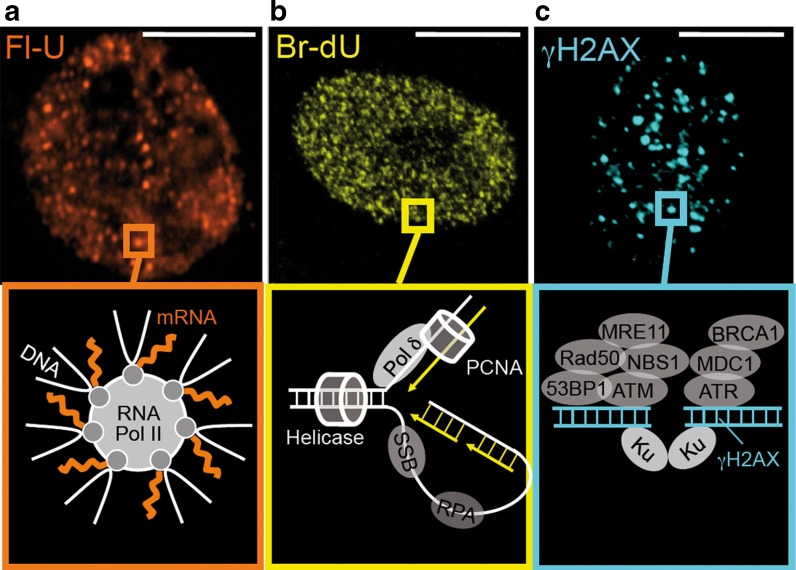
Fig. 3.
DNA and RNA metabolism occurs in small foci. a Transcription can be visualized after brief exposure of living cells to the nucleotide analog Fluoro-Uridine (Fl-U), follwed by immunodetection of the Fl-U epitope. b Bromo-deoxy-Uridine (BrdU) can be used in a similar incorporation assay to visualize nascent DNA during replication. c Sites of DNA double-strand break repair are detected using antibodies against a phosphorylated form of histone H2AX (γ-H2AX). Common to all of these active foci is the accumulation of many factors required to perform the biochemical activities. Size bar, 10 μm
Fluorescence fluctuation microscopy
The development of in vivo microscopy techniques employing the green fluorescent protein (GFP) has opened the door to probe cellular architecture and function in living cells (Lippincott-Schwartz et al. 2003). By analyzing macroscopic relaxation after disturbing the equilibrium state, fluorescence intensity images can be used to assess diffusion times, interactions, and binding constants of molecules. Fluorescence fluctuation microscopy (FFM) approaches have been developed to investigate a few molecules in small regions of a cell. These approaches supply dynamic spatiotemporal information by creating cellular diffusion and concentration maps (Fig. 4). A major consideration is the accessible resolution as nuclear processes can take place on a time scale ranging from microseconds to hours, and single molecules or huge macromolecular assemblies in well-defined stoichiometries can be involved.
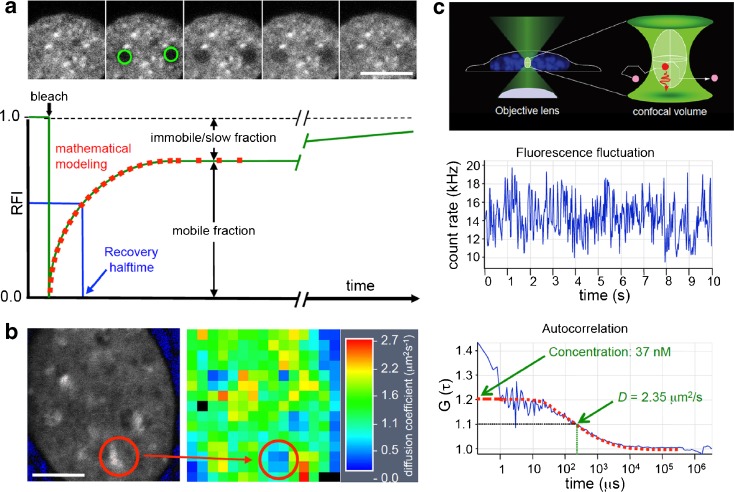
Fig. 4.
Fluorescence fluctuation microscopy. a Upper panel FRAP experiment in a nucleus expressing GFP-tagged HP1. Fluorescence was bleached within two circled areas (green) and fluorescence redistribution was monitored over time. Bottom panel Quantification of a FRAP experiment. Graphs typically show mean values from 10 to 20 FRAP experiments as relative fluorescence intensity (RFI) after normalization to prebleach levels. The FRAP curve immediately delivers information on mobile and immobile or slow populations. Ideally, the FRAP experiment is extended to allow discrimination between immobile or slowly exchanging populations. Mathematical modeling can be performed to extract biophysical parameters such as the diffusion coefficient and binding constants from fit functions (red dotted line). b RICS. GFP-HP1 was expressed in HEp-2 cells. For RICS, a time series of GFP fluorescence images (512 × 512 pixels) was acquired in a subregion of the nucleus by confocal microscopy (left panel). Subregions (64 × 64 pixels) within this time series are than extracted and correlation spectra assessed from these subregions. A diffusion coefficient map can be generated by fitting with appropriate diffusion models (middle panel) from which diffusion coefficients of GFP-HP1 can be determined at different subnuclear positions. c In FCS, a confocal volume is generated by focusing of a laser beam through an appropriate objective (upper panel). Fluorescent molecules emit photons during their movement through the confocal volume leading to a fluorescence fluctuation over time (middle panel). The fluctuation data are then subjected to autocorrelation (bottom panel) from which fluorescent molecule concentration and diffusion coefficients can be extracted after fitting of the data with appropriate mathematical diffusion models. Size bar, 5 μm
FFM allowed for the first time to not only visualize protein dynamics and chromatin interactions but also to quantitatively determine biophysical properties of proteins in the nucleus of intact cells (Erdel et al. 2010, this volume). FFM approaches include time-lapse microscopy (Heun et al. 2001), fluorescence redistribution after photobleaching (FRAP, FLIP, iFRAP, etc.) (Bancaud et al. 2009; van Royen and Houtsmuller 2010; this volume), fluorescence correlation spectroscopy (FCS) (Haustein and Schwille 2003), CP (Wachsmuth et al. 2003), raster image correlation spectroscopy (RICS) (Digman and Gratton 2009), SPT (Levi and Gratton 2008), FRET and FLIM (Wallrabe and Periasamy 2005), and MFIS (Weidtkamp-Peters et al. 2009). Figure 4 highlights some of these techniques demonstrating their potential to study nuclear protein dynamics.
Dynamic and stable interactions contribute to the assembly and maintenance of subnuclear compartments
FRAP analyses of subnuclear domains such as nucleoli, speckles, and Cajal bodies have revealed that their component parts rapidly exchange with nucleoplasmic pools (Misteli 2001a, 2008). Typical residence times of proteins in these compartments are in the seconds range. Similarly, most (but not all, see below) factors acting at transcription, replication, and repair foci show rapid exchange at chromatin (Houtsmuller et al. 1999; Dundr et al. 2002; Sporbert et al. 2002). These observations have led to the conclusion that nuclear proteins undergo repeated and rapid cycles of association and dissociation between the nucleoplasm and the compartment they are mainly working in. As a consequence, nuclear bodies and factories are in perpetual flux. Their structure is determined by the ratio of on-rate versus off-rate of its components, clearly suggesting self-assembly and/or self-organization as the mechanism of their assembly (Misteli 2001b; Matera et al. 2009). To further test this hypothesis, we have compiled FRAP data of 264 nuclear proteins collected over the last 16 years. Data are presented as individual sheets of a single table (ESM 1) containing: chromatin-binding proteins, transcription factors, DNA metabolism (covering replication, repair, recombination), RNA metabolism (covering speckles, splicing, RNA modification), centromeres, nucleolus, nuclear envelope, Cajal bodies, PML bodies, and “others.” Surprisingly, while indeed most nuclear proteins are highly dynamic, we found a small but nevertheless important set of proteins showing either very slow turnover or complete immobility (ESM 1). Figure 5 provides an overview of the FRAP data with respect to the various subnuclear structures. In many cases, very slow FRAP recoveries probably reflect relatively long residence times on chromatin. However, it is also possible in principle that slow recoveries can arise from short residence times if the average fluorescent molecule undergoes many binding events as it progresses to the center of the bleach spot (Sprague et al. 2004). Accurate residence time estimates require a test for the role of diffusion in the FRAP and a proper mathematical model for fitting (Sprague and McNally 2005). This approach should be considered for the slowly recovering proteins discussed below.
Fig. 5.
(Im)mobilities in the nucleus. Illustration of subnuclear compartments and overview of their component‘s residence times (t res) determined by FRAP
Most nucleosomes are stable for long periods of time
FRAP analyses of GFP-tagged core histones have shown that far more than 50% of H2A, H2B, H3, and H4 are stably incorporated into chromatin and do not exchange with soluble pools (ESM 1; Kimura and Cook 2001; Gautier et al. 2004). These observations are consistent with the idea of a structural foundation of genome architecture and maintenance based on stable nucleosome arrays. The transiently incorporated sub-fraction of core histones is most likely attributable to chromatin remodeling events associated with transcription, replication or repair. All four core histones are present in the same nucleosome, yet their stabilities vary. H3 and H4 are loaded into nucleosomes during DNA replication, after which >80% remains permanently bound (Kimura and Cook 2001). In contrast, ∼50% of H2A and H2B exhibit substantial turnover (Kimura and Cook 2001; Gautier et al. 2004), compatible with the assembly properties of nucleosomes in vitro and in vivo (Annunziato 2005). While the immobile fractions might be associated with heterochromatin, the stability of the central H3 and H4 coupled with the lability of the outer H2A and H2B suggest simple ways of maintaining epigenetic marks (Kimura and Cook 2001). This is consistent with many posttranslational modifications found at the N-terminal ends of H3 and H4 providing long-term memories (Margueron and Reinberg 2010).
