Abstract
Rationale: Chronic obstructive pulmonary disease (COPD), characterized by airflow limitation, is a disorder with high phenotypic and genetic heterogeneity. Pulmonary emphysema is a major but variable component of COPD; familial data suggest that different components of COPD, such as emphysema, may be influenced by specific genetic factors.
Objectives: To identify genetic determinants of emphysema assessed through high-resolution chest computed tomography in individuals with COPD.
Methods: We performed a genome-wide association study (GWAS) of emphysema determined from chest computed tomography scans with a total of 2,380 individuals with COPD in three independent cohorts of white individuals from (1) a cohort from Bergen, Norway, (2) the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Study, and (3) the National Emphysema Treatment Trial (NETT). We tested single-nucleotide polymorphism associations with the presence or absence of emphysema determined by radiologist assessment in two of the three cohorts and a quantitative emphysema trait (percentage of lung voxels less than –950 Hounsfield units) in all three cohorts.
Measurements and Main Results: We identified association of a single-nucleotide polymorphism in BICD1 with the presence or absence of emphysema (P = 5.2 × 10−7 with at least mild emphysema vs. control subjects; P = 4.8 × 10−8 with moderate and more severe emphysema vs. control subjects).
Conclusions: Our study suggests that genetic variants in BICD1 are associated with qualitative emphysema in COPD. Variants in BICD1 are associated with length of telomeres, which suggests that a mechanism linked to accelerated aging may be involved in the pathogenesis of emphysema.
Clinical trial registered with www.clinicaltrials.gov (NCT00292552).
Keywords: emphysema, chronic obstructive pulmonary disease, BICD1, single-nucleotide polymorphism
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Chronic obstructive pulmonary disease (COPD) is a disease with high phenotypic and genetic heterogeneity. Familial studies showed that different components of COPD, such as emphysema, aggregated independently within families of individuals with COPD. However, the involved genetic factors have yet to be elucidated.
What This Study Adds to the Field
This is the largest genome-wide association study (GWAS) of a radiographic phenotype in lung disease and an important phenotype in COPD. We identified a novel gene, BICD1, for emphysema susceptibility in COPD. Variants of BICD1 are associated with length of telomeres, which suggests that a mechanism linked to accelerated aging may be involved in the pathogenesis of emphysema.
Chronic obstructive pulmonary disease (COPD) is characterized by the progressive development of airflow limitation that is not fully reversible (1). COPD is intimately linked with cigarette smoking and is expected to be the third leading cause of mortality and the fifth leading cause of morbidity in the world by the year 2020 (2). Significant familial aggregation of COPD and spirometric measures have been observed (3, 4), suggesting that genetic factors play an important role in disease susceptibility and severity. These studies have motivated research efforts to identify genetic variants that predispose to COPD. However, the only widely accepted genetic risk factor for COPD is a severe deficiency of α1-antitrypsin (5), which is a rare disorder and present in only 1–2% of individuals with COPD. Genome-wide association studies of COPD affection status have identified single-nucleotide polymorphisms (SNPs) near the gene HHIP on chromosome 4, FAM13A on chromosome 4, and in a locus containing the CHRNA3, CHRNA5, and IREB2 genes on chromosome 15 as likely locations of additional COPD susceptibility loci (6–8).
A major challenge in finding true COPD susceptibility variants is the heterogeneous nature of the disease. Patients with COPD often have a combination of emphysema and small airway disease, both of which make independent contributions to airflow obstruction (9). It is possible that a given genetic variant may confer susceptibility to a specific COPD-related phenotype. One study showed evidence that airway wall thickening and emphysema assessed by chest computed tomography (CT) show independent aggregation within families of individuals with COPD (10). However, the involved genetic factors remain obscure.
Emphysema is characterized by permanent expansion of airspaces distal to the terminal bronchioles. Potential mechanisms involved in the pathogenesis of emphysema include inflammation, oxidative stress, and apoptosis (11). The presence of emphysema is associated with more severe chronic airflow obstruction (12). Among those with similar degrees of airflow obstruction, emphysema is associated with reduced body mass index and fat-free mass (13), reduced diffusion capacity (14), increased arterial stiffness (15), and increased risk of lung cancer (16). To date, genetic studies on emphysema have been performed primarily with candidate genes selected from protease–antiprotease and oxidant–antioxidant pathways (17). These studies have generally been limited by small sample sizes. We hypothesized that novel genetic determinants of emphysema, assessed through high-resolution chest computed tomography, could be identified through genome-wide association studies (GWAS) in three independent COPD cohorts of white individuals. Some of the results of this study have been reported previously in the form of an abstract (18).
METHODS
Details of the study cohorts and all methods are provided in the online supplement.
