Abstract
Understanding causal relationships, or effective connectivity, between parts of the brain is of utmost importance because a large part of the brain’s activity is thought to be internally generated and, hence, quantifying stimulus response relationships alone does not fully describe brain dynamics. Past efforts to determine effective connectivity mostly relied on model based approaches such as Granger causality or dynamic causal modeling. Transfer entropy (TE) is an alternative measure of effective connectivity based on information theory. TE does not require a model of the interaction and is inherently non-linear. We investigated the applicability of TE as a metric in a test for effective connectivity to electrophysiological data based on simulations and magnetoencephalography (MEG) recordings in a simple motor task. In particular, we demonstrate that TE improved the detectability of effective connectivity for non-linear interactions, and for sensor level MEG signals where linear methods are hampered by signal-cross-talk due to volume conduction.
Keywords: Information theory, Effective connectivity, Causality, Information transfer, Electroencephalography, Magnetoencephalography
Introduction
Science is about making predictions. To this aim scientists construct a theory of causal relationships between two observations. In neuroscience, one of the observations can often be manipulated at will, i.e. a stimulus in an experiment, and the second observation is measured, i.e. neuronal activity. If we can correctly predict the behavior of the second observation we have identified a causal relationship between stimulus and response. However, identifying causal relationships between stimuli and responses covers only part of neuronal dynamics—a large part of the brain’s activity is internally generated and contributes to the response variability that is observed despite constant stimuli (Arieli et al. 1996). For the case of internally generated dynamics it is rather difficult to infer a physical causality because a deliberate manipulation of this aspect of the system is extremely difficult. Nevertheless, we can try to make predictions based on the concept of causality as it was introduced by Wiener (1956). In Wiener’s definition an improvement of the prediction of the future of a time series X by the incorporation of information from the past of a second time series Y is seen as an indication of a causal interaction from Y to X. Such causal interactions across brain structures are also called ‘effective connectivty’ (Friston 1994) and they are thought to reveal the information flow associated to neuronal processing much more precisely than functional connectivity, which only reflects the statistical covariation of signals as typically revealed by cross-correlograms or coherency measures. Therefore, we must identify causal relationships between parts of the brain, be they single cells, cortical columns, or brain areas.
Various measures of causal relationships, or effective connectivity, exist. They can be divided into two large classes: those that quantify effective connectivity based on the abstract concept of information of random variables (e.g. Schreiber 2000), and those based on specific models of the processes generating the data. Methods in the latter class are most widely used to study effective connectivity in neuroscience, with Granger causality (GC, Granger 1969) and dynamic causal modeling (DCM, Friston et al. 2003) arguably being most popular. In the next two paragraphs we give a short overview over the data generation models in GC and DCM and their specific consequences so that the reader can appreciate the fundamental differences between these model based approaches and the information theoretic approach presented below:
Standard implementations of GC use a linear stochastic model for the intrinsic dynamics of the signal and a linear interaction.1 Therefore, GC is only well applicable when three prerequisites are met: (a) The interaction between the two units under observation has to be well approximated by a linear description, (b) the data have to have relatively low noise levels (see e.g. Nalatore et al. 2007), and (c) cross-talk between the measurements of the two signals of interest has to be low (Nolte et al. 2008). Frequency domain variants of GC such as the partial directed coherence or the directed transfer function fall in the same category (Pereda et al. 2005).
DCM assumes a bilinear state space model (BSSM). Thus, DCM covers non-linear interactions—at least partially. DCM requires knowledge about the input to the system, because this input is modeled as modulating the interactions between the parts of the system (Friston et al. 2003). DCM also requires a certain amount of a priori knowledge about the network of connectivities under investigation, because ultimately DCM compares the evidence for several competing a priori models with respect to the observed data. This a priori knowledge on the input to the system and on the potential connectivity may not always be available, e.g. in studies of the resting-state. Therefore, DCM may not be optimal for exploratory analyses.
Based on the merits and problems of the methods described in the last paragraph we may formulate four requirements that a new measure of effective connectivity must meet to be a useful addition to already established methods:
It should not require the a priori definition of the type of interaction, so that it is useful as a tool for exploratory investigations.
It should be able to detect frequently observed types of purely non-linear interactions. This is because strong non-linearities are observed across all levels of brain function, from the all-or none mechanism of action potential generation in neurons to non-linear psychometric functions, such as the power-law relationship in Weber’s law or the inverted-U relationship between arousal levels and response speeds described in the Yerkes-Dodson law (Yerkes and Dodson 1908).
It should detect effective connectivity even if there there is a wide distribution of interaction delays between the two signals, because signaling between brain areas may involve multiple pathways or transmission over various axons that connect two areas and that vary in their conduction delays (Swadlow and Waxman 1975; Swadlow et al. 1978).
It should be robust against linear cross-talk between signals. This is important for the analysis of data recorded with electro- or magnetoencephalography, that provide a large part of the available electrophysiological data today.
The fact that a potential new method should be as model free as possible naturally leads to the application of information theoretic techniques. Information theory (IT) sets a powerful framework for the quantification of information and communication (Shannon 1948). It is not surprising then that information theory also provides an ideal basis to precisely formulate causal hypotheses. In the next paragraph, we present the connection between the quantification of information and communication and Wiener’s definition of causal interactions (Wiener 1956) in more detail because of its importance for the justification of using IT methods in this work.
In the context of information theory, the key measure of information of a discrete2 random variable is its Shannon entropy (Shannon 1948; Reza 1994). This entropy quantifies the reduction of uncertainty obtained when one actually measures the value of the variable. On the other hand, Wiener’s definition of causal dependencies rests on an increase of prediction power. In particular, a signal X is said to cause a signal Y when the future of signal Y is better predicted by adding knowledge from the past and present of signal X than by using the present and past of Y alone (Wiener 1956). Therefore, if prediction enhancement can be associated to uncertainty reduction, it is expected that a causality measure would be naturally expressible in terms of information theoretic concepts.
First attempts to obtain model-free measures of the relationship between two random variables were based on mutual information (MI). MI quantifies the amount of information that can be obtained about a random variable by observing another. MI is based on probability distributions and is sensitive to second and all higher order correlations. Therefore, it does not rely on any specific model of the data. However, MI says little about causal relationships, because of its lack of directional and dynamical information: First, MI is symmetric under the exchange of signals. Thus, it cannot distinguish driver and response systems. And second, standard MI captures the amount of information that is shared by two signals. In contrast, a causal dependence is related to the information being exchanged rather than shared (for instance, due to a common drive of both signals by an external, third source). To obtain an asymmetric measure, delayed mutual information, i.e. MI between one of the signals and a lagged version of another has been proposed. Delayed MI results in an asymmetric measure and contains certain dynamical structure due to the time lag incorporated. Nevertheless, delayed mutual information has been pointed out to contain certain flaws such as problems due to a common history or shared information from a common input (Schreiber 2000).
A rigorous derivation of a Wiener causal measure within the information theoretic framework was published by Schreiber under the name of transfer entropy (Schreiber 2000). Assuming that the two time series of interest X = x t and Y = y t can be approximated by Markov processes, Schreiber proposed as a measure of causality to compute the deviation from the following generalized Markov condition
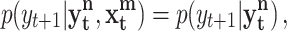 |
1 |
where 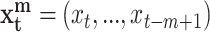 ,
, 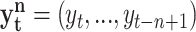 , while m and n are the orders (memory) of the Markov processes X and Y, respectively. Notice that Eq. 1 is fully satisfied when the transition probabilities or dynamics of Y is independent of the past of X, this is in the absence of causality from X to Y. To measure the departure from this condition (i.e. the presence of causality), Schreiber uses the expected Kullback-Leibler divergence between the two probability distributions at each side of Eq. 1 to define the transfer entropy from X to Y as
, while m and n are the orders (memory) of the Markov processes X and Y, respectively. Notice that Eq. 1 is fully satisfied when the transition probabilities or dynamics of Y is independent of the past of X, this is in the absence of causality from X to Y. To measure the departure from this condition (i.e. the presence of causality), Schreiber uses the expected Kullback-Leibler divergence between the two probability distributions at each side of Eq. 1 to define the transfer entropy from X to Y as
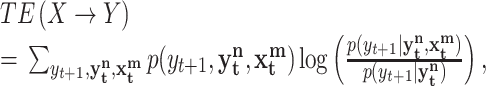 |
2 |
Transfer entropy naturally incorporates directional and dynamical information, because it is inherently asymmetric and based on transition probabilities. Interestingly, Paluš has shown that transfer entropy can be rewritten as a conditional mutual information (Paluš 2001; Hlavackova-Schindler et al. 2007).
The main convenience of such an information theoretic functional designed to detect causality is that, in principle, it does not assume any particular model for the interaction between the two systems of interest, as requested above. Thus, the sensitivity of transfer entropy to all order correlations becomes an advantage for exploratory analyses over GC or other model based approaches. This is particularly relevant when the detection of some unknown non-linear interactions is required.
Here, we demonstrate that transfer entropy does indeed fulfill the above requirements 1–4 and is therefore a useful addition to the available methods for the quantification of effective connectivity, when used as a metric in a suitable permutation test for independence. We demonstrate its ability to detect purely non-linear interactions, its ability to deal with a range of interaction delays, and its robustness against linear cross-talk on simulated data. This latter point is of particular interest for non-invasive human electrophysiology using EEG or MEG. The robustness of TE against linear cross-talk in the presence of noise, has to our knowledge not been investigated before. We test transfer entropy on a variety of simulated signals with different signal generation dynamics, including biologically plausible signals with spectra close to 1/f. We also investigate a range of linear and purely non-linear coupling mechanisms. In addition, we demonstrate that transfer entropy works without specifying a signal model, i.e. that requirement 1 is fulfilled. We extend earlier work (Hinrichs et al. 2008; Chávez et al. 2003; Gourvitch and Eggermont 2007) by explicitly demonstrating the applicability of transfer entropy for the case of linearly mixed signals.
Methods
The method section is organized in four main parts. In the first part we describe how to compute TE numerically. As several estimation techniques could be applied for this purpose we quickly review these possibilities and give the rationale for our particular choice of estimator. In the second part, we describe two particular problems that arise in neuroscience applications—delayed interactions, and observation of the signals of interest by measurements that only represent linear mixtures of these signals. The third part provides details on the simulation of test cases for the detection of effective connectivity via TE. The last part contains details of the MEG recordings in a self-paced finger-lifting task that we chose as a proof-of-concept for the analysis of neuroscience data.