Genome maintenance relies on hyper-stable protein complexes at the centromere
Cell division is a highly dynamic process in which the chromosomes are segregated in a coordinated fashion. The centromere is the genetic locus required for precise and accurate chromosome segregation and provides a platform on which the kinetochore multiprotein complex assembles (Przewloka and Glover 2009). Accurate chromosome segregation is essential for cell survival and genome maintenance. Aberrant mitotic segregation can result in aneuploidy, cell death, or cancer (Kops et al. 2005). Intuitively and confirmed by FRAP data, stable chromatin–protein and protein–protein interactions are required during mitosis when sister chromatids are pulled along microtubules into daughter cells (ESM 1). However, the kinetochore/centromere complex could in principle disassemble completely during interphase, since a stable epigenetic mark such as a unique histone modification would be sufficient to transmit centromere identity through interphase. Shortly before mitosis starts, the epigenetic mark would be used as a landing platform for cell cycle-regulated factors that initiate de novo kinetochore assembly. This, however, is not the case as FRAP of centromeric proteins performed at any time during cell cycle show that at least Cenp-A and Cenp-I (ESM 1) stay ably associated with centromeres for the complete cell cycle (Hemmerich et al. 2008; Hellwig et al. 2008).
Because there is absolutely no dynamic exchange of Cenp-A between centromeres and freely diffusible nucleoplasmic pools, the question arises how the binding sites created by centromere replication are occupied. This issue has been addressed by FRAP experiments which clearly showed that Cenp-A is assembled during the G1 phase of the cell cycle via a “loading only” mechanism (Hemmerich et al. 2008): while the first FRAP experiment showed fluorescence recovery, a subsequent FRAP showed no recovery of CENP-A indicating an undetectably small off-rate. This shows that CENP-A is “loaded only,” without exchange at its binding sites. Interestingly, Cenp-A is incorporated via the “loading only” mechanism exclusively in early G1 (Hemmerich et al. 2008) corroborating earlier SNAP-tagging results (Jansen et al. 2007). The epigenetic marking of the centromere is believed to be conveyed by Cenp-A, because it is required for the association of probably all other kinetochore proteins (Buscaino et al. 2010) and because of its sustained presence at centromeres without dynamic exchange (Hemmerich et al. 2008; ESM 1). Although Cenp-A and Cenp-I are stably bound, many rapidly exchanging centromere components have also been identified (ESM 1). Taken together, the identification by FRAP of proteins which bind to the centromeres with dynamic exchange but also without dynamic exchange for longer than one cell cycle clearly indicates that in addition to organization and self-assembly, also heredity mechanisms contribute to centromere structure and function and hence genome maintenance.
A fraction of stably bound cohesin glues sister chromatids together
Sister chromatid cohesion after replication is mediated by the multiprotein complex cohesin which acts as a topological linker (Wong 2010). Gerlich et al. (2006) performed FRAP analyses identifying two distinct binding modes of cohesin to chromatin during the cell cycle. One pool of cohesin dynamically exchanged on and off chromatin throughout the entire interphase, moving through the nucleus by diffusion. This pool was absent in metaphase. About one third of the subunits SA1 and Scc1 are stably fixed to chromosomes after replication with a residence time of 6 to 7 h (ESM 1). This pool persisted until just before chromosome segregation (Gerlich et al. 2006). These analyses provided the first experimental evidence in living cells for a permanent and stable link between sister-chromatids. This stable link may functionally contribute to homologous recombination repair of DNA double-strand breaks during the G2 phase of the cell cycle.
HA95 and lamin A immobility in the nucleoplasm is consistent with the idea of a stable nuclear matrix
The existence of a nuclear matrix was proposed by Zbarskii and Debov (1948) based on their observation that high-salt extractions of purified nuclei produced microscopically visible lattice-like residual structures. These observations were confirmed in the Berezney lab using similar extraction protocols (Berezney and Coffey 1974). The lack of direct, particularly in vivo evidence, however, raised considerable skepticism about the existence of a stable nuclear structure (Pederson 2000). Homologous to A-kinase anchoring protein 95 (HA95) is a nuclear protein harboring two zinc fingers and a putative nuclear localization signal (Ørstavik et al. 2000). HA95 binds to chromatin and the nuclear lamina network where it has a role in anchoring nuclear membranes and lamins to chromatin in interphase and in releasing membranes from chromatin at mitosis. Astonishingly, bleaching experiments revealed no detectable recovery of the diffusely localized GFP-tagged HA95 in the nucleus of living 293 T fibroblasts (Martins et al. 2000). Roughly one quarter of the nuclear pool of lamin A resides outside the nuclear lamina and distributes as a diffuse veil throughout the nucleoplasm (Moir et al. 2000). FRAP analysis of the nucleoplasmic GFP-lamin A pool revealed a recovery halftime of more than 3 h, clearly indicating a stable lamin-containing structure in living cells (Moir et al. 2000). Although overexpression artifacts of the GFP fusion proteins cannot be ruled out, the immobile properties of HA95 and lamin A in the cell nucleus provide compelling evidence for the existence of a nuclear matrix in living cells. It has been suggested that HA95 provides a stable platform at the chromatin/interchromatin interface that could regulate nuclear envelope–chromatin interactions during the cell cycle (Martins et al. 2003). Recent evidence suggests that HA95 is also functionally involved in pre-mRNA splicing (Kvissel et al. 2007). Thus, HA95 could also serve as a stable platform on chromatin, linking RNA transcription with RNA processing. It will be essential to analyze if nuclear stability and particular nuclear functions are affected in HA95-depleted cells.
Constitutive heterochromatin: more stable than initially thought?
The main function of constitutive heterochromatin is to serve as a kinetochore attachment site in mitosis, where its particular chromatin structure is thought to ensure chromosome segregation. Pericentric heterochromatin also plays important roles in organizing transcription-repressive compartments of the nucleus and to ensure regulation of particular genes (Probst and Almouzni 2008). Constitutive heterochromatin is characterized by three repressive epigenetic marks: tri-methylation of H3-K9, mono-methylation of H3-K27, and tri-methylation of H4-K20. The histone methyltransferases Suv39H1/2 and Suv420H1/2 play crucial roles in the initial steps of constitutive heterochromatin formation in mammals. Mice deficient for the Suv enzyme family members display impaired pericentric heterochromatin function leading to chromosome missegregation and increase of sister-chromatid exchange. H3-K9me3 marks placed by Suv39H activities stabilize heterochromatin protein 1 (HP1) binding to heterochromatin. HP1 proteins self-oligomerize and interact with both Suv39H and Suv420H (Fodor et al. 2010).
According to its structural function, heterochromatin had been perceived as a relatively densely packed, rigid, and inert region. Surprisingly, however, FRAP had demonstrated that GFP-tagged HP1 isoforms display dynamic binding properties, suggesting that heterochromatin is a dynamic and plastic domain, in which access to the underlying DNA would not necessarily be prevented (Cheutin et al. 2003; Festenstein et al. 2003; Schmiedeberg et al. 2004). In contrast to HP1, Suv39H1 consists of both a mobile and an immobile population in heterochromatin where ∼30% of molecules do not exchange over a time period of 7 min (ESM 1; Krouwels et al. 2005). The tight association of Suv39H1 with chromatin is obviously a more common feature because a large fraction of Suv420H2 is also tightly bound at pericentric heterochromatin (ESM 1). FRAP analyses has revealed that more than 90% of GFP-tagged Suv420H2 is immobile at these domains over several minutes (Souza et al. 2009). It is, therefore, possible that the residence times of Suv39H1 and Suv420H2 in heterochromatin are even in the hours range, indicating that these enzymes may, in addition to their catalytic activity, also serve a structural role in chromatin and act as a stable interaction platform for other heterochromatin proteins. Further structural support for heterochromatin architecture may also be provided by the heterochromatin-binding protein Ki-67 which, by FRAP, also shows a cell cycle dependent turnover in heterochromatin (ESM 1; Saiwaki et al. 2005). While Ki-67 dynamically binds to mitotic chromatin, it becomes very tightly bound to peri-nucleolar heterochromatin during G1 phase. Approximately 60% of GFP-Ki-67 is immobile over 10 min during FRAP analyses in interphase cells (Saiwaki et al. 2005). These data suggest that Ki-67 may have very long residence times on chromatin.