Study Cohorts
Subjects with COPD who took part in this study were from three cohorts, the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Study (19, 20), a cohort from Bergen, Norway (21–24), and the National Emphysema Treatment Trial (NETT) (25). In the ECLIPSE and Norway cohorts, spirometric criteria for COPD included postbronchodilator FEV1/FVC less than 0.7 and FEV1 less than 80% predicted; subjects from NETT had more severe airflow obstruction (FEV1 ≤ 45% predicted) (26). In total, 1,672 subjects from ECLIPSE, 488 from the Norway cohort, and 382 from NETT were used in the analysis.
Measurement of Emphysema
The extent of radiographic emphysema was measured in two ways. First, each CT scan was reviewed by radiologist(s) and assigned an ordinal, qualitative score. Second, the extent of emphysema was assessed by density mask analysis, using the percentage of lung voxels with attenuation less than −950 Hounsfield units (HU). Previous work has noted a substantial, but incomplete, correlation between visual scoring and density mask analysis, suggesting that both types of information may be independently valuable for analysis (27).
In the ECLIPSE cohort, included subjects underwent low-dose volumetric CT scan (120 kV peak [kVp], 40 mA, and 1.00- or 1.25-mm slice thickness) at full inspiration. Two radiologists, blinded to the individual's lung function, independently scored each CT scan. Emphysema was reported as none, trivial, mild, moderate, severe, or very severe if it affected 0, less than 5%, 5–25%, more than 25–50%, more than 50–75%, and more than 75% of the lungs, respectively. Data from the baseline measurement were used in the current analysis. In the Norway cohort study, chest CT scans were performed with a GE LightSpeed Ultra CT scanner (120 kVp, 200 mA; GE Healthcare, Milwaukee, WI), at suspended full inspiration (apex to base), using a 1-mm slice thickness at 20-mm intervals, producing an average of 13.4 ± 1.6 slices per subject. Qualitative radiologist assessment as well as quantitative densitometric analysis were performed according to similar procedures as described for the ECLIPSE cohort. All scans in ECLIPSE and the Norway cohort were evaluated at the central imaging unit at the University of British Columbia (Vancouver, BC, Canada).
In the NETT study, CT scans were performed on one of three types of scanners (General Electric, Fairfield, CT; Siemens, Malvern, PA; or Picker International, Toronto, ON, Canada) with a range of 2- to 8-mm slice thickness, with 75% of the scan data from 4 to 5 mm. Densitometric measures were performed with the Pulmonary Analysis Software Suite (PASS, Iowa City, IA). Although qualitative radiologist scoring of emphysema was performed for NETT, all subjects had emphysema, and a different visual scoring system was used; therefore, NETT was not included in the qualitative GWAS. Details of the emphysema measurements were described previously for the Norway cohort study (28) and NETT (27, 29, 30).
Genotyping Quality Control and Assessment of Population Stratification
Sample genotyping and quality control procedures have been described previously (8). Briefly, samples were genotyped on the Illumina HumanHap 550 (Norway and ECLIPSE) and Quad 610 (NETT) genotyping arrays (Illumina, Inc, San Diego, CA). Initial sample and SNP quality control (QC) was done in BeadStudio, and genotype calls were generated. This was followed by assessment for cryptic relatedness and Hardy-Weinberg equilibrium. Population stratification was modeled by principal components analysis as implemented in EIGENSOFT version 2.0 (31) in each set of study samples.
Statistical Analysis
For qualitative emphysema by radiologist scoring, we tested the binary outcome of presence (≥5% involvement) or absence (<5%) of emphysema in the ECLIPSE and Norway cohorts for association with genotype, using a logistic regression adjusting for age, sex, pack-years of smoking, current smoking status, and principal components for genetic ancestry under an additive genetic model. As a secondary analysis, we also tested for association using a set of cases more narrowly defined as having at least 25% involvement of emphysema. For quantitative emphysema by densitometry (−950 HU), we performed linear regression adjusting for age, sex, pack-years of smoking, current smoking status, and body mass index (BMI) (32) in all three cohorts (ECLIPSE, Norway, and NETT), and repeated the regression without the adjustment for BMI. All regression analyses were performed in PLINK version 1.05 (33).
We combined effect sizes and P values across studies, using a fixed-effect meta-analysis and weighting by inverse variance as implemented in the meta package (version 1.1–4) in R (http://www.r-project.org), and the weighted Z-score meta-analysis method as implemented in METAL (http://www.sph.umich.edu/csg/abecasis/metal/). To further refine our association results, we performed genotype imputation in selected genomic regions using MACH version 1.0 (34). We used a threshold of P ≤ 5 × 10−7 used by the Wellcome Trust Case–Control Consortium (35) and a more stringent threshold of P ≤ 7.2 × 10−8 (36) to declare a pooled effect as genome-wide significant.