Computation of transfer entropy
Transfer entropy for two observed time series x t and y t can be written as
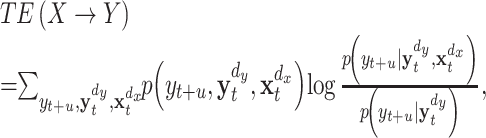 |
3 |
where t is a discrete valued time-index and u denotes the prediction time, a discrete valued time-interval. 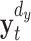 and
and 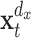 are d
x- and d
y-dimensional delay vectors as detailed below. An estimator of the transfer entropy can be obtained via different approaches (Hlavackova-Schindler et al. 2007). As with other information-theoretic functionals, any estimate shows biases and statistical errors which depend on the method used and the characteristics of the data (Hlavackova-Schindler et al. 2007; Kraskov et al. 2004). In some applications the magnitude of such errors is so large that it prevents any meaningful interpretation of the measure. To our purposes, it is crucial then to use a proper estimator that is as accurate as possible under the specific and severe constraints that most neuronal data-sets present and to complement it with an appropriate statistical test. In particular, a quantifier of transfer entropy apt for neuroscience applications should cope with at least three difficulties. First, the estimator should be robust to moderate levels of noise. Second, the estimator should rely only on a very limited number of data samples. This point is particularly restrictive since relevant neuronal dynamics typically unfolds over just a few hundred of milliseconds. And third, due to the need to reconstruct the state space from the observed signals, the estimator should be reliable when dealing with high-dimensional spaces. Under such restrictive conditions, to obtain a highly accurate estimator of TE is probably impossible without strong modelling assumptions. Unfortunately, strong modelling assumptions require specific information which is typically not available for neuroscience data. Nevertheless, some very general and biophysically motivated assumptions are available that enable the use of particular kernel-based estimators (Victor 2002). Here, we build on this framework to derive a data-efficient estimator, detailed below. Even using this improved estimator inaccuracies in estimation are unavoidable, specially for the restrictive conditions commented above, and it is necessary to evaluate the statistical significance of the TE measures, i.e. we use TE as a statistic measuring dependency of two time series and test against the null hypothesis of independent time series. Since no parametric distribution of errors is known for TE, one needs suitable surrogate data to test the null hypothesis of independent time series (‘absence of causality’). Suitable in this context means that the surrogate data should be prepared such that the causal dependency of interest is destroyed by constructing the surrogates but trivial dependencies of no interest are preserved. It is the particular combination of a data efficient estimator and a suitable statistical test that forms the core part of this study and its contribution to the field of effective connectivity analysis.
are d
x- and d
y-dimensional delay vectors as detailed below. An estimator of the transfer entropy can be obtained via different approaches (Hlavackova-Schindler et al. 2007). As with other information-theoretic functionals, any estimate shows biases and statistical errors which depend on the method used and the characteristics of the data (Hlavackova-Schindler et al. 2007; Kraskov et al. 2004). In some applications the magnitude of such errors is so large that it prevents any meaningful interpretation of the measure. To our purposes, it is crucial then to use a proper estimator that is as accurate as possible under the specific and severe constraints that most neuronal data-sets present and to complement it with an appropriate statistical test. In particular, a quantifier of transfer entropy apt for neuroscience applications should cope with at least three difficulties. First, the estimator should be robust to moderate levels of noise. Second, the estimator should rely only on a very limited number of data samples. This point is particularly restrictive since relevant neuronal dynamics typically unfolds over just a few hundred of milliseconds. And third, due to the need to reconstruct the state space from the observed signals, the estimator should be reliable when dealing with high-dimensional spaces. Under such restrictive conditions, to obtain a highly accurate estimator of TE is probably impossible without strong modelling assumptions. Unfortunately, strong modelling assumptions require specific information which is typically not available for neuroscience data. Nevertheless, some very general and biophysically motivated assumptions are available that enable the use of particular kernel-based estimators (Victor 2002). Here, we build on this framework to derive a data-efficient estimator, detailed below. Even using this improved estimator inaccuracies in estimation are unavoidable, specially for the restrictive conditions commented above, and it is necessary to evaluate the statistical significance of the TE measures, i.e. we use TE as a statistic measuring dependency of two time series and test against the null hypothesis of independent time series. Since no parametric distribution of errors is known for TE, one needs suitable surrogate data to test the null hypothesis of independent time series (‘absence of causality’). Suitable in this context means that the surrogate data should be prepared such that the causal dependency of interest is destroyed by constructing the surrogates but trivial dependencies of no interest are preserved. It is the particular combination of a data efficient estimator and a suitable statistical test that forms the core part of this study and its contribution to the field of effective connectivity analysis.
In the next subsection we detail both, how to obtain an data-efficient estimation of Eq. 3 from the raw signals, and a statistical significance analysis based on surrogate data.
Reconstructing the state space
Experimental recordings can only access a limited number of variables which are more or less related to the full state of the system of interest. However, sensible causality hypotheses are formulated in terms of the underlying systems rather than on the signals being actually measured. To partially overcome this problem several techniques are available to approximately reconstruct the full state space of a dynamical system from a single series of observations (Kantz and Schreiber 1997).
In this work, we use a Takens delay embedding (Takens 1981) to map our scalar time series into trajectories in a state space of possibly high dimension. The mapping uses delay-coordinates to create a set of vectors or points in a higher dimensional space according to
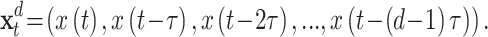 |
4 |
This procedure depends on two parameters, the dimension d and the delay τ of the embedding. While there is an extensive literature on how to choose such parameters, the different methods proposed are far away from reaching any consensus (Kantz and Schreiber 1997). A popular option is to take the delay embedding τ as the auto-correlation decay time (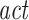 ) of the signal or the first minimum (if any) of the auto-information. To determine the embedding dimension, the Cao criterion offers an algorithm based on false neighbors computation (Cao 1997). However, alternatives for non-deterministic time-series are available (Ragwitz and Kantz 2002).
) of the signal or the first minimum (if any) of the auto-information. To determine the embedding dimension, the Cao criterion offers an algorithm based on false neighbors computation (Cao 1997). However, alternatives for non-deterministic time-series are available (Ragwitz and Kantz 2002).
The parameters d and τ considerably affect the outcome of the TE estimates. For instance, a low value of d can be insufficient to unfold the state space of a system and consequently degrade the meaning of any TE measure, as will be demonstrated below. On the other hand, a too large dimensionality makes the estimators less accurate for a given data length and significantly enlarges the computing time. Consequently, while we have used the recipes described above to orient our search for good embedding parameters, we have systematically scanned d and τ to optimize the performance of TE measures.
Estimating the transfer entropy
After having reconstructed the state spaces of any pair of time series, we are now in a position to estimate the transfer entropy between their underlying systems. We proceed by first rewriting Eq. (3) as sum of four Shannon entropies according to
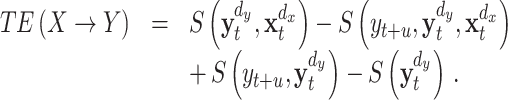 |
5 |
Thus, the problem amounts to computing the different joint and marginal probability distributions implicated in Eq. (5). In principle, there are many ways to estimate such probabilities and their performance strongly depends on the characteristics of the data to be analyzed. See Hlavackova-Schindler et al. (2007) for a detailed review of techniques. For discrete processes, the probabilities involved can be easily determined by the frequencies of visitation of different states. For continuous processes, the case of main interest in this study, a reliable estimation of the probability densities is much more delicate since a continuous density has to be approximated from a finite number of samples. Moreover, the solution of coarse-graining a continuous signal into discrete states is hard to interpret unless the measure converges when reducing the coarsening scale. In the following, we reason for our choice of the estimator and describe its functioning.
A possible strategy for the design of an estimator relies on finding the parameters that best fit the sample probability densities into some known distribution. While computationally straightforward such approach amounts to assuming a certain model for the probability distribution which without further constraints is difficult to justify. From the nonparametric approaches, fixed and adaptive histogram or partition methods are very popular and widely used. However, other nonparametric techniques such as kernel or nearest-neighbor estimators have been shown to be more data efficient and accurate while avoiding certain arbitrariness stemming from binning (Victor 2002; Kaiser and Schreiber 2002). In this work we shall use an estimator of the nearest-neighbor class.
Nearest-neighbor techniques estimate smooth probability densities from the distribution of distances of each sample point to its k-th nearest neighbor. Consequently, this procedure results in an adaptive resolution since the distance scale used changes according to the underlying density. Kozachenko-Leonenko (KL) is an example of such a class of estimators and a standard algorithm to compute Shannon entropy (Kozachenko and Leonenko 1987). Nevertheless, a naive approach of estimating TE via computing each term of Eq. 5 from a KL estimator is inadequate. To see why, it is important to notice that the probability densities involved in computing TE or MI can be of very different dimensionality (from 1 + d x up to 1 + d x + d y for the case of TE). For a fixed k, this means that different distance scales are effectively used for spaces of different dimension. Consequently, the biases of each Shannon entropy arising from the non-uniformity of the distribution will depend on the dimensionality of the space, and therefore, will not cancel each other.
To overcome such problems in mutual information estimates, Kraskov, Stögbauer, and Grassberger have proposed a new approach (Kraskov et al. 2004). The key idea is to use a fixed mass (k) only in the higher dimensional space and project the distance scale set by this mass into the lower dimensional spaces. Thus, the procedure designed for mutual information suggests to first determine the distances to k-th nearest neighbors in the joint space. Then, an estimator of MI can be obtained by counting the number of neighbors that fall within such distances for each point in the marginal space. The estimator of MI based on this method displays many good statistical properties, it greatly reduces the bias obtained with individual KL estimates, and it seems to become an exact estimator in the case of independent variables. For these reasons, in this work we have followed a similar scheme to provide an data-efficient sample estimate for transfer entropy (Gomez-Herrero et al. 2010). Thus, we have obtained an estimator that permits us, at least partially, to tackle some of the main difficulties faced in neuronal data sets mentioned in the beginning of the Methods section. In summary, since the estimator is more data efficient and accurate than other techniques (especially those based on binning), it allows to analyze shorter data sets possibly contaminated by small levels of noise. At the same time, the method is especially geared to handle the biases of high dimensional spaces naturally occurring after the embedding of raw signals.
As to computing time, this class of methods spends most of resources in finding neighbors. It is then highly advisable to implement an efficient search algorithm which is optimal for the length and dimensionality of the data to be analyzed (Cormen et al. 2001). For the current investigation, the algorithm was implemented with the help of OpenTSTool (Version1.2 on Linux 64 bit; Merkwirth et al. 2009). The full set of methods applied here is available as an open source MATLAB toolbox (Lindner et al. 2009).
In practice, it is important to consider that this kernel estimation method carries two parameters. One is the mass of the nearest-neighbors search (k) which controls the level of bias and statistical error of the estimate. For the remainder of this manuscript this parameter was set to k = 4, as suggested in Kraskov et al. (2004), unless stated otherwise. The second parameter refers to the Theiler correction which aims to exclude autocorrelation effects from the density estimation. It consists of discarding for the nearest-neighbor search those samples which are closer in time to a reference point than a given lapse (T). Here, we chose 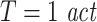 , unless stated otherwise. In general, it means that even though TE does not assume any particular model, its numerical estimation relies on at least five different parameters; the embedding delay (τ) and dimension (d), the mass of the nearest neighbor search (k), the Theiler correction window (T), and the prediction time (u). The latter accounts for non-instantaneous interactions. Specifically it reflects that in that case an increment of predictability of one signal thanks to the incorporation of the past of others should only occur for a certain latency or prediction time. Since axonal conduction delays among remote areas can amount to tens of milliseconds (Swadlow and Waxman 1975; Swadlow 1994), its incorporation for a sensible causality analysis of neuronal data sets is important for the results as we shall see below.
, unless stated otherwise. In general, it means that even though TE does not assume any particular model, its numerical estimation relies on at least five different parameters; the embedding delay (τ) and dimension (d), the mass of the nearest neighbor search (k), the Theiler correction window (T), and the prediction time (u). The latter accounts for non-instantaneous interactions. Specifically it reflects that in that case an increment of predictability of one signal thanks to the incorporation of the past of others should only occur for a certain latency or prediction time. Since axonal conduction delays among remote areas can amount to tens of milliseconds (Swadlow and Waxman 1975; Swadlow 1994), its incorporation for a sensible causality analysis of neuronal data sets is important for the results as we shall see below.
Significance analysis
To test the statistical significance of a value for TE obtained we used surrogate data. In general, generating surrogate data with the same statistical properties as the original data but selectively destroying any causal interaction is difficult. However, when the data set has a trial structure it is possible to reason that shuffling trials generates suitable surrogate data sets for the absence of causality hypothesis if stationarity and trial independency are assured. On these data we have then used a permutation test (~19,000 permutations) on the unshuffled and shuffled trials to obtain a p-value. P-values below 0.05 were considered significant. Where necessary a correction of this threshold for multiple comparisons was applied using the false discovery rate (FDR, q < 0.05; Genovese et al. 2002).
Particular problems in neuroscience data: instantaneous mixing and delayed interactions
Neuroscience data have specific characteristics that challenge a simple analysis of effective connectivity. First, the interaction may involve large time delays of unknown duration and, second, the data generated by the original processes may not be available but only measurements that represent linear mixtures of the original data—as is the case in EEG and MEG. In this section we describe a number of additional tests that may help to interpret the results obtained by computing TE values from these types of neuroscience data.