Some transcription factors can become stably bound to chromatin: pRB, E2F, Mdm2, VHL, TFIIH, HSF, and Gal4
Most of the time, transcription factors diffuse throughout the cell nucleus, encounter gene promoters in a random fashion, and bind to them for a very short time (Misteli 2001b). This, however, does not hold for all situations.
pRB and E2F
The retinoblastoma protein (pRB) is a key cell-cycle protein that regulates the critical G1-S phase transition through interaction with E2F family members. The latter are cell-cycle transcription factors that repress transcription of genes required for G1-S checkpoint transition. pRB activity is regulated through networks sensing intracellular and extracellular signals which block or permit phosphorylation (inactivation) of the Rb protein (Poznic 2009). Quite unexpectedly, in bleaching experiments, fluorescence recovery of active (hypophosphorylated) GFP-RB and E2F is minimal (Angus et al. 2003). This behavior is in stark contrast to the mobility of most other transcription factors which are usually highly mobile (ESM 1). Only after inactivation by phosphorylation, pRB and E2F become highly mobile in chromatin. The regulation of the affinity of pRB to chromatin may constitute a new mechanism of transcriptional control.
Mdm2 and VHL
Complete immobilization, in this case within the nucleolus, was also observed for the ubiquitin ligases Mdm2 and VHL in FRAP experiments (ESM 1; Mekhail et al. 2005). Upon an activation stimulus, the nucleolus rapidly releases these enzymes from static detention, thereby restoring their high mobility profiles. These observations provide strong evidence that cells have evolved mechanisms to regulate molecular networks by reversibly switching proteins between mobile and static states (Mekhail et al. 2005).
TFIIH
In tissue-derived primary cells such as post-mitotic neurons, hepatocytes, and cardiac myocytes, the transcription factor TFIIH is effectively immobilized on the chromatin for hours during transcription, whereas in proliferative cells, it has the same highly dynamic behavior as in cultured cells (Giglia-Mari et al. 2009). It was proposed that static chromatin binding of TFIIH may be established during differentiation- and cell lineage-specific transcriptional programs. Interestingly, stable chromatin binding was not irreversible in the post-mitotic cells because induction of local DNA lesions remobilized TFIIH, probably based on its requirement in DNA repair (Giglia-Mari et al. 2009).
HSF
In Drosophila salivary glands, the heat shock factor (HSF; the transcription activator of hsp70) trimerizes and translocates from the nucleoplasm to chromosomal loci after heat shock (Yao et al. 2006). FRAP experiments showed a rapid exchange of HSF at chromosomal loci under non-heat shock conditions (t 1/2 = 15 s) but a very slow exchange after heat shock (t 1/2 > 6 min). Five minutes after the FRAP bleach pulse, ∼50% of HSF was still immobilized on chromatin. The unbound pool of HSF was found by FCS to diffuse freely, indicating that this pool was not able to compete for chromatin binding (Yao et al. 2006).
Gal4
Nalley and colleagues developed a new variant of the chromatin immunoprecipitation assay that allowed a direct time-resolved observation of exchange between the transcription factor Gal4 and its DNA binding site on native promoters in yeast (Nalley et al. 2006). The assay revealed that after transcriptional activation, the Gal4–promoter complex was highly stable with a half-life of approximately 1 h. Again, this stability was established although unbound, competition–competent Gal4 molecules were present in excess amounts. During immobilization on its promoter, expression of the reporter gene occurred indicating that Gal4 functions via long-lived complexes with promoters during transcriptional induction (Nalley et al. 2006).
The nuclear envelope contains large pools of immobile proteins
Proteins of the nuclear lamina interconnect chromosomes with the nuclear envelope. The lamina harbors specific integral proteins including the lamin B receptor (LBR), lamina-associated polypeptides (LAPs), emerin, nurim, and MAN1. LBR, LAPs, and emerin bind to lamins A/C and B in vitro while LBR and LAP2b also bind chromatin via interactions with HP1 and the small DNA-binding protein BAF (Mekhail and Moazed 2010). Thus, a highly interconnected protein network provides multiple anchoring sites between chromatin and the nuclear envelope. Many of these proteins have meanwhile been subjected to FRAP analyses revealing stable incorporation of most of them into the nuclear lamina (ESM 1). Although mathematical modeling has not been employed yet, the extremely long recovery halftimes (i.e., more than 3 h for lamin B, Moir et al. 2000) of nuclear lamina constituents suggest residence times in the hours range. This is particularly true for the nuclear pore complexes, some components of which persist for long periods of time (residence times up to 70 h) within well-defined spatial regions without dynamic exchange (ESM 1; Rabut et al. 2004). Because the nucleus also undergoes dramatic morphological changes, a completely static nuclear lamina is not beneficial. Transitions between a stable and a flexible lamina might be regulated by small pools of the dynamically exchanging lamina constituents (ESM 1). Posttranslational modifications may serve as an additional mechanism to alter nuclear lamina stiffness.
Nuclear bodies contain stable components
FRAP analysis of subnuclear domains has revealed that most of their component parts rapidly exchange with nucleoplasmic pools with residence times in the seconds range (Misteli 2007; Matera et al. 2009). As a consequence, it was thought that these structures are determined by the ratio of on-rate versus off-rate of their constituents (Misteli 2008). This assembly mechanism is certainly true for speckles and nucleoli (ESM 1), which usually form in response to particular functions (splicing factor assembly and rRNA synthesis, respectively). However, FRAP experiments and mathematical modeling have revealed residence times of up to 1 h for Cajal and PML body components (ESM 1).
There are six nuclear isoforms of the PML protein in human cells from which PML V showed an extremely long residence time (48 min) (ESM 1). All other isoforms show substantially faster exchange, although they share the same nuclear body-binding domains with PML V (Weidtkamp-Peters et al. 2008). When analyzed in nuclei devoid of any endogenous PML protein (PML−/− MEFs), fractions of PML II becomes completely immobile within nuclear bodies and the residence time of PML IV is extended to 1.5 h (Brand et al. 2010). These observations indicate that probably all PML isoforms have the potential to build a stable scaffold within nuclear bodies, and that during evolution PML V was selected to execute this function (Weidtkamp-Peters et al. 2008). In Cajal bodies, p80 coiling was identified as the component with the longest residence time both, in human cells (Dundr et al. 2004) and in Xenopus germinal vesicles (Handwerger et al. 2003; Deryusheva and Gall 2004). These data indicate that the architecture of PML bodies and Cajal bodies may be primarily dictated by protein interactions forming stable scaffolds (Handwerger et al. 2003; Deryusheva and Gall 2004; Brand et al. 2010). On the other hand, their functionality certainly involves stochastic encounter of specific, fast exchanging components (Dundr et al. 2004; Weidtkamp-Peters et al. 2008).
Replication and repair factories contain a few stable components
RNA transcription sites and DNA repair foci have been visualized in mammalian cells, and initial FRAP studies indicated that they are stochastically assembled de novo in each round of transcription or repair from freely diffusible components (ESM 1; Houtsmuller et al. 1999; Dundr et al. 2002). DNA repair factor recruitment dynamics point towards diffusion-based DNA lesion recognition by individual components or small subcomplexes as the primary mode of DNA repair, instead of the recruitment of large preassembled holoenzymes (Luijsterburg et al. 2007; Dinant et al. 2009). Nevertheless, FRAP analyses also indicate that some replication and repair factors can reside on chromatin for extended periods of time (ESM 1). Data for ERCC1 for example suggest that this component is assembled at sites of DNA damage with other constituents of the nucleotide excision repair (NER) machinery into immobile holocomplexes during one repair event with a mean residence time of 4 min which is the estimated time of a single repair event (Luijsterburg et al. 2010). Furthermore, strip-FRAP on globally irradiated cells showed incomplete recovery of DDB2 for several hours after 16 J/m2 UV exposure, indicative of a significant immobile fraction over a period of hours at damaged chromatin (Luijsterburg et al. 2007). Twenty-four hours after exposure, most UV-induced lesions have been removed by NER, suggesting full release of bound molecules when repair is completed. The UV-induced immobilization was also shown for the NER proteins TFIIH, XPA, and ERCC1-XPF but was not observed for RAD52 group proteins (ESM 1; Houtsmuller et al. 1999; Essers et al. 2002; Politi et al. 2005). In UV-treated cells, the fraction of lesion-bound XPA molecules increases to maximally 35%, and the increase is directly proportional to the DNA damage load. The immobile pool of XPA at damaged chromatin remains over a period of 2 to 4 h post-UV exposure (Rademakers et al. 2003). While cells in living tissues may never experience DNA damage loads comparable to the cell culture studies, the immobilization of DNA repair factors ERCC1, XPA, and DDB1 on chromatin suggests that stable protein scaffolds may functionally contribute to DNA repair pathways.
Proliferating cell nuclear antigen (PCNA) is a further example of stable chromatin attachment. FRAP revealed elevated residence times for this factor at sites of DNA replication and repair (Sporbert et al. 2002; Solomon et al. 2004a, b; Mortusewicz et al. 2006; ESM 1). As a consequence, it was proposed that PCNA does not “bring” other factors to sites of replication or repair. Instead, PCNA appears to act as a stationary loading platform that is reused over multiple DNA synthesis or repair cycles with PCNA-binding proteins associating transiently and subsequently dissociating rather than being part of one stable, multifunctional, processive replication machinery (Mortusewicz et al. 2008).
An extension of models for the assembly of macromolecular structures in the nucleus: the “induced stability” model
Current models of the functional organization of the cell nucleus mainly consider short-lived interactions between freely diffusing components (Misteli 2001b, 2007). Undoubtedly, these events play crucial roles in gene regulation and genome maintenance (Darzacq et al. 2005; Lukas et al. 2005; Dinant et al. 2009; Hübner and Spector 2010). However, the observation of many stable interactions in the nucleus (ESM 1), even at transcription start sites on chromatin (TFIIH, HSF, Gal4, see above), leads us to propose an extension of the model for the formation of macromolecular complexes in the cell nucleus (Fig. 6).