RESULTS
A total of 499,578 SNP markers passed quality control in all cohorts; a total of 1,586, 435, and 362 subjects with COPD from the ECLIPSE, Norway, and NETT cohorts, respectively, passed quality control and were not outliers by genetic ancestry. Baseline characteristics for these subjects are shown in Table 1. CT scan quality was insufficient to quantify the lung density in a substantial number of subjects (13%) from ECLIPSE. Those subjects with missing scores for quantitative emphysema were excluded from the GWAS analysis of the quantitative emphysema phenotype.
TABLE 1.
CHARACTERISTICS OF SUBJECTS WITH CHRONIC OBSTRUCTIVE PULMONARY DISEASE USED FOR GENOME-WIDE ASSOCIATION STUDY OF EMPHYSEMA QUALITATIVE AND QUANTITATIVE TRAITS
| ECLIPSE | Norway | NETT | |
|---|---|---|---|
| Subjects, n | 1,586 | 435 | 362 |
| Age, yr (SD) | 63.4 (7.0) | 63.8 (9.6) | 67.4 (5.8) |
| Female, % | 34.8 | 37.5 | 35.4 |
| Post-FEV1, % predicted (SD) | 47.7 (15.7) | 52.6 (17.0) | 28.1 (7.4) |
| Pack-years of smoking (SD) | 50.7 (27.6) | 30.9 (18.1) | 66.1 (30.9) |
| Current smokers, % | 35.4 | 48.7 | 0 |
| Emphysema −950 (SD)* | 18.4 (12.1) | 11.5 (11.8) | 16.9 (10.8) |
Definition of abbreviations: ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; NETT = National Emphysema Treatment Trial.
NETT subject data were used only for quantitative emphysema analysis.
Quantitative emphysema trait: percentage of lung voxels less than −950 Hounsfield units.
GWAS on Qualitative Emphysema
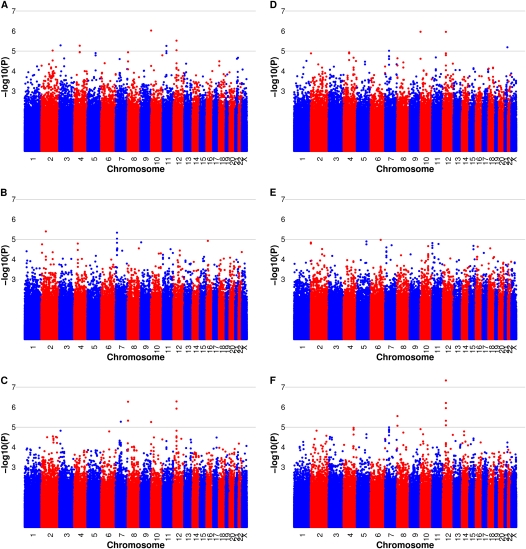
Our primary association analysis on the presence or absence of emphysema (defined by visual assessment of ≥5% vs. <5% affected) included 1,167 subjects with emphysema and 390 subjects with nonemphysematous COPD in ECLIPSE as well as 288 subjects with emphysema and 144 subjects with nonemphysematous COPD in the Norway cohort. Genomic control values were 1.00 in ECLIPSE and 1.01 in Norway after adjustment with principal components for genetic ancestry, suggesting that population stratification was well controlled. None of the SNPs from individual GWAS in ECLIPSE or Norway generated genome-wide significant association (Figure 1). The most significant association signal from meta-analysis was for rs10844154 on chromosome 12p11.2 with a meta-analysis P value of 5.2 × 10−7 and odds ratio (OR) of 1.46, located within the gene BICD1 [bicaudal D homolog 1 (Drosophila)] (Table 2). Our secondary analysis using the same nonemphysematous subjects but a more stringent definition of emphysema cases (>25% emphysema), resulted in 826 subjects with emphysema in ECLIPSE and 172 in the Norway cohort. In this severe emphysema analysis, rs10844154 remained the most significant finding and showed increased statistical significance and effect size with a meta-analysis P value of 4.8 × 10−8 and OR of 1.56 (Table E1 in the online data supplement). This P value reached the most stringent criteria for genome-wide significance (36).
Figure 1.
Manhattan plots of –log10 P values for association analysis of emphysema radiologist scores, including cases defined as at least 5% emphysema versus chronic obstructive pulmonary disease (COPD) without emphysema control subjects in ECLIPSE, the Norway cohort, and meta-analysis (A–C), and cases defined as greater than 25% emphysema versus COPD without emphysema control subjects in ECLIPSE, the Norway cohort, and meta-analysis (D–F).
TABLE 2.