Tests for instantaneous linear mixing and for multiple noisy observations of a single source
Instantaneous, linear mixing of the original signals by the measurement process as is always present in MEG and EEG data. This may result in two problems: First, linear mixing may reduce signal asymmetry and, thus, make it more difficult to detect effective connectivity of the underlying sources. This problem is mainly one of reduced sensitivity of the method and maybe dealt with, e.g. by increasing the amount of data. A second problem arises when a single source signal with an internal memory structure is observed multiple times on different channels with individual channel noise. As demonstrated before (Nolte et al. 2008) this latter case can result in false positive detection of effective connectivity for methods based on Wiener’s definition of causality (Wiener 1956). This problem is more severe, because it reduces the specificity of the method. As an example of this problem think of an AR process of order m, s(t)
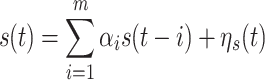 |
6 |
that is mixed with a mixing parameter ε onto two sensor signals X′,Y′ in the following way
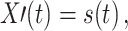 |
7 |
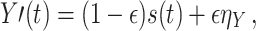 |
8 |
where the dynamics for Y′ can be rewritten as
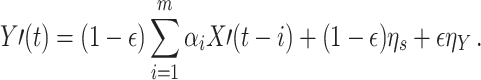 |
9 |
In this case TE will identify a causal relationship between X′ and Y′ as it detects the relationship between the past of X′ and the present X′ that is contained in Y′ as (1 − ε)η
s. Therefore, we implemented the following additional test (‘time-shift test’) to avoid false positive reports for the case of instantaneous, linear mixing: We shifted the time series for X′ by one sample into the past 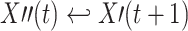 such that a potential instantaneous mixing becomes lagged and thereby causal in Wiener’s sense. For instantaneous mixing processes TE values increase for the interaction from the shifted time series X′′(t) to Y′ compared to the interaction from the original time series X′(t) to Y′. Therefore, an increase of this kind may indicate the presence of instantaneous mixing. The actual shift test implements the null hypothesis of instantaneous mixing and the alternative hypothesis of no instantaneous mixing in the following way:
such that a potential instantaneous mixing becomes lagged and thereby causal in Wiener’s sense. For instantaneous mixing processes TE values increase for the interaction from the shifted time series X′′(t) to Y′ compared to the interaction from the original time series X′(t) to Y′. Therefore, an increase of this kind may indicate the presence of instantaneous mixing. The actual shift test implements the null hypothesis of instantaneous mixing and the alternative hypothesis of no instantaneous mixing in the following way:
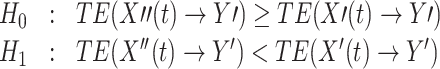 |
10 |
If the null hypothesis of instananeous mixing is not discarded by this test, i.e. if TE values for the original data are not significantly larger than those for the shifted data, then we have to discard the hypothesis of a causal interaction from X′ to Y′. Therefore, when data potentially contained instantaneous mixing, we tested for the presence of instantaneous mixing before proceeding to test the hypothesis of effective connectivity. More specifically, this test was applied for the instantaneously mixed simulation data (Figs. 4, 5, 6) and the MEG data (Fig. 8). In general, we suggest to use this test, whenever the data in question may have been obtained via a measurement function that contained linear, instantaneuos mixing.
Fig. 4.
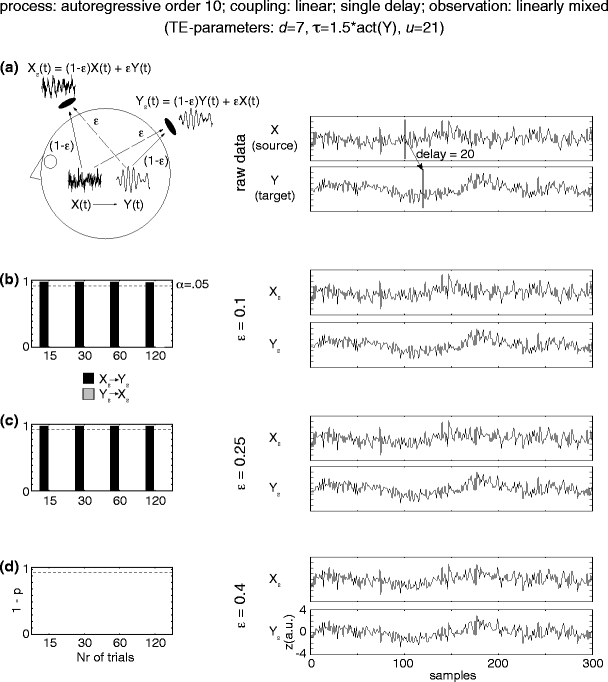
Simulation results for linearly mixed measurements (X ε, Y ε ) of two unidirectionally and linearly coupled underlying source signals (X →Y). (a) Mixing model and original autoregressive source time courses X, Y. (b–d) Effective connectivity between sensor-level signals X ε, Y ε. Left statistics of permutation tests of TE values for the original sensor level data against trial-shuffled surrogate data after application of the additional time-shift test. The plots contain values of (1-p) in dependence of the number of investigated number of trials. Black bars indicate values for the effective connectivity from the sensor dominated by the driving source signal (X ε) to the sensor dominated by the receiving source signal (Y ε). Light grey bars indicate the reverse direction of effective connectivity. The dashed line corresponds to siginificant effective connectivity (p < 0.05). Right time- courses of signals X ε and Y ε for a single trial
Fig. 5.
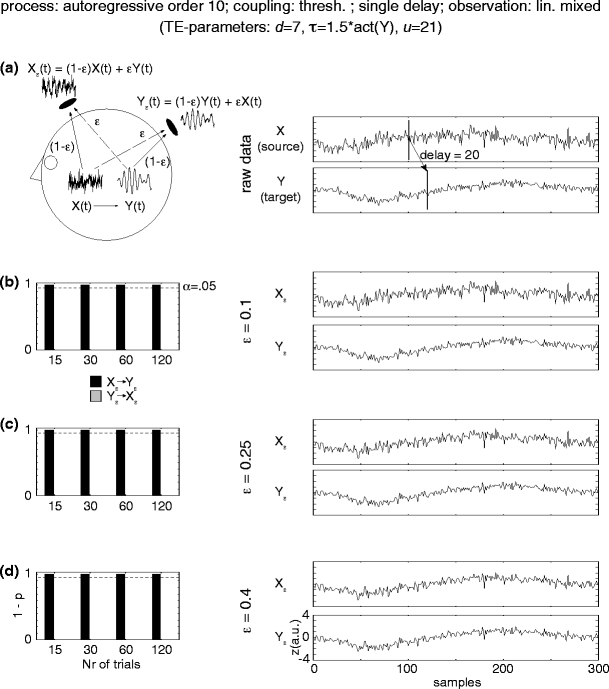
Simulation results for linearly mixed measurements (X ε, Y ε ) of two unidirectionally coupled underlying source signals (X →Y) coupled via a threshold function. (a) Mixing model and original autoregressive source time courses X, Y. (b–d) Effective connectivity between sensor-level signals X ε, Y ε. Left statistics of permutation tests of TE values for the original sensor level data against trial-shuffled surrogate data after application of the additional time-shift test. Black bars indicate values for the effective connectivity from the sensor dominated by the driving source signal (X ε) to the sensor dominated by the receiving source signal (Y ε). Light grey bars indicate the reverse direction of effective connectivity. The dashed line corresponds to siginificant effective connectivity (p < 0.05). Right time-courses of signals X ε and Y ε for a single trial
Fig. 6.
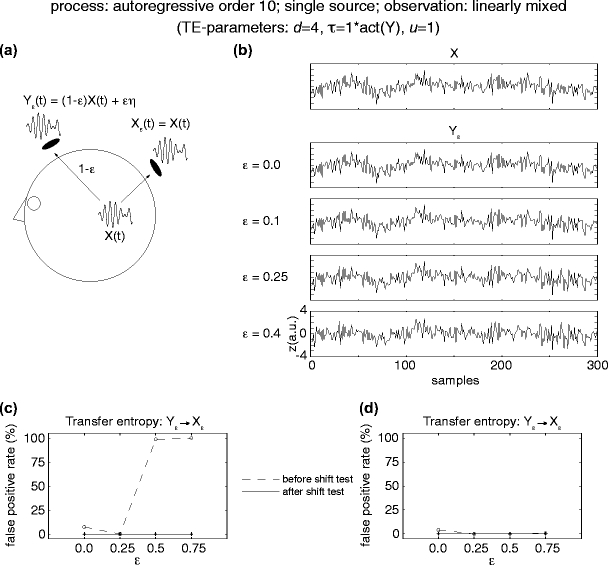
False positive rates for the detection of effective connectivity when observing one source via two EEG or MEG sensors. (a) Signal generation by an autoregressive order ten process X(t) and simultaneous observation of this source signal on two sensor signals X ε, Y ε. One of the signals is a copy of the source signal (X ε (t) = X(t)); the other, (Y ε), is dampened by a factor of (1 − ε) and corrupted by white noise εη. (b) Resulting signal time courses for the source signal X(t) and the observed sensor signals Y ε for different values of ε. (c) False positive detection rate for effective connectivity from the noise free sensor signal X ε to the noise corrupted signal Y ε before (dashed line) and after (solid line) the additional time-shift test for instantaneous mixing. In accordance with (Nolte et al. 2008) TE without the additional test yields a certain amount of false positive results. (d) False positive detection rate for effective connectivity from the noise corrupted signal Y ε to the noise free sensor signal X ε. Lines as in (c). No false positives were observed after the additional time shifting test
Fig. 8.
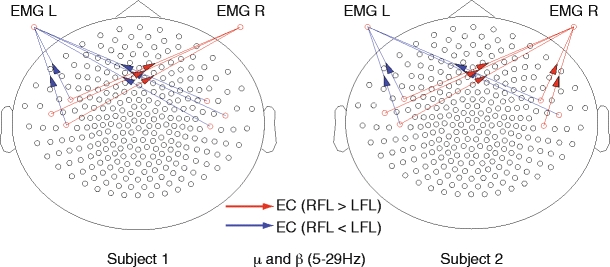
Differences in effective connectivity (EC) between lifting of the right (RFL) and left index finger (LFL) for subject 1 (left) and subject 2 (right). The investigated frequency band was 5–29 Hz encompassing the μ and β rhythms, and avoiding 50 Hz contamination. Red lines indicate links where effective connectivity as quantified by TE was significantly larger for lifting of the right index finger, compared to left. Blue lines indicate links where effective connectivity as quantified by TE was significantly larger for lifting of the left index finger, compared to right. Connectivity from contra- and ipsilateral motor cortices to muscles (EMG L, EMG R) of the moved finger is stronger than to the passive finger
A less conservative approach to the same problem would be to discard data for TE analysis only when we have significant evidence for the presence of instantaneous mixing. In this case the hypotheses would be:
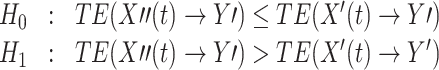 |
11 |
In this case we would proceed analysing the data if we did not have to reject H 0. For the remainder of this manuscript, however, we stick to testing the more conservative null hypothesis presented in Eq. (10).
Delayed interactions, Wiener’s definition of causality, and choice of embedding parameters
This paragraph introduces a difficulty related to Wiener’s definition of causality. As described above, non-zero TE values can be directly translated into improved predictions in Wiener’s sense by interpreting the terms in Eq. 2 as transition probabilities, i.e. as information that is useful for prediction. TE quantifies the gain in our knowledge about the transition probabilities in one system Y, that we obtain if we condition these probabilities on the past values of another system X. It is obvious that this gain, i.e. the value of TE, can be erroneously high, if the transition probabilities for system Y alone are not evaluated correctly. We now describe a case where this error is particularly likely to occur: Consider two processes with lagged interactions and long autocorrelation times. We assume that system X drives Y with an interaction delay δ (Fig. 1). A problem arises if we test for a causal interaction from Y to X, i.e. the reverse direction compared to the actual coupling, and do not take enough care to fully capture the dynamics of X via embedding. If for example the embedding dimension d or the embedding delay τ was chosen too small, then some information contained in the past of X is not used although it would improve (auto-) prediction. This information is actually transferred to Y via the delayed interaction from X to Y. It is available in Y with a delay δ, and therefore, at time-points were data from Y is used for the prediction of X. As stated before this information is useful for the prediction of X. Thus, inclusion of Y will improve prediction. Hence, TE values will be non-zero and we will wrongly conclude that process Y drives process X.