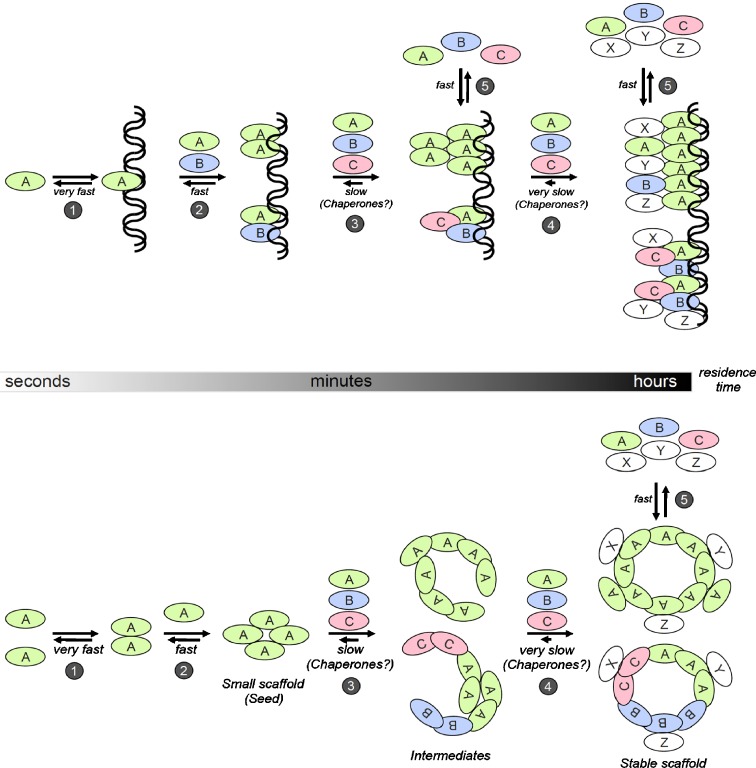
Fig. 6.
Model for the assembly of macromolecular complexes in the cell nucleus. a Formation of a chromatin-binding protein complex. Protein A binds transiently to DNA with very short residence times (1). By dimer formation or interaction with a second DNA-binding protein the overall residence time increases (2). Chromatin-association may be further increased by homo- or hetero-multimerization leading to various degrees of stability of the complex (3 and 4). The assembly of these complexes may be supported by chaperones. At these more or less stable protein/chromatin complexes, further binding partners (X, Y, Z) but also sub-fractions of the endogenous components (A, B, C) may exchange with short residence times (5). b Formation of a macromolecular protein complex. Protein A with homo-oligomerization properties assembles into small scaffolds (1, 2) which may serve as seeds for attachment of additional binding partners (3). The resulting intermediates may consist of the initial scaffolding protein only or may also contain additional components with scaffolding properties (B, C). Stable scaffolds may arise through accumulation of additional binding partners (4). The assembly of intermediates and stable scaffolds may be supported by chaperones. Stable scaffolds acquire functional properties by transient retention of a specific set of nuclear proteins (X, Y, Z) (5). Even a subpopulation of the scaffold components may exchange rapidly at the macromolecular structure depending on the number and strength of binding events upon contact (5)
The revised model is based on initial rapid binding and unbinding of proteins on DNA. Subsequent formation of dimers or multimers or interactions with additional DNA-binding proteins can induce an increase in the overall residence time of the complex on chromatin (Fig. 6). A similar effect may be realized by cooperative binding of several DNA-binding domains within the same protein, as recently demonstrated for linker histones (Stasevich et al. 2010). The more stable protein/chromatin complex may now serve as a platform for further binding partners. As the complex “grows,” the binding partners may increase their residence time on chromatin through multiplication of binding contacts. Chaperones may support the addition/incorporation of specific components as has been shown for the centromere complex (Foltz et al. 2009; Shuaib et al. 2010) or nucleosome (dis)assembly (De Koning et al. 2007; Park and Luger 2008a, b). Complex-induced alterations in chromatin structure may also contribute to increased stability (not shown in the model). All components, including the initial proteins, may exchange with short residence times at the stable core (Fig. 6a), as in chemical self-organizing systems (Aniansson and Wall 1974; Diekmann 1979). This model describes active transcription, replication, and repair complexes but also heterochromatin, cohesin, telomere, and centromere assembly.
An analogous model may also hold for the assembly of macromolecular protein complexes in the interchromatin space. A protein with homo-oligomerization properties, or interactions between different proteins, can lead to the assembly into small scaffolds onto which additional binding partners or further scaffolding proteins may attach (Fig. 6). The accumulation of additional binding partners may lead to larger, more stable scaffolds. These larger structures could acquire functional properties by transient retention of a specific set of nuclear proteins. As above, a subpopulation of scaffold components may exchange rapidly at the macromolecular structure depending on number and strength of binding events upon contact (Fig. 6). This model likely holds for the assembly of PML nuclear bodies, Cajal bodies, snRNPs, speckles, pre-nucleolar bodies, and the nuclear envelope including nuclear pore complexes. Chaperones may also be involved in the assembly process. For example, the survival of motor neurons complex can act as a chaperone for the assembly of snRNPs (Battle et al. 2006), although in vitro U1-snRNPs self-assemble (Hamm et al. 1987). Unlike Cajal bodies where most, if not all components are part of a large interaction network capable of Cajal body induction (Kaiser et al. 2008), formation of PML nuclear bodies require PML protein since typical PML body components are disperse in PML-negative cells (Ishov et al. 1999). Even quantitative expression of Sp100 failed to recruit PML body components to Sp100 nuclear bodies in PML-negative cells (Schwanitz and Hemmerich, unpublished observations).
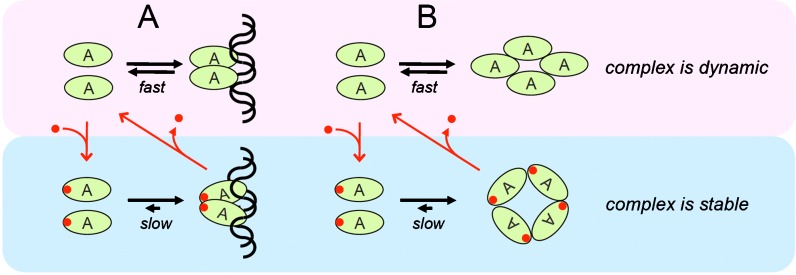
The formation of stable complexes also raises problems. Immobilization requires tight control because it can lead to “pathological immobility” as shown for nuclear inclusions seeded by the polyQ proteins ataxins or huntingtin, which immobilize not only themselves but also important nuclear proteins, such as CBP (Chai et al. 2002; von Mikecz 2009). Therefore, the models described above raise the fundamental question how the stability of a complex is controlled. The regulation of the stability of a complex in the nucleus may be conducted through posttranslational modifications or a “stabilizing” factor (Fig. 7). Such alterations of the complex may be regulated in a cell cycle-dependent manner as proposed for many components of the centromere/kinetochore complex (Hemmerich et al. 2008). We, therefore, propose an “induced stability” mechanism which allows controlled switching between dynamic and stable complex formation (Fig. 7).
Fig. 7.
Switching between dynamic and stable interactions: induced stability. The binding of proteins to chromatin (a) or the interaction between proteins (b) usually occurs dynamically. A posttranslational modification or binding of a “stabilizing” factor (red dot) may induce high-affinity interactions that allow stable complex formation
Self-assembly, self-organization, and assisted assembly in the nucleus
Are the slowly recovering FRAPs for some nuclear components evidence for self-assembled nuclear structures? Self-assembly and self-organization are the main driving forces to create living systems from single molecules (Kirschner et al. 2000; Fraden and Kamien 2000; Gabora 2006; Pelesko 2007; Karsenti 2008). With respect to nuclear organization and function, self-organization was proposed to be the major underlying mechanism (Misteli 2001a, b, 2009; Matera et al. 2009; Rajapakse et al. 2010). Self-assembly and self-organization are often used interchangeably and sometimes in association. In current literature, the boundary between these two mechanisms is rather elusive and elastic (Bensaude-Vincent 2009).
Self-assembly implies spatial structuring as a result of free energy minimization in a closed system (John and Bär 2005; also called “static self-assembly,” Pelesko 2007). Thus, self-assembly drives the system towards the thermodynamic equilibrium, as for example in phase separation of lipids or the stable assembly of virus particles from coat protein monomers (Makowski 1980). Tobacco mosaic virus can self-assemble in a test tube starting only with its basic protein and nucleic acid components (Fraenkel-Conrat and Williams 1955; Butler and Klug 1978). Also, the bacterial ribosome self-assembles. However, the large and the small subunit must be allowed to assemble separately first. Once both subunits have assembled in separate test tubes, they can be combined forming a fully functional ribosome (Nomura 1973). Self-assembly is a process in which components, either separate or linked, spontaneously form ordered aggregates (Whitesides and Boncheva 2004; Bensaude-Vincent 2009), although every single step is reversible. Nuclear structures that can be generated spontaneously by self-assembly in vitro include complete nuclei, the nuclear envelope, nuclear pore complexes, subcomplexes of the replication machinery, kinetochore subcomplexes, U1-snRNPs, NuMa scaffolds, prenucleolar bodies, and PML bodies (Table 1). In addition to the classical four forces (H-bonds, van der Waals forces, hydrophobic, and charge interactions), nonspecific (entropic) forces also influence these self-assembly processes. Entropic forces can induce the formation of human chromosome territories and position them appropriately in the nucleus (Finan et al. 2010). Since self-assembled structures form spontaneously and are stable, FRAP of their components might be expected to yield slow recoveries.