ECLIPSE/NORWAY COHORT META-ANALYSIS OF EMPHYSEMA QUALITATIVE TRAIT
| Closest Gene | Risk/Nonrisk Allele | Risk Allele Frequency | ECLIPSE |
Norway |
Meta-analysis P Value* | Meta-analysis OR (P Value)† | Heterogeneity I2 (%)† | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Type | OR | P Value | OR | P Value | ||||||
| rs10844154 | 12 | Intron | BICD1 | C/A | 0.58 | 1.50 | 3.0 × 10−6 | 1.35 | 0.06 | 5.2 × 10−7 | 1.46 (5.5 × 10−7) | 0 |
| rs641525 | 8 | Intergenic | CSMD1 | T/G | 0.95 | 2.17 | 1.2 × 10−5 | 2.24 | 0.02 | 5.4 × 10−7 | 2.19 (5.3 × 10−7) | 0 |
| rs161981 | 12 | Intron | BICD1 | C/T | 0.56 | 1.47 | 9.2 × 10−6 | 1.37 | 0.05 | 1.2 × 10−6 | 1.44 (1.2 × 10−6) | 0 |
| rs341672 | 8 | Intergenic | CSMD1 | T/C | 0.92 | 1.82 | 3.3 × 10−5 | 1.71 | 0.05 | 4.8 × 10−6 | 1.80 (4.7 × 10−6) | 0 |
| rs1012036 | 7 | Intergenic | AC006320.2 | C/T | 0.77 | 1.43 | 2.1 × 10−4 | 1.63 | 6.0 × 10−3 | 5.4 × 10−6 | 1.47 (5.23 × 10−6) | 0 |
| rs2999399 | 10 | Intergenic | ADARB2 | T/C | 0.76 | 1.60 | 9.5 × 10−7 | 1.08 | 0.66 | 5.6 × 10−6 | 1.47 (5.9 × 10−6) | 73 |
Definition of abbreviations: Chr = chromosome; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; OR = odds ratio; SNP = single-nucleotide polymorphism.
Cases defined as at least 5% emphysema versus COPD without emphysema (<5%) control subjects: loci with meta-analysis P value < 10−5.
Based on weighted Z-score method.
Based on fixed-effects meta-regression; I2: I2 index used to assess magnitude of heterogeneity of ORs among studies.
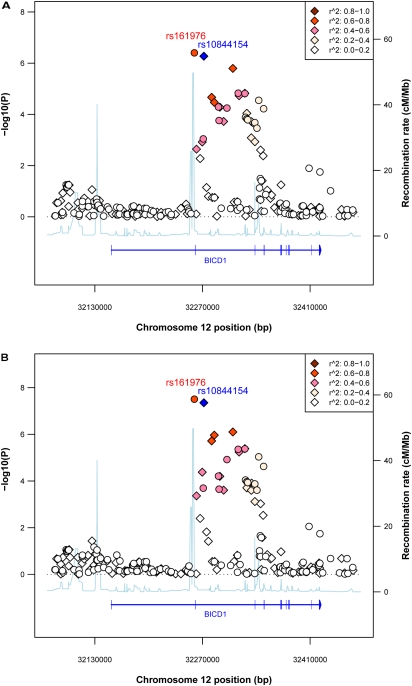
To refine this candidate region that may contain emphysema loci, we imputed SNPs in a region 200 kb from each side of rs10844154 in both sample sets with HapMap III phased data as reference (37). A stronger association signal was found for rs161976, typed in ECLIPSE and imputed in the Norway samples, with meta-analysis P values of 3.92 × 10−7 and 3.09 × 10−8 in subjects with emphysema versus nonemphysematous subjects and subjects with severe emphysema versus nonemphysematous subjects, respectively (Figure 2). The most significantly associated genotyped SNP (rs10844154) and imputed SNP (rs161976) lie in two introns flanking exon 2 of BICD1. In both sample sets, the risk alleles of these two SNPs increased in frequency in subject groups with increased amounts of emphysema (Table 3).
Figure 2.
Plot of –log10 P values for association meta-analysis of emphysema qualitative trait for BICD1 region, (A) cases defined as at least 5% emphysema, and (B) cases defined as greater than 25% emphysema versus chronic obstructive pulmonary disease (COPD) without emphysema. “Round”: imputed; “Diamond”: typed. Color indicates r2 with the most significant and typed SNP (blue diamond). Recombination rate data were based on using Build 36 coordinates, which was downloaded from HapMap.
TABLE 3.
RISK ALLELE FREQUENCY OF TOP TWO SINGLE-NUCLEOTIDE POLYMORPHISMS IN DIFFERENT SUBJECT GROUPS
| Risk Allele Frequency |
||||
|---|---|---|---|---|
| Affected (%) | n | rs10844154 | rs161976* | |
| ECLIPSE | ≤5 | 390 | 0.485 | 0.479 |
| >5–25 | 341 | 0.572 | 0.556 | |
| >25 | 826 | 0.604 | 0.594 | |
| Norway cohort | ≤5 | 144 | 0.583 | 0.552 |
| >5–25 | 116 | 0.599 | 0.581 | |
| >25 | 172 | 0.671 | 0.642 | |
Allele frequency in the Norway cohort on rs161976 was estimated on the basis of imputed genotype data.