Fig. 1.
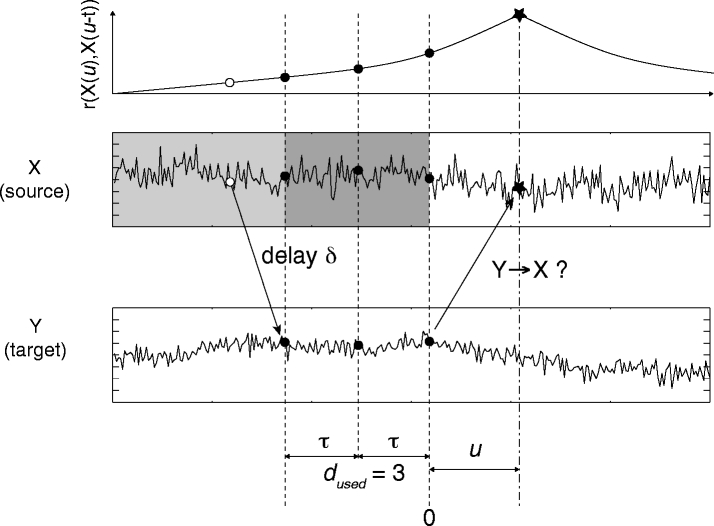
Illustration of false positive effective connectivity due to insufficient embedding for delayed interactions. Source signal X drives target signal Y with a delay δ. The internal memory of process X is reflected in the slowly decaying autocorrelation function (top). For the evaluation of TE from Y to X, X is embedded for auto-prediction with d = 3 and τ, as indicated by the dark gray box. The data point of X that is to be predicted with prediction time u is indicated by the star shaped symbol. Data points used for auto-prediction are indicated by filled circles on signal X. Data points used for cross-prediction from Y to X are indicated by filled circles on signal Y. Due to the delayed interaction from X to Y information about X earlier than the embedding time gets transferred from X to Y where it gets included in the embedding (open circle). Y contains information the history of X that is useful for predicting X (see open circle, autocorrelation curve) but not contained in the embedding used on X. Hence, inclusion of Y will improve the prediction of X and false positive effective connectivity is found. Introducing a larger embedding dimension or or larger embedding delay, incorporates this information into the embedding of X. Examples of this effect can be found in Tables 1 and 2
Simulated data
We used simulated data to test the ability of TE to uncover causal relations under different situations relevant to neuroscience applications. In particular, we always considered two interacting systems and simulated different internal dynamics (autoregressive and 1/f characteristics), effective connectivity (linear, threshold and quadratic coupling), and interaction delays (single delay and a distribution of delays). In addition, we simulated linear instantaneous mixing processes during measurement, because of their relevance for EEG and MEG.
Internal signal dynamics
We have simulated two types of complex internal signal dynamics. In the first case, an autoregressive process of order 10, AR(10), is generated for each system. The dynamics is then given by
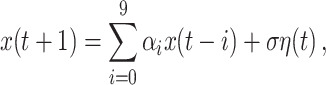 |
12 |
where the coefficients α i are drawn from a normalized Gaussian distribution, the innovation term η represents a Gaussian white noise source, and σ controls the relative strength of the noise contribution. Notice, that we use here the typical notation in dynamical systems where the innovation term η(t) is delayed one unit with respect the output x(t + 1).
As a second case, we have considered signals with a 1/f θ profile in their power spectra. To produce such signals we have followed the approach in Granger (1980). Accordingly, the 1/f θ time series are generated as the aggregation of numerous AR(1) processes with an appropriate distribution of coefficients. Mathematically, each 1/f θ signal is then given by
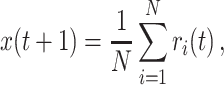 |
13 |
where we aggregate over N = 500 AR(1) processes each described as
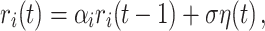 |
14 |
with the coefficients α
i randomly chosen according to the probability density function 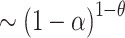 .
.
Types of interaction
To simulate a causal interaction between two systems we added to the internal dynamics of one process (Y) a term related to the past dynamics of the other (X). Three types of interaction or effective connectivity were considered; linear, quadratic, and threshold. In the linear case, the interaction is proportional to the amount of signal at X. The last two cases represent strong non-linearities which challenge approaches of detection based on linear or parametric methods. The effective connectivity mediated by the threshold function is of special relevance in neuroscience applications due to the approximated all or none character of the neuronal spike generation and transmission. Mathematically, the update of y(t) is then modeled by the addition of an interaction term such that the full dynamics is described as
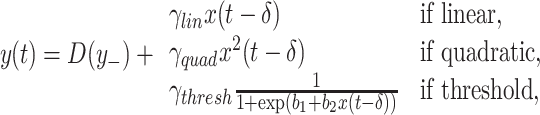 |
where D(.) represents the internal dynamics (AR(10) or 1/f) of y and y − represents past values of y. In the last case, the threshold function is implemented through a sigmoidal with parameters b 1 and b 2 which control the threshold level and its slope, respectively. Here, b 1 was set to 0 and b 2 was set to 50. In all cases, δ represents a delay which typically arises from the finite speed of propagation of any influence between physically separated systems. Note that since we deal with discrete time models (maps) in our modeling δ takes only positive integer values.
In case that two systems interact via multiple pathways it is possible that different latencies arise in their communication. For example, it is known that the different characteristics of the axons joining two brain areas typically lead to a distribution of axonal conduction delays (Swadlow et al. 1978; Swadlow 1985). To account for that scenario we have also simulated the case where δ instead of a single value is a distribution. Accordingly, for each type of interaction we have considered the case where the interaction term is
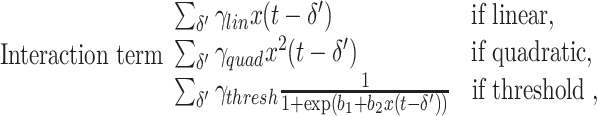 |
where the sums are extended over a certain domain of positive integer values. In the results section we consider the case in which δ′ takes values on a uniform distribution of width 6 centered around a given delay.
The coupling constants γ lin, γ quad, γ thresh were always chosen such that the variance of the interaction term was comparable to the variance of y(t) that would be obtained in the absence of any coupling.
Linear mixing
Linear instantaneous mixing is present in human non-invasive electrophysiological measurements such as EEG or MEG and has been shown to be problematic for GC (Nolte et al. 2008). The problem we encounter for linearly and instantaneously mixed signals is twofold: On the one hand, instantaneous mixing from coupled source signals onto sensor signals by the measurement process degrades signal asymmetry (Tognoli and Scott Kelso 2009), it will therefore be harder to detect effective connectivity. On the other hand—as shown in Nolte et al. (2008)—instantaneous presence of a single source signal in two measurements of different signal to noise ratio may be interpreted as effective connectivity erroneously. To test the influence of linear instantaneous mixing we created two test cases:
-
(A)The first test case consisted in unidirectionally coupled signal pairs X →Y generated from coupled AR(10) processes as described above and then transformed into two linear instantaneous mixtures X ε,Y ε in the following way:
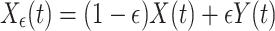
15
Here, ε is a parameter that describes the amount of linear mixing or ‘signal cross-talk’. A value of ε of 0.5 means that the mixing leads to two identical signals and, hence, no significant TE should be observed. We then investigated for three different values of ε = (0.1,0.25,0.4) how well TE detects the underlying effective connectivity from X to Y if only the linear mixtures X ε,Y ε are available.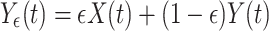
16 -
(B)The second test case consisted in generating measurement signals X ε,Y ε in the following way:
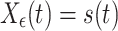
17 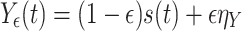
18 Here, s(t) is the common source, a mean-free AR(10) process with unit variance. s(t) is measured twice: once noise free in X ε and once dampened by a factor (1 − ε) and corrupted by independent Gaussian noise of unit variance, η Y, in Y ε. Here, we tested the ability of our implementation of TE to reject the hypothesis of effective connectivity. This second test case is of particular importance for the application of TE to EEG and MEG measurements where often a single source may be observed on two sensors that have different noise characteristics, i.e. due to differences in contact resistance of the EEG electrodes or the characteristics of the MEG-SQUIDS.
Choice of embedding parameters for delayed interactions
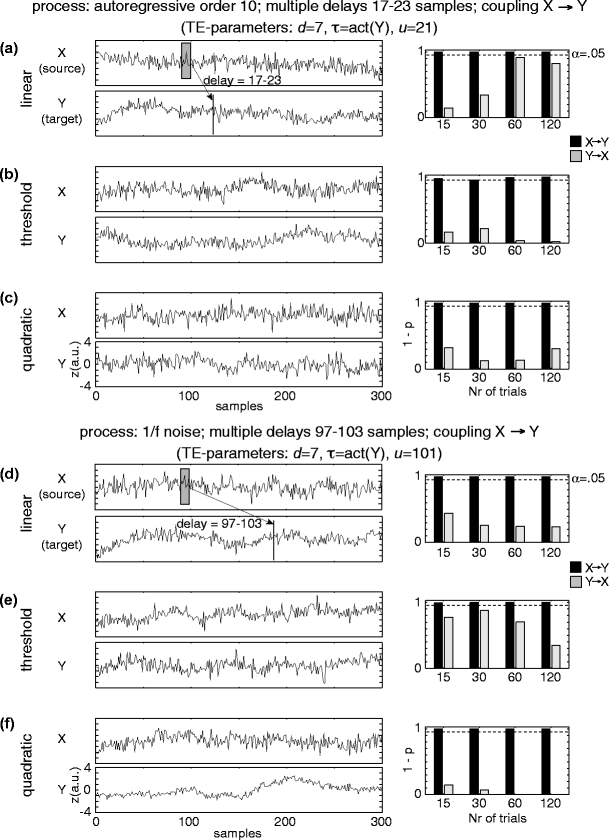
To demonstrate the effects of suboptimal embedding parameters for the case of delayed interactions we simulated processes with autoregressive order 10 (AR(10)) dynamics, three different interaction delays (5, 20, 100 samples) and all three coupling types (linear, threshold, quadratic). The two processes were coupled unidirectionally X →Y. 15, 30, 60, and 120 trials were simulated. We tested for effective connectivity in both possible directions using permutation testing. All coupled processes were investigated with three different prediction times u of 6, 21, and 101 samples. The remaining analysis parameters were: d = 7, 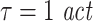 , k = 4,
, k = 4, 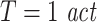 . In addition, we simulated processes with 1/f dynamics, an interaction delay δ of 100 samples and a unidirectional, quadratic coupling. 30 trials were simulated and we tested for effective connectivity in both directions. These coupled processes were investigated with all possible combinations of three different embedding dimensions d = 4, 7, 10, two different embedding delays
. In addition, we simulated processes with 1/f dynamics, an interaction delay δ of 100 samples and a unidirectional, quadratic coupling. 30 trials were simulated and we tested for effective connectivity in both directions. These coupled processes were investigated with all possible combinations of three different embedding dimensions d = 4, 7, 10, two different embedding delays 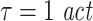 or
or 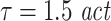 and three different prediction times u = 6, 21, 101 samples. The remaining analysis parameters were: k = 4,
and three different prediction times u = 6, 21, 101 samples. The remaining analysis parameters were: k = 4, 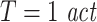 . Results are presented in Tables 1 and 2.
. Results are presented in Tables 1 and 2.
Table 1.