Table 1.
Evidence for self-assembly of nuclear structures
| Self-assembled nuclear structure | Biochemical assay | Reference |
|---|---|---|
| Nucleus-like structures | Xenopus cell free extract | Forbes et al. 1983; Zhao et al. 2000 |
| Nucleosome | Biochemically purified comonents | Ruiz-Carrillo et al. 1979 |
| Noll et al. 1980; Daban and Cantor 1982 | ||
| Nucleosome | Xenopus cell free extract | Newport 1987; Sheehan et al. 1988 |
| Chromatin fibers | Biochemical purification | Leforestier et al. 2001 |
| Mitotic chromosomes | Xenopus cell free extract | Lohka and Masui 1983 |
| Nuclear lamina | Biochemical purification | Aebi et al. 1986; Glass and Gerace 1990 |
| Nuclear envelope | Xenopus cell free extract | Newport 1987; Sheehan et al. 1988 |
| Nuclear envelope | CHO cell free extracts | Burke and Gerace 1986 |
| Nuclear pore complex | Xenopus cell free extract | Newport 1987; Sheehan et al. 1988 |
| Nuclear pore complex | Bacterially expressed components | Lutzmann et al. 2002 |
| Replication machinery | Xenopus cell free extract | Blow and Laskey 1986; Newport 1987 |
| Replication factor C | Bacterially expressed | Uhlmann et al. 1996 |
| Kinetochore subcomplexes | Bacterially expressed | Gestaut et al. 2010 |
| Dam1 complex | Purified components | Westermann et al. 2005 |
| Mis12 complex | Bacterially expressed components | Kline et al. 2006 |
| U1-snRNP | Xenopus cell free extract | Hamm et al. 1987 |
| NuMa scaffolds | Bacterially expressed | Harborth et al. 1999 |
| Prenucleolar bodies | Xenopus cell free extract | Bell et al. 1992 |
| PML-like bodies | Bacterially expressed | Kentsis et al. 2002 |
Self-organization requires a situation far away from thermodynamic equilibrium in a steady state and is possible only in open systems with an external energy source (Gerhart and Kirschner 1997; Toussaint and Schneider 1998; John and Bär 2005; Halley and Winkler 2008). Self-organization may also be regarded as “dynamic self-assembly” (Pelesko 2007). Self-organizing complexes and self-assembly processes can also be controlled efficiently either by posttranslational modifications or by chaperone action. The assembly of NuMa scaffolds in the nucleoplasm for example is tightly controlled by phosphorylation (Saredi et al. 1997). Similarly, nuclear lamina self-assembly is regulated by phosphorylation and methylation of lamins (Gerace and Blobel 1980; Chelsky et al. 1989). SUMO-attachment to PML (which harbors three SUMOylation sites) is a requirement for PML nuclear body assembly (Ishov et al. 1999; Duprez et al. 1999; Shen et al. 2006), and the number of SUMO modifications regulates the residence time of PML molecules at nuclear bodies, ranging from seconds to hours (Weidtkamp-Peters et al. 2008; Brand et al. 2010). Gradients are used in a number of biological systems to transmit spatial information over a range of distances. Intracellular gradients utilize intrinsic auto-regulatory feedback loops and diffusion to establish regions of activity, for example within the mitotic cytosol (reviewed in Fuller 2010). These intracellular gradients bear several hallmarks of self-organizing biological systems that designate spatial information during pattern formation. Since self-organized systems are dynamic, FRAP of their components might be expected to yield fast recoveries. However, our model for nuclear assembly discussed in the preceding section proposes that initially dynamic assemblies can become more stable over time or during specific phases of the cell cycle, yielding eventually slower FRAP recoveries for some components. Thus, slow FRAP recoveries could also arise from self-organization or a mixture of self-organization and self-assembly.
In biological systems, it is obviously very difficult to clearly separate self-assembly from self-organization. In vitro, self-assembly and self-organization can act in concert to create functional large molecule aggregates (Cuccia et al. 2002; Drain 2002). For example, purified DNA can be assembled in the test tube into structures that closely resemble cell nuclei. By combining naked DNA, cytosol, and light membrane fractions from Xenopus egg extracts, complete nuclei can be formed, including nuclear membranes with pore complexes, and these reconstituted nuclei are capable of normal nuclear processes (Laskey and Leno 1990; Cross and Powers 2008). The assembled nuclei are morphologically indistinguishable from normal eukaryotic nuclei. Nuclear assembly involves discrete intermediate steps, including nucleosome assembly, scaffold assembly, and nuclear envelope assembly, indicating that during reconstitution nuclear organization is assembled one level at a time (Newport 1987). The nuclear envelope contains functional, self-assembled nuclear pore complexes (Sheehan et al. 1988). After envelope assembly, these artificial nuclei are even capable of DNA replication (Newport 1987). Notably, nuclear lamina and envelope assembly in vitro do not require energy (Burke and Gerace 1986), a hallmark of the self-assembly process. The inherent ability of a primitive genome to induce its own inclusion in a membrane has even led to a model of the origin of eukaryotic cells solely based on self-assembly mechanisms (de Roos 2006).
We suggest that both self-assembly and self-organization contribute to the formation and function of nuclear networks (Table 2). The cellular thermodynamic ground state is dictated by minimizing the free energy of intracellular matrices based on self-assembly mechanisms (Fraden and Kamien 2000). Depletion attraction may support self-assembly. Depletion attraction occurs when aggregation of large complexes increases the entropy of the system through an increase of small crowding molecules within the system (Marenduzzo et al. 2006a). It has been demonstrated in vitro, that depletion attraction can be used to control the self-assembly of colloidal particles (Yodh et al. 2001). Theoretical approaches and experimental evidence suggest that the same mechanism helps to organize chromatin architecture in living cells (Marenduzzo et al. 2006b; Misteli 2009; Finan et al. 2010, this issue). Our proposal is supported by in silico analyses: When the formation of protein patterns near membranes of living cells is analyzed by mathematical modeling, self-assembly and self-organization eventually lead to similar patterns (John and Bär 2005). However, their evolution occurs on different length and time scales. Self-assembly produced periodic protein accumulations below 0.1 μm in a few seconds followed by extremely slow coarsening, whereas self-organization resulted in a pattern of 100 μm within a few minutes (John and Bär 2005). These observations support a model in which both processes contribute to particular nuclear functions depending on size and required activity. Self-assembly is required for the formation of kinetochore subcomplexes (Kline et al. 2006; Gestaut et al. 2010), which are then incorporated into the centromere by self-organization (Westermann et al. 2005). Similarly, nucleoporins self-assemble to pre-complexes before stable incorporation into the nuclear pore (Lutzmann et al. 2002). Self-assembly in living cells may also be crucial to create localized sites of specific enzymatic activity. For example, enzyme self-assembly is observed in vitro (Gao et al. 2010) as well as in vivo (Noree et al. 2010). Self-assembly may, therefore, control enzymatic activity depending on the metabolic status. This may hold for PML nuclear bodies at which SUMO and ubiquitin pathways locally and, therefore, probably efficiently merge in the nucleus (Häkli et al. 2005; Lallemand-Breitenbach et al. 2008; Sharma et al. 2010).
Table 2.
Potential assembly mechanisms in living cells
| Self-assembly | Self-organization | Assisted assembly |
|---|---|---|
| Nuclear lamina | Chromosome territory | Nucleosome |
| Nuclear pore complex | DNA Replication sites | Centromere |
| Nuclear envelope | DNA Repair sites | |
| PML nuclear bodies | Transcription sites | |
| Prenucleolar bodies | Heterochromatin | |
| snRNP pre-complexes | Telomere | |
| Centromere pre-complexes | Nucleolus | |
| NuMa | Cajal bodies | |
| Speckles |
For essential cellular processes which require precise timing at well-defined locations, stochastic self-assembly and self-organizing reactions might not be sufficiently effective. In these cases, assisting or helper molecules seem to be required to secure function. Nucleosomes spontaneously self-assemble in vitro from biochemically purified components (Ruiz-Carrillo et al. 1979). The in vivo assembly of nucleosomes (Corpet and Almouzni 2009) and the (re)positioning of nucleosomes at transcriptional start sites and gene boundaries (Mito et al. 2007; Dion et al. 2007) are regulated by chaperones, loading factors, chromatin re-modeling enzymes, and histone modifiers (Fig. 8) (Park and Luger 2008a, b). Recent research dedicated to the question how and when histone variants are deposited into nucleosomes has for example revealed that Cenp-A incorporation into centromeric chromatin requires specific loading factors (Furuyama et al. 2006; Shuaib et al. 2010). In addition, some processes like the transport of larger proteins through nuclear pores will not happen stochastically but require molecular support by helper molecules like importins (Harel and Forbes 2004). Being mechanistically overlapping but clearly different from self-assembly and self-organization we would like to introduce the term “assisted assembly” for those complex formation processes in the nucleus that rely on helper molecules (Table 2). This model, of course, raises fundamental questions: When do helper molecules attach to their substrate during the cell cycle? Do they interact in the nucleoplasm or at the complex where the substrate will be incorporated in? Will depletion of the helper molecule destabilize the complex? The first two questions will be answered by fluorescence fluctuation microscopy of GFP-tagged chaperones in the future. The third question has been solved already for Cenp-A. At least one of its chaperones, HJURP, only appears at the centromere in early G1 when Cenp-A is incorporated into centromeric chromatin. Depletion of HJURP by siRNA abrogates Cenp-A incorporation, leading to kinetochore malfunction (Dunleavy et al. 2009; Foltz et al. 2009; Shuaib et al. 2010).