GWAS on Quantitative Emphysema in ECLIPSE, Norway, and NETT
We performed a genome-wide association analysis for quantitative densitometric emphysema, combining the results in ECLIPSE, Norway, and NETT by meta-analysis. In contrast to the results of the qualitative emphysema analysis, none of the SNPs showed genome-wide significant association either in our primary analysis, that is, with correction for BMI (Table 4; also see Figure E1 in the online supplement), or in our secondary analysis, that is, without correction for BMI (Table E2 and Figure E1). In meta-analysis, SNP rs7905537, located in an intergenic region in 10p11.2, had the most significant P value of 6.82 × 10−7 with correction for BMI.
TABLE 4.
ECLIPSE/NORWAY COHORT/NETT META-ANALYSIS OF EMPHYSEMA QUANTITATIVE TRAIT WITH CORRECTION FOR BODY MASS INDEX: LOCI WITH META-ANALYSIS P VALUE < 10−5
| Closest Gene | Risk/Nonrisk Allele | Risk Allele Frequency | ECLIPSE |
Norway |
NETT |
Meta P Value* | Meta β (P Value)† | Hetero-geneity I2 (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Type | β | P Value | β | P Value | β | P Value | ||||||
| rs7905537 | 10 | Intergenic | PARD3 | A/C | 0.75 | 2.22 | 4.0 × 10−6 | 1.14 | 0.17 | 1.40 | 0.10 | 6.8 × 10−7 | 1.85 (8.1 × 10−7) | 0 |
| rs9292394 | 5 | Intergenic | CDH6 | T/G | 0.78 | 1.35 | 6.6 × 10−3 | 2.96 | 5.3 × 10−4 | 2.21 | 0.02 | 3.5 × 10−6 | 1.83 (2.1 × 10−6) | 31 |
| rs7911712 | 10 | In noncoding gene‡ | AL139119.2 | A/G | 0.15 | 1.60 | 4.4 × 10−3 | 3.51 | 6.5 × 10−4 | 2.11 | 0.07 | 5.4 × 10−6 | 2.05 (5.5 × 10−6) | 25 |
| rs224731 | 10 | Intergenic | PARD3 | A/G | 0.71 | 1.75 | 1.4 × 10−4 | 1.50 | 6.7 × 10−2 | 1.36 | 0.10 | 6.0 × 10−6 | 1.63 (6.4 × 10−6) | 0 |
Definition of abbreviations: Chr = chromosome; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; NETT = National Emphysema Treatment Trial; SNP = single-nucleotide polymorphism; β = regression coefficient under an additive genetic model.
Based on weighted Z-score method.
Based on fixed-effects meta-regression; I2: I2 index used to assess magnitude of heterogeneity of b among studies.
The SNP is located within a noncoding gene; the transcript of the gene is not translated into a protein and is classified as “processed transcript” in Ensembl.
To evaluate the effect of BMI on SNP–trait associations, we selected SNPs with meta-analysis P values less than 5 × 10−5 from the analysis either with or without correction for BMI and subsequently examined their P value and the effect size defined by the β in the linear regression model. Twenty-one of the selected SNPs had more than a 10-fold change in P values when BMI was used for correction, and 13 SNPs had more than a 20% change in estimated effect sizes (see Table E3), suggesting that BMI did impact the observed association between these SNPs and the quantitative emphysema. To test whether or not the difference in estimated SNP effects was caused by association between these SNPs and BMI, we then tested SNP–BMI associations. No significant association was found (4 of the 56 SNPs had P values < 0.05 and the most significant P value was 0.008; none of them was significant after Bonferroni correction for multiple comparisons).
DISCUSSION
This multicohort study is the largest reported genetic association analysis of radiographic emphysema in COPD to date, and the first genome-wide association study for this disease phenotype. Our most significant finding was at the BICD1 locus with the presence of emphysema as a radiologist-assessed qualitative trait; this locus has not been previously implicated in the pathogenesis of emphysema.