Detection of true and false effective connectivity for a fixed embedding dimension d of 7, and an embedding delay τ of 1 autocorrelation time
| Dynamics | δ | Coupling | u | X →Y | Y →X |
|---|---|---|---|---|---|
| True | False | ||||
| AR(10) | 5 | Lin | 6 | 1 | 1 |
| AR(10) | 5 | Lin | 21 | 1 | 0 |
| AR(10) | 5 | Lin | 101 | 0 | 0 |
| AR(10) | 5 | Threshold | 6 | 1 | 1 |
| AR(10) | 5 | Threshold | 21 | 1 | 0 |
| AR(10) | 5 | Threshold | 101 | 0 | 0 |
| AR(10) | 5 | Quadratic | 6 | 1 | 1 |
| AR(10) | 5 | Quadratic | 21 | 1 | 0 |
| AR(10) | 5 | Quadratic | 101 | 0 | 0 |
| AR(10) | 20 | Lin | 6 | 1 | 1 |
| AR(10) | 20 | Lin | 21 | 1 | 0 |
| AR(10) | 20 | Lin | 101 | 1 | 0 |
| AR(10) | 20 | Threshold | 6 | 0 | 0 |
| AR(10) | 20 | Threshold | 21 | 1 | 0 |
| AR(10) | 20 | Threshold | 101 | 0 | 0 |
| AR(10) | 20 | Quadratic | 6 | 0 | 0 |
| AR(10) | 20 | Quadratic | 21 | 1 | 0 |
| AR(10) | 20 | Quadratic | 101 | 0 | 0 |
| AR(10) | 100 | Lin | 6 | 1 | 0 |
| AR(10) | 100 | Lin | 21 | 1 | 0 |
| AR(10) | 100 | Lin | 101 | 1 | 0 |
| AR(10) | 100 | Threshold | 6 | 0 | 0 |
| AR(10) | 100 | Threshold | 21 | 0 | 0 |
| AR(10) | 100 | Threshold | 101 | 1 | 0 |
| AR(10) | 100 | Quadratic | 6 | 1 | 0 |
| AR(10) | 100 | Quadratic | 21 | 1 | 0 |
| AR(10) | 100 | Quadratic | 101 | 1 | 0 |
Given is the detected effective connectivity in dependence of the parameter prediction time u for data with different interaction delays δ of 5, 20, and 100 samples. Data were simulated with autoregressive order ten dynamics and unidirectional coupling X →Y via three different coupling functions (linear, threshold, quadratic). Simulation results based on 120 trials. Note: false positives emerge for short interaction delays δ, i.e. the inclusion of more recent samples of X, i.e. samples that are just before the earliest embedding time-point; false positives in these cases are suppressed using a larger prediction time, i.e. moving the embedding of X and the samples of X that are transferred to Y further into the past; short interaction delays can robustly be detected with prediction times that are longer than the interaction delay, if the difference is not excessive
Table 2.
Detection of true and false effective connectivity in dependence of the parameters embedding delay τ, embedding dimension d, and prediction time u for data with unidirectional coupling X →Y via a quadratic function, 1/f dynamics and an interaction delay δ of 100 samples
| Dynamics | δ | d | u | τ [ACT] | X →Y | Y →X |
|---|---|---|---|---|---|---|
| 1/f | 100 | 4 | 21 | 1 | 0 | 0 |
| 1/f | 100 | 4 | 101 | 1 | 1 | 1 |
| 1/f | 100 | 7 | 21 | 1 | 0 | 1 |
| 1/f | 100 | 7 | 101 | 1 | 1 | 0 |
| 1/f | 100 | 10 | 21 | 1 | 0 | 0 |
| 1/f | 100 | 10 | 101 | 1 | 1 | 0 |
| 1/f | 100 | 4 | 21 | 1.5 | 0 | 0 |
| 1/f | 100 | 4 | 101 | 1.5 | 1 | 0 |
| 1/f | 100 | 7 | 21 | 1.5 | 0 | 0 |
| 1/f | 100 | 7 | 101 | 1.5 | 1 | 0 |
| 1/f | 100 | 10 | 21 | 1.5 | 0 | 0 |
| 1/f | 100 | 10 | 101 | 1.5 | 1 | 0 |
Simulation results based on 30 trials. Note how a larger τ eliminates false positive TE results for effective connectivity. Also note how the delocalization in time provided by the embedding enables the detection of effective connectivities also for interaction delays larger than the prediction time
MEG experiment
Rationale
In order to demonstrate the applicability of TE to neuroscience data obtained non-invasively we performed MEG recordings in a motor task. Our aim was to show that TE indeed gave the results that were expected based on prior, neuroanatomical knowledge. To verify the correctness of results in experimental data is difficult because no knowledge about the ultimate ground truth exists when data are not simulated. Therefore, we chose an extremely simple experiment—self-paced finger lifting of the index fingers in a self-chosen sequence—where very clear hypotheses about the expected connectivity from the motor cortices to the finger muscles exist.
Subjects and experimental task
Two subjects (S1, m, RH, 38 yrs; S2, f, RH, 23 yrs) participated in the experiment. Subjects gave written informed consent prior to the recording. Subjects had to lift the right and left index finger in a self-chosen randomly alternating sequence with approximately 2s pause between successive finger liftings. Finger movements were detected using a photosensor. In addition, an electromyogram (EMG) response was recorded from the extensor muscles of the the right and left index fingers.
Recording and preprocessing
MEG data were recorded using a 275 channel whole head system (OMEGA2005, VSM MedTech Ltd., Coquitlam, BC, Canada) in a synthetic 3rd order gradiometer configuration. Additional electrocardiographic, -occulographicc and -myographic recordings were made to measure the electrocardiogram (ECG), horizontal and vertical electrooculography (EOG) traces, and the electromyogram (EMG) for the extensor muscles of the right and left index fingers. Data were hardware filtered between 0.5 and 300 Hz and digitized at a sampling rate of 1.2 kHz. Data were recorded in two continuous sessions lasting 600 s each. For the analysis of effective connectivity between scalp sensors and the EMG, data were preprocessed using the Fieldtrip open-source toolbox for MATLAB (http://fieldtrip.fcdonders.nl/; version 2008-12-10). Data were digitally filtered between 5 and 200 Hz and then cut in trials from −1,000 ms before to 90 ms after the photosensor indicated a lift of the left or right index finger. This latency range ensured that enough EMG activity was included in the analysis. We used the artifact rejection routines implemented in Fieldtrip to discard trials contaminated with eye-blinks, muscular activity and sensor jumps.
Analysis of effective connectivity at the MEG sensor level using transfer entropy
Effective connectivity was analyzed using the algorithm to compute transfer entropy as described above. The algorithm was implemented as a toolbox (Lindner et al. 2009) for Fieldtrip data structures (http://fieldtrip.fcdonders.nl/) in MATLAB. The nearest neighbour search routines were implemented using OpenTSTool (Version1.2 on Linux 64 bit; Merkwirth et al. 2009). Parameters for the analysis were chosen based on a scanning of the parameter space, to obtain maximum sensitivity. In more detail we computed the difference between the transfer entropy for the MEG data and the surrogate data for all combinations of parameters chosen from: 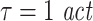 , u ∈ [10,16,22,30,150], d ∈ [4,5,7], k ∈ [4,5,6,7,8,9,10]. We performed the statistical test for a significant deviation from independence for each of these parametersets. This way a multiple testing problem arose, in addition to the multiple testing based on the multiple directed intercations between the chosen sensors (see next paragraph). We therefore performed a correction for multiple comparisons using the false discovery rate (FDR, q < 0.05, Genovese et al. 2002). The parameter values with optimum sensitivity, i.e. most sginificant results across sensor pairs after corrcetion for multiple comparison were: embedding dimensions d = 7, embedding delay
, u ∈ [10,16,22,30,150], d ∈ [4,5,7], k ∈ [4,5,6,7,8,9,10]. We performed the statistical test for a significant deviation from independence for each of these parametersets. This way a multiple testing problem arose, in addition to the multiple testing based on the multiple directed intercations between the chosen sensors (see next paragraph). We therefore performed a correction for multiple comparisons using the false discovery rate (FDR, q < 0.05, Genovese et al. 2002). The parameter values with optimum sensitivity, i.e. most sginificant results across sensor pairs after corrcetion for multiple comparison were: embedding dimensions d = 7, embedding delay 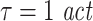 , forward prediction time u = 16 ms, number of neighbors considered for density estimations k = 4, time window for exclusion of temporally correlated neighbors
, forward prediction time u = 16 ms, number of neighbors considered for density estimations k = 4, time window for exclusion of temporally correlated neighbors 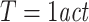 . In addition we required that prediction should be possible for at least 150 samples, i.e. individual trials where the combination of a long autocorrelation time and the embedding dimension of 7 did not leave enough data for prediction were discarded. We required that at least 30 trials should survive this exclusion step for a dataset to be analyzed.
. In addition we required that prediction should be possible for at least 150 samples, i.e. individual trials where the combination of a long autocorrelation time and the embedding dimension of 7 did not leave enough data for prediction were discarded. We required that at least 30 trials should survive this exclusion step for a dataset to be analyzed.
Even a simple task like self-paced lifting of the left or right index finger potentially involves a very complex network of brain areas related to volition, self-paced timing, and motor execution. Not all of the involved causal interactions are clearly understood to date. We therefore focused on a set on interactions where clear-cut hypothesis about the direction of causal interactions and the differences between the two conditions existed: We examined TE from the three bilateral sensor pairs displaying the largest amplitudes in the magnetically evoked fields (MEFs) (compare Fig. 7) before onset of the two movements (left or right finger lift) to both EMG channels. This also helped to reduce computation time, as for an all-to-all analysis of effective connectivity at the MEG and EMG sensor level would involve the analysis of 277 ×276 directed connections. We then tested connectivities in both conditions against each other by comparing the distributions of TE values in the two conditions using a permutation test. For this latter comparison a clear lateralization effect was expected, as task related causal interactions common to both conditions should cancel. Activity in at least three different frequency bands has been found in the motor cortex and it has been proposed that each of these different frequency bands subserves a different function:
A slow rhythm (6–10 Hz) has been postulated to provide a common timing for agonist/antagonist muscles pairs in slow movements and is thought to arise from from synchronization in a cerebello-thalamo-cortical loop (Gross et al. 2002). The coupling of cortical (primary motor cortex M1, primary somatosensory cortex S1) activity to muscular activity was proposed to be bidirectional (Gross et al. 2002) in this frequency range. The coupling may also depend on oscillations in spinal stretch reflex loops (Erimaki and Christakos 2008).
Activity in the beta range (~20 Hz) has been suggested to subserve the maintenance of current limb position (Pogosyan et al. 2009) and strong cortico-muscular coherence in this band has been found in isometric contraction accordingly (Schoffelen et al. 2008). Coherent activity in the beta band has also been demonstrated between bilateral motor cortices (Mima et al. 2000; Murthy and Fetz 1996).
In contrast, motor-act related activity in the gamma band (>30 Hz) is reported less frequently and its relation to motor control is less clearly understood to date (Donoghue et al. 1998). We therefore focused our analysis on a frequency interval from 5–29 Hz.
Note that we omitted the frequently proposed preprocessing of the EMG traces by rectification (Myers et al. 2003), as TE should be able to detect effective connectivity without this additional step.
Fig. 7.

Neuromagnetic fields in a finger lifting task. (a) Single-trial raw traces of magnetic fields (thin line) measured by two MEG sensors over left (MLT24) and right (MRT24) motor cortex (also compare (d) for the position of these sensors). In this trial the right finger was lifted. (b) Corresponding single trial EMG traces obtained from the left (EMG L) and right (EMG R) forearm. Time ‘0’ indicates the sample when the light barrier switch detected the finger lift. (c) Topography of magnetic fields averaged over trials at −50 ms before the registration of a right index finger lift. Note the dipolar pattern over left central cortex. (d) Layout of the MEG sensors. Sensors used for analysis of effective connectivity are indicated by solid circles. Lines with arrowheads indicate the investigated connections
Results
Overview
In this section we first present the analysis of effective connectivity in pairs of simulated signals {X,Y}. All signal pairs were unidirectionally coupled from X to Y. We used three coupling functions: linear, threshold and a purely non-linear quadratic coupling. We simulated two different signal dynamics, AR(10) processes and processes with 1/f spectra, that were close to spectra observed in biological signals. The two signals of a pair always had similar characteristics. We always analyzed both directions of potential effective connectivity: X →Y and Y →X to quantify both, sensitivity and specificity of our method.
In addition to this basic simulation we investigated the following special cases: coupling via multiple coupling delays for linear and threshold interactions, linearly mixed observation of two coupled signals for linear and threshold coupling, and observation of a single signal via two sensors with different noise levels. In this last case no effective connectivity should be detected. The absence of false positives in this latter case is of particular importance for EEG and MEG sensor-level analysis.