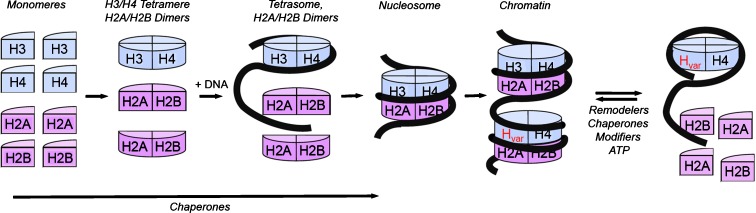
Fig. 8.
Nucleosome dynamics based on “assisted assembly.” During replication nascent DNA is loaded with histones into nucleosomes. The first step involves formation of an H3/H4 tetramer and two H2A/H2B dimers. In the second step ca. 80 bp of DNA stably associates with the H3/H4 tetramer together forming the tetrasome. Nucleosome assembly is completed when two H2A/H2B dimers combine with the tetrasome and ca. 40 bp of DNA (Ruiz-Carrillo et al. 1979). Adjacent nucleosomes then self-assemble to form chromatin (Noll et al. 1980). The rigid structure of the nucleosome needs to be dissociated when active biochemistry, such as transcription, repair or replication occurs, or if variant histones (H var) need to be incorporated. These processes require remodeling enzymes, chaperones, modifiers, and energy (ATP)
Conclusions
Chromatin-binding proteins are highly dynamic; they roam the nucleus in an energy-independent manner in search for high-affinity binding sites, and their residence times on chromatin are typically in the order of several seconds. This dynamic behavior is thought to play a major role in generating combinatorial protein complexes on chromatin which provide a mechanism to fine-regulate transcription, chromatin organization, and genomic plasticity (Misteli 2001a, b, 2008). By analyzing FRAP data of about 264 nuclear proteins, we identified nuclear factors that do not follow this rule of highly transient interactions, instead some of them exhibit residence times in the hours range (Fig. 5). Thus, although dynamic transient interactions are a general and important property of nuclear proteins, they are not universal. Stable complexes may arise from dynamic ones through a mechanism we coined “induced stability.” The demonstration in living cells of two proteins (HA95 and lamin B) which are almost completely immobile throughout the nucleoplasm may even reanimate research into the nuclear matrix. In particular cases, helper proteins support correct complex assembly, a phenomenon we have termed “assisted assembly.” The idea of immobile nuclear entities in the nucleus was already put forward by Bazett-Jones and colleagues in 2001 (Hendzel et al. 2001). The combined action of self-assembly and self-organization processes may lead to nuclear structures which are stable for hours. It should be considered in the future that stable interactions also contribute to the outcome of physiological cues impacting on nuclear structure and function. Therefore, “immobile populations” inferred from FRAP experiments should be studied in more detail. More examples of stable nuclear protein networks may surface as the community continues to perform fluorescence fluctuation microscopy on hitherto unexplored nuclear components.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
(XLS 242 kb)
Acknowledgements
Research in the labs of PH and SD are supported by the DFG and the BMBF. We would like to thank Karolin Klement and Sandra Münch for FISH images of chromosome territory 1 and anti-telomere immunfluorescence data, respectively. We apologize to all researchers whose FRAP data on nuclear proteins were not considered in this paper but strongly encourage them to communicate their data to us in order to complete and extend the FRAP data table on nuclear proteins started here (ESM 1).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- CB
Cajal body
- CBP
C-AMP response element binding protein (CREB) binding protein
- CT
Chromosome territory
- CP
Continuous fluorescence photobleaching
- FFM
Fluorescence fluctuation microscopy
- FLIM
Fluorescence lifetime imaging
- FLIP
Fluorescence loss in photobleaching
- FRAP
Fluorescence recovery after photobleaching
- FRET
Förster resonance energy transfer
- FCS
Fluorescence correlation spectroscopy
- Gem
Gemini body
- GFP
Green fluorescent protein
- HA95
Homologous to A-kinase anchoring protein 95
- HSF
Heat shock factor
- iFRAP
Inverse fluorescence recovery after photobleaching
- LAP
Lamin-associated polypeptides
- LBR
Lamin B receptor
- MFIS
Multiparameter fluorescence lifetime image spectroscopy
- NER
Nucleotide excision repair
- PcG proteins
Polycomb group proteins
- PCNA
Proliferating cell nuclear antigen
- pRB
Retinoblastoma protein
- PML
Promyelocytic leukemia
- RICS
Raster image correlation spectroscopy
- snRNP
Small ribonucleoprotein particle
- SPT
Single particle tracking
Contributor Information
Peter Hemmerich, Email: phemmer@fli-leibniz.de.
Stephan Diekmann, Email: diekmann@fli-leibniz.de.
References
- Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Angus SP, Solomon DA, Kuschel L, Hennigan RF, Knudsen ES. Retinoblastoma tumor suppressor: analyses of dynamic behavior in living cells reveal multiple modes of regulation. Mol Cell Biol. 2003;23:8172–8188. doi: 10.1128/MCB.23.22.8172-8188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniansson EAG, Wall SN. Kinetics of step-wise micelle association. J Phys Chem. 1974;78:1024–1030. [Google Scholar]
- Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- Bancaud A, Huet S, Rabut G, Ellenberg J. Fluorescence perturbation techniques to study mobility and molecular dynamics of proteins in live cells: FRAP, photoactivation, photoconversion, and FLIP. In: Swedlow J, Goldman R, Spector D, editors. Live cell imaging: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Press; 2009. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- Bell P, Dabauvalle MC, Scheer U. In vitro assembly of prenucleolar bodies in Xenopus egg extract. J Cell Biol. 1992;118:1297–1304. doi: 10.1083/jcb.118.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude-Vincent B. Self-assembly, self-organization: nanotechnology and vitalism. Nanoethics. 2009;3:31–42. [Google Scholar]
- Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60:1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Brand P, Lenser T, Hemmerich P. Assembly dynamics of PML nuclear bodies in living cells. PMC Biophysics. 2010;3:3. doi: 10.1186/1757-5036-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Gerace L. A cell free system to study reassembly of the nuclear envelope at the end of mitosis. Cell. 1986;44:639–652. doi: 10.1016/0092-8674(86)90273-4. [DOI] [PubMed] [Google Scholar]
- Buscaino A, Allshire R, Pidoux A. Building centromeres: home sweet home or a nomadic existence? Curr Opin Genet Dev. 2010;20:118–126. doi: 10.1016/j.gde.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Butler PJG, Klug A. The assembly of a virus. Sci Am. 1978;242:62–69. doi: 10.1038/scientificamerican1178-62. [DOI] [PubMed] [Google Scholar]
- Chai Y, Shao J, Miller VM, Williams A, Paulson HL. Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc Natl Acad Sci USA. 2002;99:9310–9315. doi: 10.1073/pnas.152101299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D, Sobotka C, O’Neill CL. Lamin B methylation and assembly into the nuclear envelope. J Biol Chem. 1989;264:7637–7643. [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- Cook PR. A model for all genomes: the role of transcription factories. J Mol Biol. 2010;395:1–10. doi: 10.1016/j.jmb.2009.10.031. [DOI] [PubMed] [Google Scholar]
- Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2009;19:29–41. doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories - a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Cross M, Powers M. In vitro nuclear assembly using fractionated xenopus egg extracts. J Vis Exp. 2008;19:908. doi: 10.3791/908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccia LA, Ruiz E, Lehn JM, Homo JC, Schmutz M. Helical self-organization and hierarchical self-assembly of an oligoheterocyclic pyridine-pyridazine strand into extended supramolecular fibers. Chemistry. 2002;8:3448–3457. doi: 10.1002/1521-3765(20020802)8:15<3448::AID-CHEM3448>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Daban JR, Cantor CR. Structural and kinetic study of the self-assembly of nucleosome core particles. J Mol Biol. 1982;156:749–769. doi: 10.1016/0022-2836(82)90140-1. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Singer RH, Shav-Tal Y. Dynamics of transcription and mRNA export. Curr Opin Cell Biol. 2005;17:332–339. doi: 10.1016/j.ceb.2005.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- de Roos AD. The origin of the eukaryotic cell based on conservation of existing interfaces. Artif Life. 2006;12:513–523. doi: 10.1162/artl.2006.12.4.513. [DOI] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG. Dynamics of coilin in Cajal bodies of the Xenopus germinal vesicle. Proc Natl Acad Sci USA. 2004;101:4810–4814. doi: 10.1073/pnas.0401106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S. On the micellation kinetics of sodium-docylsulphate. Ber Bunsenges Phys Chem. 1979;83:528–532. [Google Scholar]
- Diekmann S, Hemmerich P, editors. Visions of the cell nucleus. Los Angeles: American Scientific; 2005. [Google Scholar]