The GWAS results showed little overlap between the extent of emphysema assessed using quantitative assessment of low attenuation areas and radiologist qualitative scores. For the top 254 SNPs with meta-analysis P values less than 5 × 10−4 from association analysis of qualitative emphysema, only 4 and 5 of them have meta-analysis P values less than 5 × 10−4 for association with quantitative emphysema with and without adjustment for BMI, respectively. These differences are likely due to several factors. First, although the correlation between qualitative (radiologist-derived) and quantitative (low attenuation areas) analysis is statistically significant, the magnitude of this correlation suggests that there are still substantial differences in the extent of emphysema determined by these two methods (27, 38); in the present study, 11.5 and 2.3% of subjects in the ECLIPSE and Norway cohorts, respectively, who had absent or trivial emphysema by radiologist score, had −950 HU values above the bottom quartile of quantitative emphysema measurements based on subjects with severe emphysema by radiologist score. Second, the NETT study was included in the quantitative, but not the qualitative, analysis; although theoretically inclusion of this study should increase power, differences in the study population and the measurement of emphysema could have contributed to increased heterogeneity and thus reduced the power of our analysis. Third, it is known that BMI, radiation dose, and slice thickness affect the apparent X-ray attenuation values in the CT scan and, therefore, quantitative analysis of low-attenuation areas (39, 40).
The locus most highly associated with qualitative emphysema is in a region that covers the second exon of the gene BICD1. BICD1 is one of two human homologs of Drosophila bicaudal-D (BicD). The function of BICD protein has been well characterized—it forms a complex with dynein–dynactin and plays a critical role in mediating dynein function. Dynein is involved in many cellular processes, including mitosis, nuclear migration and mRNA transport, and transport of a variety of axonal and dendritic vesicles. The second exon of BICD1 encodes the sequence located in the coiled-coil domain at the N terminus of the protein, which directly interacts with dynein (41–43). Mangino and colleagues, following up a linkage study, identified SNPs in the first intron of BICD1 gene associated with telomere length in leukocytes (44, 45); the variants were associated with BICD1 mRNA levels and the estimated effect of the variants was equivalent to between 15 and 20 years of age-related attrition in telomere length (44). Although the identified SNPs in our study were located in a different linkage disequilibrium block, their findings suggest that the effect of BICD1 in emphysema could be related to shortened telomeres, which has been previously observed in patients with COPD (46). Directly studying the association among variants in BICD1, telomere length, and emphysema is a reasonable next step for investigation.
Our second-ranked locus in the qualitative emphysema analysis did not reach genome-wide significance; this locus is at 8p23, a genomic region showing linkage with postbronchodilator FEV1 in families ascertained through early-onset COPD (47) and 55 kb downstream from CSMD1, a tumor suppressor gene known to be important in lung cancer (48). In our analysis of quantitative emphysema, the most significant association with correction for BMI was in chromosome 10, approximately 170 kb downstream of PARD3 [par-3 partitioning defective 3 homolog (C. elegans)], a gene important in maintaining tight junctions, with homozygous deletions reported in lung cancer cell lines (49). Without correction for BMI (Table E2), our most significant association was at rs1348350, 115 kb downstream from MBL2 [mannose-binding lectin (protein C) 2], a pattern recognition receptor important for innate immunity, genetic variants of which have been associated with hospitalization for COPD (50), and with age of first Pseudomonas aeruginosa infection and speed of pulmonary function decline in children with cystic fibrosis (51). Another potentially interesting signal was at rs4905179, 6 kb upstream from SERPINA6 [serpin peptidase inhibitor, clade A (α1-antiproteinase, antitrypsin), member 6], and 37 and 49 kb downstream from SERPINA2 and SERPINA1 (the gene encoding α1-antitrypsin), respectively; SERPINA6 is also known as corticosteroid-binding globulin, and differential expression of the mouse orthologous gene has been reported between strains differentially susceptible to cigarette smoke (52). Whether these associations are indeed true susceptibility loci for emphysema remains to be determined.
COPD is a complex disease, and the chronic airflow limitation that is characteristic of COPD results from a combination of pathological processes, including emphysema. The contribution that each of these factors makes in an individual subject cannot be determined by lung function measurements alone. In addition, assessment of emphysema severity may have important prognostic and therapeutic implications. Chest CT scans provide a sensitive and noninvasive method to identify emphysema in patients with COPD. Because phenotypic heterogeneity is one of the key issues in identifying consistent genetic association results in COPD, objective classification of patients with COPD based on emphysema measurements using CT may help reduce this heterogeneity. In addition to being the largest reported study in which CT analysis was conducted to objectively define emphysema, our subjects represented a range of severity as assessed by lung function.
Our study also had several limitations. The CT scans performed in each cohort did not use identical CT acquisition protocols or scanner types, and these differences might have led to differences in the measurement of the extent of emphysema; however, protocols were standardized within each study. Moreover, our strongest signal is from radiologist-defined emphysema, which is arguably less affected by the previously described factors. Although we obtained more significant results by radiologist scoring, our findings do not necessarily indicate the superiority of one method over another for future analyses. Radiologist scoring and density mask analysis methods each have their advantages and disadvantages, and although our study is large for a CT-based study, it is relatively small and underpowered to detect effect sizes typically found by genome-wide association studies.