As a proof of principle we then applied the analysis of effective connectivity via TE to MEG signals recorded in a self-paced finger lifting task. Here the aim was to recover the known connectivity from contralateral motor cortices to the muscles of the moved limb, via a comparison of effective connectivty for left and right finger lifting.
Simulation study
Detection of non-linear interactions for various signal dynamics
Transfer entropy in combination with permutation testing correctly detected effective connectivity (X →Y) for both, autoregressive order 10 and 1/f signal dynamics and all three simulated coupling types (linear, threshold, quadratic) if at least 30 trials were used to compute statistics (Fig. 2). No false positives, i.e. significant results for the direction Y →X, were observed. We note that the cross-correlation function between the signals X and Y were flat when coupled non-linearly, which indicates that linear approaches may be insufficient to detect a significant interaction in those cases.
Fig. 2.
Detection of effective connectivity by TE for two unidirectionally coupled signals (X →Y). (a–c) Signals generated from an autoregressive order ten process and coupled via (a) linear, (b) threshold, and (c) quadratic coupling. (d–f) Signals generated with dynamics of a 1/f noise process and coupled via (d) linear, (e) threshold, and (f) quadratic coupling. A single interaction delay of 20 samples was used. Time courses of source (X) and target (Y) signals on the left and results of permutation testing for a varying number of trials (15, 30, 60, 120) on the right. Black bars indicate (1-p) values for coupling X →Y (true coupling direction), gray bars indicate values of (1-p) for coupling Y →X. The dashed line corresponds to significant effective connectivity (p < 0.05)
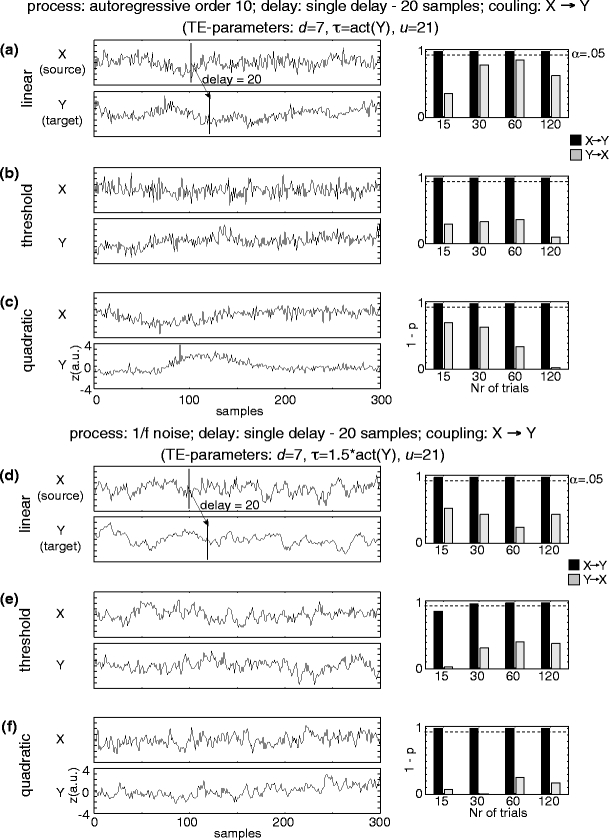
Detection of interactions with multiple interaction delays
The statistical evaluation of TE values robustly detected the correct direction of effective connectivity (X→Y) for the two unidirectionally coupled AR(10) time series (X,Y), coupled via a range of delays δ from 17–23 samples, and for the two unidirectionally coupled 1/f time series, coupled via a range of delays δ from 97-103 samples. The correct coupling direction (X →Y) was found for all three investigated coupling functions (linear, threshold, quadratic), even if only 15 trials were investigated (Fig. 3). For these analysis we used a prediction time u of 21 samples for the case of a delay δ of 17–23 samples, and a prediction time u of 101 samples for the delay δ of 97–103 samples. Correct detection of effective connectivity was also possible when using a prediction time u of 21 samples for the delay δ of 97–103 samples, i.e. a prediction time that was shorter than the interaction delay (data not shown). This was expected because of the delocalization in time provided for by the delay embedding. However, no effective connectivity was detected when using a prediction time u of 101 samples for a interaction delay δ of 17–23 samples, i.e. when using a prediction time that was considerably longer than the interaction delay (data not shown; compare Table 1 for single interaction delays). No false positive effective connectivities (Y→X) were found. However, relatively high values for (1-p) for some cases indicate that the embedding parameters were not optimally chosen, as discussed below.
Fig. 3.
Detection of effective connectivity by TE for two unidirectionally coupled time series (X →Y) with a range of coupling delays as indicated by the shaded boxes in (a) and (d). (a–c) autoregressive order ten processes; interaction delays 17–23 samples. (a) Linear interaction, (b) threshold coupling, and (c) quadratic coupling. (d–f) 1/f processes; interaction delays 97–103 samples. (d) Linear interaction, (e) threshold coupling, and (f) quadratic coupling. Time series are plotted on the left, results of permutation testing for different numbers of simulated trials (15, 30, 60, 120) on the right. Black bars indicate values of (1-p) for coupling X →Y (true coupling direction), gray bars indicate values of (1-p) for coupling Y →X. The dashed line corresponds to significant effective connectivity (p < 0.05)
Detection of effective connectivity from linearly mixed measurement signals
In order to investigate the application of TE to EEG and MEG sensor signals, where the signals from the processes in question can only be observed after linear mixing processes, we simulated two unidirectionally coupled AR(10) signals (X→Y with linear or threshold coupling). These signals then underwent a symmetric linear mixing process in dependence of a parameter ε in the range from 0.1 to 0.4, where a value of ε = 0.5 would indicate identical mixed signals (see Eqs. 15, 16). For the case of linearly coupled source signals TE indicated effective connectivity in direction from the sensor signal X ε that had a higher contribution from the driving process (X) to the sensor Y ε dominated by the receiving process (Y) for all investigated cases of linearly mixed measurement signals except for the case of ε = 0.4. In this case TE detected the correct direction of the interaction and did not result in false positive detection, however, the time-shift test indicated the presence of instantaneous mixing and the result could not be counted as a correct detection of effective connectivity. For the case of source signals that were coupled via a threshold function TE in combination with the time-shift test correctly identified effective connectivty and did not result in false positive detection for all of the investigated linear mixing strengths. These observations held even if only 15 trials were evaluated (Figs. 4 and 5).
Robustness against instantaneous mixing
To quantify the false positive rates when applying transfer entropy to multiple observations of the same signal, but with differential noise, we simulated an autoregressive order 10 process and two observation of this process: one noise free observation, X ε, and a second observation, Y ε, corrupted by a varying amount of white noise (Fig. 6(a) and (b)). Similar to the performance of GC in this case (Nolte et al. 2008), the application of TE resulted in a considerable number of false positive detections of effective connectivity from the noise free sensor signal to the noise-corrupted sensor signal (Fig. 6(c)). However, application of the time-shifting test as proposed in the methods section removed all false positive cases.
Choosing embedding parameters for delayed interactions
To demonstrate the importance of correct embedding we simulated unidirectionally coupled signals with various interaction delays and analyzed effective connectivity with different choices for the embedding dimension d, the embedding delay τ and the prediction time u (Tables 1 and 2). As expected because of theoretical considerations (see Fig. 1), false positive effective connectivity is reported for short interaction delays (5, 20 samples) in combination with short prediction times (six samples) and insufficient embedding (d = 4, 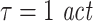 ). In contrast, if we try to detect long interactions delays (δ = 20, 100) with too short prediction times (u = 6), again with insufficient embedding, the method looses its sensitivity, as expected. This indicates that for given analysis parameters (d,τ,u) the range of interaction delays δ that can be investigated reliably is limited (Table 1). The above problem is solved naturally by increasing embedding dimensions and embedding delays as demonstrated in Table 2—although this may not be possible in practical terms sometimes. In our simulations we generally found an embedding delay of
). In contrast, if we try to detect long interactions delays (δ = 20, 100) with too short prediction times (u = 6), again with insufficient embedding, the method looses its sensitivity, as expected. This indicates that for given analysis parameters (d,τ,u) the range of interaction delays δ that can be investigated reliably is limited (Table 1). The above problem is solved naturally by increasing embedding dimensions and embedding delays as demonstrated in Table 2—although this may not be possible in practical terms sometimes. In our simulations we generally found an embedding delay of 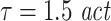 in combination with embedding dimensions between 7 and 10 to be more appropriate than smaller (d = 4, also see Table 2) or larger embedding dimensions (d = 13, 16, 19, data not shown) or a shorter embedding delay (
in combination with embedding dimensions between 7 and 10 to be more appropriate than smaller (d = 4, also see Table 2) or larger embedding dimensions (d = 13, 16, 19, data not shown) or a shorter embedding delay (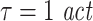 ). While it is often proposed to use
). While it is often proposed to use 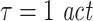 for embedding our data suggest that for the evaluation of TE it is particularly important to cover most or all of the memory inherent in both, source and target signals. For our data this could be be achieved by choosing
for embedding our data suggest that for the evaluation of TE it is particularly important to cover most or all of the memory inherent in both, source and target signals. For our data this could be be achieved by choosing 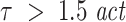 to prevent against false positive detection of causality in the presence of delayed interactions. We also observed that values of the prediction time u close to the actual interaction delay δ made the analysis of TE both, more sensitive and more robust against false positives, even for suboptimal choices of d and τ (Tables 1 and 2). Hence, a choice of u close to δ, e.g. based on prior (e.g. anatomical) knowlegde, may yield a method that is more robust in the face of unkown and hard to determine values for d and τ.
to prevent against false positive detection of causality in the presence of delayed interactions. We also observed that values of the prediction time u close to the actual interaction delay δ made the analysis of TE both, more sensitive and more robust against false positives, even for suboptimal choices of d and τ (Tables 1 and 2). Hence, a choice of u close to δ, e.g. based on prior (e.g. anatomical) knowlegde, may yield a method that is more robust in the face of unkown and hard to determine values for d and τ.
Effective connectivity at the MEG sensor level
Motor evoked fields
Self paced lifting of the right or left index fingers in a self chosen sequence resulted in robust motor evoked fields, that were compatible with motor evoked fields reported in the literature (Mayville et al 2005; Weinberg et al. 1990; Nagamine et al. 1996; Pedersen et al. 1998) (Fig. 7). We observed a slow readiness field at sensors over contralateral motor cortices starting approximately 350 ms before onset of EMG activity and a pronounced reversal of field polarity during movement execution (data not shown).
Movement related effective connectivity
As expected, effective connectivity from sensors over contralateral motor cortices was significantly larger to EMG electrodes over the muscle of the moved finger than to the EMG electrode over the muscle of the non-moved finger (Fig. 8). Unexpectedly however, effective connectivity from ipsilateral motor cortices was also significantly larger to the EMG electrodes over the muscle of the moved finger than to the EMG electrode over the muscle of the non-moved finger. Effective connectivity was never larger from any sensor over motor cortices to the EMG electrodes over the muscle of the non-moved finger.
Discussion
Transfer entropy as a tool to quantify effective connectivity
In the present study we aimed to demonstrate that TE is a useful addition to existing methods for the quantification of effective connectivity. We argued that existing methods like GC, that are based on linear stochastic models of the data, may have difficulties detecting purely non-linear interactions, such as inverted-U relationships. Here, we could show that transfer entropy reliably detected effective connectivity correctly when two signals were coupled by a quadratic, i.e. purely non-linear, function (Fig. 2). Particularly relevant for neural interactions, we have also shown that couplings mediated by threshold or sigmoidal functions are correctly captured by TE.
Furthermore, we extended the original definition of TE to deal with long interaction delays and demonstrated that TE detected effective connectivity correctly when the coupling of two signals was mediated by multiple interactions that spanned a range of latencies (Fig. 3).