- Digman MA, Gratton E. Analysis of diffusion and binding in cells using the RICS approach. Microsc Res Tech. 2009;72:323–332. doi: 10.1002/jemt.20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C, Luijsterburg MS, Höfer T, von Bornstaedt G, Vermeulen W, Houtsmuller AB, van Driel R. Assembly of multiprotein complexes that control genome function. J Cell Biol. 2009;185:21–26. doi: 10.1083/jcb.200811080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Drain CM. Self-organization of self-assembled photonic materials into functional devices: photo-switched conductors. PNAS. 2002;99:5178–5182. doi: 10.1073/pnas.062635099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de Thé H, Hay RT, Freemont PS. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci. 1999;112:381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- Erdel F, Müller-Ott K, Baum M, Wachsmuth M, Rippe K (2010) Dissecting chromatin interactions in living cells from protein mobility maps. Chrom Res doi:10.1007/s10577-010-9155-6 [DOI] [PubMed]
- Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers JH, Kanaar R. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, Kioussis D. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- Finan K, Cook PR, Marenduzzo D (2010) Non-specific (entropic) forces as major determinants of the structure of mammalian chromosomes. Chromsome Res doi:10.1007/s10577-010-9150-y [DOI] [PubMed]
- Fodor BD, Shukeir N, Reuter G, Jenuwein T. Mammalian Su(var) Genes in Chromatin Control. Annu Rev Cell Dev Biol. 2010;26:471–501. doi: 10.1146/annurev.cellbio.042308.113225. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, III, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes DJ, Kirschner MW, Newport JW. Spontaneous formation of nucleus-like structures around bacteriophage DNA microinjected into Xenopus eggs. Cell. 1983;34:13–23. doi: 10.1016/0092-8674(83)90132-0. [DOI] [PubMed] [Google Scholar]
- Fraden S, Kamien RD. Self-assembly in vivo. Biophys J. 2000;78:2189–2190. doi: 10.1016/S0006-3495(00)76767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel-Conrat H, Williams RC. Reconstruction of active Tobacco Mosaic Virus from its intrinsic protein and nucleic acids components. PNAS. 1955;41:690–698. doi: 10.1073/pnas.41.10.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol. 2001;154:499–509. doi: 10.1083/jcb.200105084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller BG. Self-organization of intracellular gradients during mitosis. Cell Div. 2010;5:5. doi: 10.1186/1747-1028-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci USA. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabora L. Self-other organization: why early life did not evolve through natural selection. J Theor Biol. 2006;241:443–450. doi: 10.1016/j.jtbi.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gao Y, Yang Z, Kuang Y, Ma ML, Li J, Zhao F, Xu B. Enzyme-instructed self-assembly of peptide derivatives to form nanofibers and hydrogels. Biopolymers. 2010;94:19–31. doi: 10.1002/bip.21321. [DOI] [PubMed] [Google Scholar]
- Gautier T, Abbott DW, Molla A, Verdel A, Ausio J, Dimitrov S. Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 2004;5:715–720. doi: 10.1038/sj.embor.7400182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L, Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. Cells, embryos, and evolution. Boston: Blackwell; 1997. [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- Gestaut DR, Cooper J, Asbury CL, Davis TN, Wordeman L. Reconstitution and functional analysis of kinetochore subcomplexes. Methods Cell Biol. 2010;95:641–656. doi: 10.1016/S0091-679X(10)95032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G, Theil AF, Mari PO, Mourgues S, Nonnekens J, Andrieux LO, de Wit J, Miquel C, Wijgers N, Maas A, Fousteri M, Hoeijmakers JHJ, Vermeulen W. Differentiation driven changes in the dynamic organization of basal transcription initiation. PLoS Biol. 2009;7(10):e1000220. doi: 10.1371/journal.pbio.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JR, Gerace L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J Cell Biol. 1990;111:1047–1057. doi: 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häkli M, Karvonen U, Jänne OA, Palvimo JJ. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp Cell Res. 2005;304:224–233. doi: 10.1016/j.yexcr.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Halley JD, Winkler DA. Consistent concepts of self-organization and self-assembly. Complexity. 2008;14:10–17. [Google Scholar]
- Hamm J, Kazmaier M, Mattaj IW. In vitro assembly of U1 snRNPs. EMBO J. 1987;6:3479–3485. doi: 10.1002/j.1460-2075.1987.tb02672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Handwerger KE, Murphy C, Gall JG. Steady-state dynamics of Cajal body components in the Xenopus germinal vesicle. J Cell Biol. 2003;160:495–504. doi: 10.1083/jcb.200212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J, Wang J, Gueth-Hallonet C, Weber K, Osborn M. Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 1999;18:1689–1700. doi: 10.1093/emboj/18.6.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Haustein E, Schwille P. Ultrasensitive investigations of biological systems by fluorescence correlation spectroscopy. Methods. 2003;29:153–166. doi: 10.1016/s1046-2023(02)00306-7. [DOI] [PubMed] [Google Scholar]
- Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19:311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Hellwig D, Münch S, Orthaus S, Hoischen C, Hemmerich P, Diekmann S. Live cell imaging reveals sustained centromere binding of CENP-T via CENP-A and CENP-B. J Biophotonics. 2008;1:254–254. doi: 10.1002/jbio.200810014. [DOI] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Kruhlak MJ, MacLean NA, Boisvert F, Lever MA, Bazett-Jones DP. Compartmentalization of regulatory proteins in the cell nucleus. J Steroid Biochem Mol Biol. 2001;76:9–21. doi: 10.1016/s0960-0760(00)00153-9. [DOI] [PubMed] [Google Scholar]
- Heun P, Taddei A, Gasser SM. From snapshots to moving pictures: new perspectives on nuclear organization. Trends Cell Biol. 2001;11:519–525. doi: 10.1016/s0962-8924(01)02174-2. [DOI] [PubMed] [Google Scholar]
- Houtsmuller AB, Rademakers S, Nigg AL, Hoogstraten D, Hoeijmakers JH, Vermeulen W. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 1999;284:958–961. doi: 10.1126/science.284.5416.958. [DOI] [PubMed] [Google Scholar]
- Hübner MR, Spector DL. Chromatin dynamics. Annu Rev Biophys. 2010;9:471–489. doi: 10.1146/annurev.biophys.093008.131348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, III, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John K, Bär M. Alternative mechanisms of structuring biomembranes: self-assembly vs. self-organization. Phys Rev Lett. 2005;95:198101–198105. doi: 10.1103/PhysRevLett.95.198101. [DOI] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Karsenti E. Self-organization in cell biology: a brief history. Nat Rev Mol Cell Biol. 2008;9:255–262. doi: 10.1038/nrm2357. [DOI] [PubMed] [Google Scholar]
- Kentsis A, Gordon RE, Borden KL. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc Natl Acad Sci USA. 2002;99:15404–15409. doi: 10.1073/pnas.202608799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Gerhart J, Mitchison T. Molecular “vitalism”. Cell. 2000;100:79–88. doi: 10.1016/s0092-8674(00)81685-2. [DOI] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Krouwels IM, Wiesmeijer K, Abraham TE, Molenaar C, Verwoerd NP, Tanke HJ, Dirks RW. A glue for heterochromatin maintenance: stable SUV39H1 binding to heterochromatin is reinforced by the SET domain. J Cell Biol. 2005;170:537–549. doi: 10.1083/jcb.200502154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvissel AK, Ørstavik S, Eikvar S, Brede G, Jahnsen T, Collas P, Akusjärvi G, Skålhegg BS. Involvement of the catalytic subunit of protein kinase A and of HA95 in pre-mRNA splicing. Exp Cell Res. 2007;313:2795–2809. doi: 10.1016/j.yexcr.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Laskey RA, Leno GH. Assembly of the cell nucleus. Trends Genet. 1990;6:406–410. doi: 10.1016/0168-9525(90)90301-l. [DOI] [PubMed] [Google Scholar]
- Leforestier A, Dubochet J, Livolant F. Bilayers of nucleosome core particles. Biophys J. 2001;81:2414–2421. doi: 10.1016/S0006-3495(01)75888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H, Sporbert A, Cardoso MC. Targeting regulatory factors to intranuclear replication sites. Crit Rev Eukaryot Gene Expr. 2000;10:127–133. [PubMed] [Google Scholar]
- Levi V, Gratton E. Chromatin dynamics during interphase explored by single-particle tracking. Chromosome Res. 2008;16:439–449. doi: 10.1007/s10577-008-1240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH. Photobleaching and photoactivation: following protein dynamics in living cells. Nat Cell Biol. 2003;2003:S7–S14. [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Goedhart J, Moser J, Kool H, Geverts B, Houtsmuller AB, Mullenders LH, Vermeulen W, van Driel R. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. J Cell Sci. 2007;120:2706–2716. doi: 10.1242/jcs.008367. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, von Bornstaedt G, Gourdin AM, Politi AZ, Moné MJ, Warmerdam DO, Goedhart J, Vermeulen W, van Driel R, Höfer T. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J Cell Biol. 2010;189:445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Bartek J, Lukas J. Imaging of protein movement induced by chromosomal breakage: tiny ‘local’ lesions pose great ‘global’ challenges. Chromosoma. 2005;114:146–154. doi: 10.1007/s00412-005-0011-y. [DOI] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L. Structural studies of the assembly of simple viruses. Prog Clin Biol Res. 1980;40:233–258. [PubMed] [Google Scholar]