In conclusion, we report the first GWAS analysis of CT-defined emphysema in patients with COPD and identified BICD1 as a potential susceptibility gene. BICD1 has been suggested to be involved in regulating telomere length, and it has been previously suggested that telomere length is altered in cigarette smoke–induced emphysema. Several loci associated with both qualitative and quantitative emphysema were also identified that did not reach genome-wide significance levels. Additional studies are needed to evaluate the role of these genes in emphysema.
Supplementary Material
Acknowledgments
The authors acknowledge the ECLIPSE steering and scientific committees and investigators (listed below). ECLIPSE Steering Committee: Harvey Coxson (Canada), Lisa Edwards (GlaxoSmithKline, USA), Katharine Knobil (Co-chair, GlaxoSmithKline, UK), David Lomas (UK), William MacNee (UK), Edwin Silverman (USA), Ruth Tal-Singer (GlaxoSmithKline, USA), Jørgen Vestbo (Co-chair, Denmark), Julie Yates (GlaxoSmithKline, USA). ECLIPSE Scientific Committee: Alvar Agusti (Spain), Peter Calverley (UK), Bartolome Celli (USA), Courtney Crim (GlaxoSmithKline, USA), Bruce Miller (GlaxoSmithKline, UK), William MacNee (Chair, UK), Stephen Rennard (USA), Ruth Tal-Singer (GlaxoSmithKline, USA), Emiel Wouters (The Netherlands), Julie Yates (GlaxoSmithKline, USA). ECLIPSE Investigators: Bulgaria: Yavor Ivanov, Pleven; Kosta Kostov, Sofia. Canada: Jean Bourbeau, Montreal, Que Mark Fitzgerald, Vancouver, BC; Paul Hernandez, Halifax, NS; Kieran Killian, Hamilton, On; Robert Levy, Vancouver, BC; Francois Maltais, Montreal, Que; Denis O'Donnell, Kingston, On. Czech Republic: Jan Krepelka, Praha. Denmark: Jørgen Vestbo, Hvidovre. Netherlands: Emiel Wouters, Horn-Maastricht. New Zealand: Dean Quinn, Wellington. Norway: Per Bakke, Bergen. Slovenia: Mitja Kosnik, Golnik. Spain: Alvar Agusti, Jaume Sauleda, Palma de Mallorca. Ukraine: Yuri Feschenko, Kiev; Vladamir Gavrisyuk, Kiev; Lyudmila Yashina, Kiev; Nadezhda Monogarova, Donetsk. UK: Peter Calverley, Liverpool; David Lomas, Cambridge; William MacNee, Edinburgh; David Singh, Manchester; Jadwiga Wedzicha, London. United States of America: Antonio Anzueto, San Antonio, TX; Sidney Braman, Providence, RI; Richard Casaburi, Torrance CA; Bart Celli, Boston, MA; Glenn Giessel, Richmond, VA; Mark Gotfried, Phoenix, AZ; Gary Greenwald, Rancho Mirage, CA; Nicola Hanania, Houston, TX; Don Mahler, Lebanon, NH; Barry Make, Denver, CO; Stephen Rennard, Omaha, NE; Carolyn Rochester, New Haven, CT; Paul Scanlon, Rochester, MN; Dan Schuller, Omaha, NE; Frank Sciurba, Pittsburgh, PA; Amir Sharafkhaneh, Houston, TX; Thomas Siler, St. Charles, MO, Edwin Silverman, Boston, MA; Adam Wanner, Miami, FL; Robert Wise, Baltimore, MD; Richard ZuWallack, Hartford, CT. NETT Study: We acknowledge the co-investigators in the NETT Genetics Ancillary Study including Joshua Benditt, Gerard Criner, Malcolm DeCamp, Philip Diaz, Mark Ginsburg, Larry Kaiser, Marcia Katz, Mark Krasna, Neil MacIntyre, Barry Make, Rob McKenna, Fernando Martinez, Zab Mosenifar, John Reilly, Andrew Ries, Paul Scanlon, Frank Sciurba, and James Utz.