Moreover, we considered the problem of volume conduction and showed that TE robustly detected effective connectivity when only linear mixtures of the original coupled signals were available (Figs. 4 and 5), if signals were not too close to being identical. In addition, if the two measurements reflected a common underlying source signal (’common drive’) but had different levels of measurement noise added, TE in combination with a test on time shifted data, correctly rejected the hypothesis of effective connectivity between the two measurement signals, in contrast to a naive application of GC (Nolte et al. 2008). Therefore, TE in combination with this test is well applicable to EEG and MEG sensor-level signals, where linear instantaneous mixing is inherent in the measurement method. However, without the additional test on time shifted data, TE had a non-negligible rate of false positives detections of effective connectivity. The origin of these false positives can be understood as follows. Theoretically transfer entropy is zero in the absence of causality, i.e. when processes are fully independent—as should be the case for surrogate data. TE is also zero for identical copies of a single signal, as required from a causality measure, when driver and response system cannot be distinguished. Here, we considered the case of volume conduction of a single signal onto two sensors in the presence of additional noise. Hence, the use of surrogate data for a test of the causality hypothesis inevitably leads to the comparison of two (noisy) zeros and false positives. Because of this difficulty we suggest to perform the time-shift test whenever multiple observations of a single source signal are likely to be present in the data, as is the case for EEG and MEG measurements.
Last but not least, we proposed TE as a tool for the exploratory investigation of effective connectivity, because it is a model-free measure based on information theory. Complicated types of coupling such as cross-frequency phase coupling (Palva et al. 2005) should be readily detectable without prior specification, e.g. the coupling via a quadratic function—as investigated here—, introduces a frequency doubled (and distorted) input to the target signal. Nevertheless it was readily detected by TE. While the argument on model-freeness holds theoretically, any practical implementation comes with certain parameters that have to be adapted to the data empirically, such as the correct choice of a delay τ and the number of dimensions d used for delay embedding. In addition, the implementation of TE proposed here incorporates a parameter for the prediction time u to adapt the analysis for cases where a long interaction delay is present. If chosen ad hoc these parameters amount to a sort of model for the data. To keep the method model-free we therefore proposed to scan a sufficiently large parameter space on pilot data before analyzing the data of interest or to scan the parameter space and to correct for the arising multiple comparison problem later on, during statistical testing.
To handle the estimation of TE, the parameter scanning and the statistical testing, including the shift-test, we implemented the proposed procedure in the form of a convenient open-source MATLAB toolbox for the Fieldtrip data format that is available from the authors (Lindner et al. 2009).
Limitations
Despite the above-mentioned merits, the TE method also has limitations that have to be considered carefully to avoid misinterpretations of the results:
We note that model-freeness is not always an advantage. In contrast to model-based methods, the detection of effective connectivity via TE does not entail information on the type of interaction. This fact has two important consequences. First, the absence of a specific model of the interaction leads to a high sensitivity for all types of depedencies between two time-series. This way, trivial (nuisance) dependencies, might be detected by testing against surrogates. This is bound to happen if these dependencies are not kept intact when creating the surrogate data. Second, the specific type of interaction must be separately assessed post hoc by using model based methods, after the presence of effective connectivity was established using transfer entropy. In principle the analysis of effective connectivity using TE, and the post-hoc comparison of signal pairs with and without significant interaction in an exploratory search of the actual mechanism of this interaction are possible in the same dataset. This is because these two questions are orthogonal. However, the relationship between siginificant effective connectivity—detected by TE—and a specific mechanism of the interaction needs to be tested on independent data.
Another limitation is that false positive reports are possible when the embedding parameters for the reconstruction of the state space are not chosen correctly. We therefore suggest to use TE with a careful choice of parameters, especially with respect to τ, and only after checking that the data to be analyzed meets certain characteristics. In the following we list a number of characteristics to be considered. First, strong non-stationarities in the data can make impossible to average over time to reliably estimate the probability densities on which TE is based. Consequently, TE should only be used on data of sufficient length that show at most weak non-stationarities. For an approach to overcome this limitation problem by using the trial structure of data sets see Gomez-Herrero et al. (2010). Second, in this work we have only assessed pairwise interactions. Although a fully multivariate extension is conceptually possible (Gomez-Herrero et al. 2010; Lizier et al. 2008), practical data lengths and computing time restrict its use. Third, TE analysis is difficult to interpret when signals have a different physical origin such as for example a chemical concentration and an electric field. The reason is that even though the signals entering the TE analysis are z-scored to obtain a certain normalization, there is no clear physical meaning of distance in the joint space of the signals, and consequently, no a priori justification to use any particular coarse-graining box in the two directions. Since the results of TE are sensitive to the use of different coarse-graining scales in the two directions, the meaning of any numerical estimate of TE for signals of different physical origin is difficult to establish. Finally, if the interaction to be captured is known to be linear, then the use of linear approaches is fully justified and usually outperforms TE in aspects such as computing time and data-efficiency. Last but not least we should comment on some general limitations related to the concept of causality as defined by Wiener. It is important to note that Wiener’s definition does not include any interventions to determine causality, i.e. it describes observational causality. Methods based on Wiener’s principle such as GC, TE share certain limitations:
The decsription of all system involved has to be causally complete, i.e. there must not be unobserved common causes that do not enter the analysis.
If two systems are related by a deterministic map, no causality can be inferred. This would exclude systems exhibiting complete synchronization, for example. Technically this is reflected in Eq. 4: For TE to be well defined the probability densities and their logarithms must exist. Therefore δ-distributions in the joint embedding space of two signals, which are equivalent to deterministic maps between these signals, are excluded.
The concept of observational causality rests on the axiom that the past and present may cause the future but the future may not cause the past. For this axiom to be useful observations must be made at a rate that allows a meaningful distinction between past, present and future with respect to the interaction delays involved. This means that interactions that take place on a timescale faster than the sampling rate must be missed in methods based on observational causality.
Application of TE to MEG recordings in a motor task
As a proof-of-principle, we applied TE to MEG data recorded during self paced finger lifting. The analysis of the effective connectivity from MEG to EMG signals was performed without the recommended rectification of the EMG signal (Myers et al. 2003) to proove that TE could perform the analysis well without this step. Our expectations of stronger effective connectivity from contralateral motor cortex to the moved finger were met for both fingers in both investigated subjects. Surprisingly, however, we also found stronger effective connectivity from ipsilateral motor cortex to the moved finger. It is not clear at present whether this effective connectivity reflected an indirect interaction: Contralateral motor cortex may drive both, ipsilateral cortex and the muscles of the moved finger, albeit with strongly differing delays. In this case, TE may erroneously detect effective connectivity from ipsilateral cortex to the muscle, as discussed above. Additional analyses, quantifying the coupling between the two motor cortices will be necessary to clarify this issue. As discussed below, these analyses should preferentially be performed using a multivariate extension of the TE method.
Comparison to existing literature
The application of non-linear methods to detect effective connectivity in neuroscience data has been suggested before: One of the earliest attempts to extend GC to the non-linear case and to apply it to neurophysiological data was presented by Freiwald et al. (1999). They used a locally linear, non-linear autoregressive (LLNAR) model where time varying autoregression coefficients were used to capture non-linearities. This model was only tested, however, on simulations of unidirectionally and linearly coupled signals and correctly identified the coupling as unidirectional and as linear. No attempt was made to validate the model on simulations of explicitly non-linear directed interactions. Application to EEG data from a patient with complex partial seizures indicated non-linear coupling of the signals measured at electrode positions C3 and C4. Another test on local field potential (LFP) data recorded in the anterior inferotemporal cortex (macaque area TE) of the macaque monkey however detected no indication of a non-linear interaction. We add to these results by demonstrating that also purely non-linear (square, threshold) interactions are reliably detected using TE in combination with appropriate statistical testing and by demonstrating that interactions can also be found in MEG and EMG data, even when omitting the usual rectification of the EMG. Chávez et al. (2003) used TE on data from an epileptic patient and also proposed a statistical test based on block-resampling of the data that is similar to the trial shuffling approach used here. They found that TE with a fixed prediction time and a fixed inclusion radius for neighbor search was able to detect the directed linear and non-linear interactions for the simulated models. Our findings are in agreement with these results. In addition, we demonstrated that TE also detects directed non-linear interactions for biologically plausible data with 1/f characteristics and a range of interaction delays instead of a single delay. Hinrichs et al. (2008) used a measure that is very similar to transfer entropy as it was investigated here. However, in contrast to our study they substituted the time-consuming estimation of probability densities by kernel-based methods with a linear method based on the data covariance matrices. As explicitely stated in the mathematical appendix of Hinrichs et al. (2008) this effectively limits the detection of directed interactions to linear ones. Here, we demonstrate that, while being relatively time consuming, a kernel based estimation of the required probability densities is feasible using the Kraskov-Stögbauer-Grassberger estimator (Kraskov et al. 2004), even for a dimensionality of five and higher. We note however, that the amount of data necessary for these estimations may not always be available and that the achievable ‘temporal resolution’ is limited by this factor. Interestingly, scanning of the prediction time u, revealed an optimal interaction delay in the MEG/EMG data of around 16 ms, in accordance with their findings.
Outlook
As demonstrated in this study TE is a useful tool to quantify effective connectivity in neuroscience data. Its ability to detect purely non-linear interactions and to operate without the specification of one or more a priori models make it particularly useful for exploratory data analysis, but its use is not limited to this application. The implementation of TE estimation used here only considered pairs of signals, i.e. it is a bivariate method. Direct and indirect interactions may, therefore, not be separated well. However, an extension to the multivariate case is possible as noted before (e.g. Chávez et al. 2003) and is currently under investigation. Its application to cellular automata by Lizier and colleagues have already revealed interesting insights into the pattern formation and information flow in these models of complex systems (Lizier et al. 2008).
The problem of direct versus indirect interactions can also be ameliorated for the case of MEG and EEG data by performing the analysis at the level of source time-courses obtained from a suitable source analysis method. Using source level time-courses will reduce the number of signals for analysis. A post hoc analysis of the obtained reduced network of effective connectivty by DCM may be possible then. Using source level time-courses will also improve the interpretability of the obtained effective connectivities compared to those at the sensor level. This is because for a given causal interaction observed at the sensor level any of the multiple sources reflected in the sensor signal may be responsible for the observed effective connectivity.
Although the estimation of TE presented here is geared at continuous data TE has found application in the analysis of spiking data as reported in Gourvitch and Eggermont (2007). The particularities to estimate TE from point processes can be found there. Thus, both macroscopic (fMRI, EEG/MEG) and more local signals (LFP, single unit activity) can be readily analized in the common framework of TE. In the future, it will be interesting to compare the effective connectivities for a variety of temporal and spatial scales as revealed by TE.
Conclusion
Transfer entropy robustly detected effective connectivity in simulated data both for complex internal signal dynamics (1/f) and for strongly non-linear coupling. Detection of effective connectivity was possible without specifying an a priori model. With the use of an additional test for linear instantaneous mixing it was robust against false positives due to simulated volume conduction. Therefore it is not only applicable for invasive electrophysiological data but also for EEG and MEG sensor-level analysis. Analysis of MEG and EMG sensor-level data recorded in a simple motor task data revealed the expected connectivity, even without rectification of the EMG signal. We therefore propose TE as a useful tool for the analysis of effective connectivity in neuroscience data.
Acknowledgements
The authors would like to thank Viola Priesemann from the Max Planck Institute for Brain Research, Frankfurt, for valuable comments on this manuscript, German Gomez Herrero from the Technical University of Tampere, Wei Wu from the Humboldt-Universität in Berlin, Mikhail Prokopenko from the CSIRO in Sydney, and Prof. Jochen Triesch from the Frankfurt Institute for Advanced Studies (FIAS) for stimulating discussions, and Sarah Straub from the Department of Psychology, University of Regensburg for assistance in data acquisition.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Historically, however, GC was formulated without explicit assumptions about the linearity of the system (Granger 1969) and was therefore closely related to Wiener’s formal definition of causality (Wiener 1956).
For a continuous random variable the natural generalization of Shannon entropy is its differential entropy. Although differential entropy does not inherit the properties of Shannon entropy as an information measure, the derived measures of mutual information and transfer entropy retain the properties and meaning they have in the discrete variable case. We refer the reader to Kaiser and Schreiber (2002) for a more detailed discussion of TE for continuous variables. In addition, measurements of physical systems typically come as discrete random variables because of the binning inherent in the digital processing of the data.