- Marenduzzo D, Finan K, Cook PR. The depletion attraction: an underappreciated force driving cellular organization. J Cell Biol. 2006;175:681–686. doi: 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenduzzo D, Micheletti C, Cook PR. Entropy-driven genome organization. Biophys J. 2006;90:3712–3721. doi: 10.1529/biophysj.105.077685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins SB, Eide T, Steen RL, Jahnsen T, Skålhegg BS, Collas P. HA95 is a protein of the chromatin and nuclear matrix regulating nuclear envelope dynamics. J Cell Sci. 2000;21:3703–3713. doi: 10.1242/jcs.113.21.3703. [DOI] [PubMed] [Google Scholar]
- Martins S, Eikvar S, Furukawa K, Collas P. HA95 and LAP2 beta mediate a novel chromatin-nuclear envelope interaction implicated in initiation of DNA replication. J Cell Biol. 2003;160:177–188. doi: 10.1083/jcb.200210026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol. 2010;11:317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. Regulation of ubiquitin ligase dynamics by the nucleolus. J Cell Biol. 2005;170:733–744. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organisation in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Misteli T. Physiological importance of RNA and protein mobility in the cell nucleus. Histochem Cell Biol. 2008;129:5–11. doi: 10.1007/s00418-007-0355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Self-organization in the genome. Proc Natl Acad Sci USA. 2009;106:6885–6886. doi: 10.1073/pnas.0902010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34:3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O, Leonhardt H, Cardoso MC. Spatiotemporal dynamics of regulatory protein recruitment at DNA damage sites. J Cell Biochem. 2008;104:1562–1569. doi: 10.1002/jcb.21751. [DOI] [PubMed] [Google Scholar]
- Nalley K, Johnston SA, Kodadek T. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature. 2006;442:1054–1057. doi: 10.1038/nature05067. [DOI] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Noll M, Zimmer S, Engel A, Dubochet J. Self-assembly of single and closely spaced nucleosome core particles. Nucleic Acids Res. 1980;8:21–42. doi: 10.1093/nar/8.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M. Assembly of bacterial ribosomes. Science. 1973;179:864–873. doi: 10.1126/science.179.4076.864. [DOI] [PubMed] [Google Scholar]
- Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010;190:541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørstavik S, Eide T, Collas P, Han IO, Taskén K, Kieff E, Jahnsen T, Skålhegg BS. Identification, cloning and characterization of a novel nuclear protein, HA95, homologous to A-kinase anchoring protein 95. Biol Cell. 2000;92:27–37. doi: 10.1016/s0248-4900(00)88761-4. [DOI] [PubMed] [Google Scholar]
- Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Half a century of “the nuclear matrix”. Mol Biol Cell. 2000;11:799–805. doi: 10.1091/mbc.11.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelesko JA. Self assembly. Boca Raton: Chapman and Hall; 2007. [Google Scholar]
- Politi A, Moné MJ, Houtsmuller AB, Hoogstraten D, Vermeulen W, Heinrich R, van Driel R. Mathematical modeling of nucleotide excision repair reveals efficiency of sequential assembly strategies. Mol Cell. 2005;19:679–690. doi: 10.1016/j.molcel.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Poznic M. Retinoblastoma protein: a central processing unit. J Biosci. 2009;34:305–312. doi: 10.1007/s12038-009-0034-2. [DOI] [PubMed] [Google Scholar]
- Probst AV, Almouzni G. Pericentric heterochromatin: dynamic organization during early development in mammals. Differentiation. 2008;76:15–23. doi: 10.1111/j.1432-0436.2007.00220.x. [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Glover DM. The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet. 2009;43:439–465. doi: 10.1146/annurev-genet-102108-134310. [DOI] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rademakers S, Volker M, Hoogstraten D, Nigg AL, Moné MJ, Van Zeeland AA, Hoeijmakers JH, Houtsmuller AB, Vermeulen W. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol Cell Biol. 2003;23:5755–5767. doi: 10.1128/MCB.23.16.5755-5767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse I, Scalzo D, Tapscott SJ, Kosak ST, Groudine M. Networking the nucleus. Mol Syst Biol. 2010;6:395. doi: 10.1038/msb.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Richter K, Nessling M, Lichter P. Experimental evidence for the influence of molecular crowding on nuclear architecture. J Cell Sci. 2007;120:1673–1680. doi: 10.1242/jcs.03440. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A, Jorcano JL, Eder G, Lurz R. In vitro core particle and nucleosome assembly at physiological ionic strength. Proc Natl Acad Sci USA. 1979;76:3284–3288. doi: 10.1073/pnas.76.7.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiwaki T, Kotera I, Sasaki M, Takagi M, Yoneda Y. In vivo dynamics and kinetics of pKi-67: transition from a mobile to an immobile form at the onset of anaphase. Exp Cell Res. 2005;308:123–134. doi: 10.1016/j.yexcr.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Saredi A, Howard L, Compton DA. Phosphorylation regulates the assembly of NuMA in a mammalian mitotic extract. J Cell Sci. 1997;110:1287–1297. doi: 10.1242/jcs.110.11.1287. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu Hoerste G, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Murillas R, Zhang H, Kuehn MR. N4BP1 is a newly identified nucleolar protein that undergoes SUMO-regulated polyubiquitylation and proteasomal turnover at promyelocytic leukemia nuclear bodies. J Cell Sci. 2010;123:1227–1234. doi: 10.1242/jcs.060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Cardoso MC, Knudsen ES. Dynamic targeting of the replication machinery to sites of DNA damage. J Cell Biol. 2004;166:455–463. doi: 10.1083/jcb.200312048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Souza PP, Völkel P, Trinel D, Vandamme J, Rosnoblet C, Héliot L, Angrand PO. The histone methyltransferase SUV420H2 and Heterochromatin Proteins HP1 interact but show different dynamic behaviours. BMC Cell Biol. 2009;10:41. doi: 10.1186/1471-2121-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol Cell. 2002;10:1355–1365. doi: 10.1016/s1097-2765(02)00729-3. [DOI] [PubMed] [Google Scholar]
- Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich TJ, Mueller F, Brown DT, McNally JG. Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO J. 2010;29:1225–1234. doi: 10.1038/emboj.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok D, Ching RW, Bazett-Jones DP. PML nuclear bodies as sites of epigenetic regulation. Front Biosci. 2009;14:1325–1336. doi: 10.2741/3311. [DOI] [PubMed] [Google Scholar]
- Toussaint O, Schneider ED. The thermodynamics and evolution of complexity in biological systems. Comp Biochem Physiol A Mol Integr Physiol. 1998;120:3–9. doi: 10.1016/s1095-6433(98)10002-8. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Cai J, Flores-Rozas H, Dean FB, Finkelstein J, O’Donnell M, Hurwitz J. In vitro reconstitution of human replication factor C from its five subunits. Proc Natl Acad Sci USA. 1996;93:6521–6526. doi: 10.1073/pnas.93.13.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Royen J, Houtsmuller AB (2010) DNA binding kinetics of nuclear proteins. Chrom Res, in press
- von Mikecz A. PolyQ fibrillation in the cell nucleus: who’s bad? Trends Cell Biol. 2009;19:685–691. doi: 10.1016/j.tcb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Wachsmuth M, Weidemann T, Müller G, Hoffmann-Rohrer UW, Knoch TA, Waldeck W, Langowski J. Analyzing intracellular binding and diffusion with continuous fluorescence photobleaching. Biophys J. 2003;84:3353–3363. doi: 10.1016/S0006-3495(03)70059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P. Dynamics of component exchange at PML nuclear bodies. J Cell Sci. 2008;121:2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Felekyan S, Bleckmann A, Simon R, Becker W, Kühnemuth R, Seidel CA. Multiparameter fluorescence image spectroscopy to study molecular interactions. Photochem Photobiol Sci. 2009;8:470–480. doi: 10.1039/b903245m. [DOI] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Whitesides GM, Boncheva M. Beyond molecules: self-assembly of mesoscopic and macroscopic components. Proc Natl Acad Sci USA. 2004;99:4769–4774. doi: 10.1073/pnas.082065899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RW. An update on cohesin function as a ‘molecular glue’ on chromosomes and spindles. Cell Cycle. 2010;9:1754–1758. doi: 10.4161/cc.9.9.11806. [DOI] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Yodh AG, Lin KH, Crocker JC, Dinsmore AD, Verma R, Kaplan PD. Entropically driven self-assembly and interaction in suspension. Philos Trans R Soc Lond A. 2001;359:921–937. [Google Scholar]
- Zbarskii IB, Debov SS. On the proteins of the cell nucleus. Dokl Akad Nauk SSSR. 1948;63:795–798. [Google Scholar]
- Zhao Y, Liu X, Wu M, Tao W, Zhai Z. In vitro nuclear reconstitution could be induced in a plant cell-free system. FEBS Lett. 2000;480:208–122. doi: 10.1016/s0014-5793(00)01938-4. [DOI] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS 242 kb)