R. Vicente, M. Wibral, and M. Lindner contributed equally.
ML was funded by the Hessian initiative for the development of scientific and economic excellence (LOEWE). RV and GP were in part supported by the Hertie Foundation and the EU (EU project GABA—FP6-2005-NEST-Path-043309).
Contributor Information
Raul Vicente, Email: vicente@fias.uni-frankfurt.de.
Michael Wibral, Email: michael.wibral@web.de.
Michael Lindner, Email: m.lindner@idea-frankfurt.eu.
Gordon Pipa, Email: pipa@mpih-frankfurt.mpg.de.
References
- Arieli A, Sterkin A, Grinvald A, Aertsen A. Dynamics of ongoing activity: Explanation of the large variability in evoked cortical responses. Science. 1996;273(5283):1868–1871. doi: 10.1126/science.273.5283.1868. [DOI] [PubMed] [Google Scholar]
- Cao L. Practical method for determining the minimum embedding dimension of a scalar time series. Physica, A, 1997;110:43–50. [Google Scholar]
- Chávez M, Martinerie J, Quyen MLV. Statistical assessment of non-linear causality: Application to epileptic eeg signals. Journal of Neuroscience Methods. 2003;124(2):113–128. doi: 10.1016/S0165-0270(02)00367-9. [DOI] [PubMed] [Google Scholar]
- Cormen, T., Leiserson, C., Rivest, R., & Stein, C. (2001). Introduction to algorithms. MIT Press and McGraw-Hill.
- Donoghue JP, Sanes JN, Hatsopoulos NG, Gal G. Neural discharge and local field potential oscillations in primate motor cortex during voluntary movements. Journal of Neurophysiology. 1998;79(1):159–173. doi: 10.1152/jn.1998.79.1.159. [DOI] [PubMed] [Google Scholar]
- Erimaki S, Christakos CN. Coherent motor unit rhythms in the 6–10 hz range during time-varying voluntary muscle contractions: Neural mechanism and relation to rhythmical motor control. Journal of Neurophysiology. 2008;99(2):473–483. doi: 10.1152/jn.00341.2007. [DOI] [PubMed] [Google Scholar]
- Freiwald WA, Valdes P, Bosch J, Biscay R, Jimenez JC, Rodriguez LM, et al. Testing non-linearity and directedness of interactions between neural groups in the macaque inferotemporal cortex. Journal of Neuroscience Methods. 1999;94(1):105–119. doi: 10.1016/S0165-0270(99)00129-6. [DOI] [PubMed] [Google Scholar]
- Friston K. Functional and effective connectivity in neuroimaging: A synthesis. Human Brain Mapping. 1994;2:56–78. doi: 10.1002/hbm.460020107. [DOI] [Google Scholar]
- Friston KJ, Harrison L, Penny W. Dynamic causal modelling. NeuroImage. 2003;19(4):1273–1302. doi: 10.1016/S1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage. 2002;15(4):870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Gomez-Herrero, G., Wu, W., Rutanen, K., Soriano, M. C., Pipa, G., & Vicente, R. (2010). Assessing coupling dynamics from an ensemble of time series. arXiv:1008.0539v1.
- Gourvitch B, Eggermont JJ. Evaluating information transfer between auditory cortical neurons. Journal of Neurophysiology. 2007;97(3):2533–2543. doi: 10.1152/jn.01106.2006. [DOI] [PubMed] [Google Scholar]
- Granger C. Long memory relationships and the aggregation of dynamic models. Journal of Econometrics. 1980;14:227–238. doi: 10.1016/0304-4076(80)90092-5. [DOI] [Google Scholar]
- Granger CWJ. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969;37:424–438. doi: 10.2307/1912791. [DOI] [Google Scholar]
- Gross J, Timmermann L, Kujala J, Dirks M, Schmitz F, Salmelin R, et al. The neural basis of intermittent motor control in humans. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(4):2299–2302. doi: 10.1073/pnas.032682099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs H, Noesselt T, Heinze HJ. Directed information flow: A model free measure to analyze causal interactions in event related eeg-meg-experiments. Human Brain Mapping. 2008;29(2):193–206. doi: 10.1002/hbm.20382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavackova-Schindler K, Palus M, Vejmelka M, Bhattacharya J. Causality detection based on information-theoretic approaches in time series analysis. Physics Reports. 2007;441:1–46. doi: 10.1016/j.physrep.2006.12.004. [DOI] [Google Scholar]
- Kaiser A, Schreiber T. Information transfer in continuous processes. Physica, D. 2002;110:43–62. doi: 10.1016/S0167-2789(02)00432-3. [DOI] [Google Scholar]
- Kantz, H., & Schreiber, T. (1997). Nonlinear time series analysis. Cambridge University Press.
- Kozachenko L, Leonenko N. Sample estimate of entropy of a random vector. Problems of Information Transmission. 1987;23:95–100. [Google Scholar]
- Kraskov, A., Stögbauer, H., & Grassberger, P. (2004). Estimating mutual information. Physical Review. E, Statistical, Nonlinear, and Soft Matter Physics, 69(6 Pt 2), 066,138. [DOI] [PubMed]
- Lindner, M., Vicente, R., & Wibral, M. (2009). Trentool—the transfer entropy toolbox. http://www.michael-wibral.de/TRENTOOL. Accessed 7 August 2010.
- Lizier, J., Prokopenko, M., & Zomaya, A. (2008). Local information transfer as a spatiotemporal filter for complex systems. Physical Review. E, 77, 026,110. [DOI] [PubMed]
- Mayville JM, Fuchs A, Kelso JAS. Neuromagnetic motor fields accompanying self-paced rhythmic finger movement at different rates. Experimental Brain Research. 2005;166(2):190–199. doi: 10.1007/s00221-005-2354-2. [DOI] [PubMed] [Google Scholar]
- Merkwirth, C., Parlitz, U., Wedekind, I., Engster, D., & Lauterborn, W. (2009). Opentstool version 1.2 (2/2009). http://www.physik3.gwdg.de/tstool/index.html. Accessed 7 August 2010.
- Mima T, Matsuoka T, Hallett M. Functional coupling of human right and left cortical motor areas demonstrated with partial coherence analysis. Neuroscience Letters. 2000;287(2):93–96. doi: 10.1016/S0304-3940(00)01165-4. [DOI] [PubMed] [Google Scholar]
- Murthy VN, Fetz EE. Oscillatory activity in sensorimotor cortex of awake monkeys: Synchronization of local field potentials and relation to behavior. Journal of Neurophysiology. 1996;76(6):3949–3967. doi: 10.1152/jn.1996.76.6.3949. [DOI] [PubMed] [Google Scholar]
- Myers LJ, Lowery M, O’Malley M, Vaughan CL, Heneghan C, Gibson ASC, et al. Rectification and non-linear pre-processing of emg signals for cortico-muscular analysis. Journal of Neuroscience Methods. 2003;124(2):157–165. doi: 10.1016/S0165-0270(03)00004-9. [DOI] [PubMed] [Google Scholar]
- Nagamine, T., Kajola, M., Salmelin, R., Shibasaki, H., & Hari, R. Movement-related slow cortical magnetic fields and changes of spontaneous meg- and eeg-brain rhythms. Electroencephalography and Clinical Neurophysiology, 99(3), 274–286. [DOI] [PubMed]
- Nalatore, H., Ding, M., & Rangarajan, G. (2007). Mitigating the effects of measurement noise on Granger causality. Physical Review. E, Statistical, Nonlinear and Soft Matter Physics, 75(3 Pt 1), 031,123. [DOI] [PubMed]
- Nolte, G., Ziehe, A., Nikulin, V. V., Schloegl, A., Kraemer, N., Brismar, T., et al. (2008). Robustly estimating the flow direction of information in complex physical systems. Physical Review Letters, 100(23), 234,101. [DOI] [PubMed]
- Paluš, M. (2001). Synchronization as adjustment of information rates: Detection from bivariate time series. Physical Review. E, 63, 046,211. [DOI] [PubMed]
- Palva JM, Palva S, Kaila K. Phase synchrony among neuronal oscillations in the human cortex. Journal of Neuroscience. 2005;25(15):4250–4254. doi: 10.1523/JNEUROSCI.4250-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JR, Johannsen P, Bak CK, Kofoed B, Saermark K, Gjedde A. Origin of human motor readiness field linked to left middle frontal gyrus by meg and pet. NeuroImage. 1998;8(2):214–220. doi: 10.1006/nimg.1998.0362. [DOI] [PubMed] [Google Scholar]
- Pereda E, Quiroga R, Bhattacharya J. Nonlinear multivariate analysis of neurophysiological signals. Progress in Neurobiology. 2005;77:1–37. doi: 10.1016/j.pneurobio.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Pogosyan A, Gaynor LD, Eusebio A, Brown P. Boosting cortical activity at beta-band frequencies slows movement in humans. Current Biology. 2009;19(19):1637–1641. doi: 10.1016/j.cub.2009.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragwitz M, Kantz H. Markov models from data by simple nonlinear time series predictors in delay embedding spaces. Physical Review. E. 2002;65:56201. doi: 10.1103/PhysRevE.65.056201. [DOI] [PubMed] [Google Scholar]
- Reza, F. (1994). An introduction to information theory. Dover.
- Schoffelen JM, Oostenveld R, Fries P. Imaging the human motor system’s beta-band synchronization during isometric contraction. NeuroImage. 2008;41(2):437–447. doi: 10.1016/j.neuroimage.2008.01.045. [DOI] [PubMed] [Google Scholar]
- Schreiber T. Measuring information transfer. Physical Review Letters. 2000;85(2):461–464. doi: 10.1103/PhysRevLett.85.461. [DOI] [PubMed] [Google Scholar]
- Shannon C. A mathematical theory of communication. Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- Swadlow H. Physiological properties of individual cerebral axons studied in vivo for as long as one year. Journal of Neurophysiology. 1985;54:1346–1362. doi: 10.1152/jn.1985.54.5.1346. [DOI] [PubMed] [Google Scholar]
- Swadlow H. Efferent neurons and suspected interneurons in motor cortex of the awake rabbit: Axonal properties, sensory receptive fields, and subthreshold synaptic inputs. Journal of Neurophysiology. 1994;71:437–453. doi: 10.1152/jn.1994.71.2.437. [DOI] [PubMed] [Google Scholar]
- Swadlow H, Rosene D, Waxman S. Characteristics of interhemispheric impulse conduction between the prelunate gyri of the rhesus monkey. Experimental Brain Research. 1978;33:455–467. doi: 10.1007/BF00235567. [DOI] [PubMed] [Google Scholar]
- Swadlow H, Waxman S. Observations on impulse conduction along central axons. Proceedings of the National Academy of Sciences. 1975;72:5156–5159. doi: 10.1073/pnas.72.12.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takens, F. (1981). Dynamical Systems and Turbulence, Warwick 1980. In Lecture Notes in Mathematics (Vol. 898, chap.). Detecting Strange Attractors in Turbulence (pp. 366–381). Springer.
- Tognoli E, Scott Kelso J. Brain coordination dynamics: True and false faces of phase synchrony and metastability. Progress in Neurobiology. 2009;12:31–40. doi: 10.1016/j.pneurobio.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor J. Binless strategies for estimation of information from neural data. Physical review. E. 2002;66:51903. doi: 10.1103/PhysRevE.66.051903. [DOI] [PubMed] [Google Scholar]
- Weinberg H, Cheyne D, Crisp D. Electroencephalographic and magnetoencephalographic studies of motor function. Advances in Neurology. 1990;54:193–205. [PubMed] [Google Scholar]
- Wiener N. The theory of prediction. In: Beckenbach EF, editor. In modern mathematics for the engineer. New York: . McGraw-Hill; 1956. [Google Scholar]
- Yerkes RM, Dodson JD. The relation of strength of stimulus to rapidity of habit-formation. Journal of Comparative Neurology and Psychology. 1908;18:459. doi: 10.1002/cne.920180503. [DOI] [Google Scholar]