Supported by GlaxoSmithKline (the Norway cohort [RES11080] and the ECLIPSE Study [Clinicaltrials.gov identifier NCT00292552; GSK Code SCO104960]) and by contracts with the National Heart, Lung, and Blood Institute (N01HR76101-N01HR76116, N01HR76118, N01HR76119), the Centers for Medicare and Medicaid Services, and the Agency for Healthcare Research and Quality (the National Emphysema Treatment Trial). This work was supported by U.S. National Institutes of Health K12HL089990 (M.H.C.), and R01HL075478, R01HL084323, and P01HL083069 (E.K.S.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201004-0541OC on August 27, 2010
Author Disclosure: X.K. is an employee of GlaxoSmithKline. M.H.C. received more than $100,001 from the NIH in sponsored grants as an NIH K12 grant. W.A. is an employee of GlaxoSmithKline and holds more than $100,001 in stock in GlaxoSmithKline. W.A.'s employer, GlaxoSmithKline, provides more than $100,001 in sponsored grants as educational support for ATS meetings. H.O.C. received up to $1,000 from GlaxoSmithKline and up to $1,000 from AstraZeneca in consultancy fees, $1,001–$5,000 from GlaxoSmithKline in advisory board fees, up to $1,000 from AstraZeneca in lecture fees, more than $100,001 from GlaxoSmithKline, $50,001–$100,000 from Spiration Inc, and more than $100,001 from Wyeth in industry-sponsored grants, and more than $100,001 from the NIH and more than $100,001 from the CIHR in sponsored grants. N.M. received more than $100,001 from GlaxoSmithKline in industry-sponsored grants. G.W. received $1,001–$5,000 from MedImmune in consultancy fees and more than $100,001 from the NHLBI in sponsored grants as a K23 Career Development Award. E.H. received $1,001–$5,000 from QI2, $1,001–$5,000 from AstraZeneca, $1,001–$5,000 from Sanofi-Aventis, and $1,001–$5,000 from Grifols in consultancy fees for imaging protocol advice and oversight, up to $1,000 from Siemens Medical Systems in advisory board fees (travel expenses only), $1,001–$5,000 from QI2, $1,001–$5,000 from AstraZeneca, $1,001–$5,000 from Sanofi-Aventis, and $1,001–$5,000 from Grifols in lecture fees, holds a patent from AMFM for texture analysis of medical images, more than $100,001 in stock ownership and options from VIDA Diagnostics as founder and shareholder, and more than $100,001 from the NIH as NIH RO1 grants and contracts. E.H.'s primary potential conflict of interest is as a founder and shareholder of VIDA Diagnostics, which is commercializing image analysis software, developed in E.H.'s university laboratory, which has been used in this study. P.B. received $1,001–$5,000 from GlaxoSmithKline and $1,001–$5,000 from AstraZeneca in lecture fees, $10,001–$50,000 from GlaxoSmithKline in industry-sponsored grants as a principal investigator. A.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.A.L. received $10,001–$50,000 from GlaxoSmithKline, $1,001–$5,000 from Novartis, up to $1,000 from Genzyme, and up to $1,000 from Amicus in consultancy fees, $10,001–$50,000 from GlaxoSmithKline, $1,001–$5,000 from Talecris, and $1,001–$5,000 from Thorax in advisory board fees, $1,001–$5,000 from GlaxoSmithKline, up to $1,000 from AstraZeneca, $1,001–$5,000 from Boehringer Ingelheim, and $1,001–$5,000 from LFB in lecture fees, more than $100,001 from GlaxoSmithKline and more than $100,001 from Merck, Sharpe & Dohme in industry-sponsored grants, more than $100,001 from the Medical Research Council, more than $100,001 from the Wellcome Trust, and more than $100,001 from the British Lung Foundation in sponsored grants, and $1,001–$5,000 from the Medical Research Council in advisory board fees. E.K.S. received $10,001–$50,000 from GlaxoSmithKline, and $10,001–$50,000 from AstraZeneca in consultancy fees, $1,001–$5,000 from GlaxoSmithKline, $5,001–$10,000 from AstraZeneca, and $1,001–$5,000 from Bayer in lecture fees, more than $100,001 from GlaxoSmithKline in industry-sponsored grants, and more than $100,001 from the NIH in sponsored grants.
References
- 1.Celli BR, MacNee W. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J 2004;23:932–946. [DOI] [PubMed] [Google Scholar]
- 2.Murray CJ, Lopez AD. Alternative projections of mortality and disability by cause 1990–2020: Global Burden of Disease Study. Lancet 1997;349:1498–1504. [DOI] [PubMed] [Google Scholar]
- 3.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O'Donnell WJ, Reilly JJ, Ginns L, Mentzer S, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med 1998;157:1770–1778. [DOI] [PubMed] [Google Scholar]
- 4.McCloskey SC, Patel BD, Hinchliffe SJ, Reid ED, Wareham NJ, Lomas DA. Siblings of patients with severe chronic obstructive pulmonary disease have a significant risk of airflow obstruction. Am J Respir Crit Care Med 2001;164:1419–1424. [DOI] [PubMed] [Google Scholar]
- 5.Silverman EK, Sandhaus RA. α1-Antitrypsin deficiency. N Engl J Med 2009;360:2749–2757. [DOI] [PubMed] [Google Scholar]
- 6.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009;5:e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilk JB, Chen Th, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet 2009;5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet 2010;42:200–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim WJ, Silverman EK, Hoffman E, Criner GJ, Mosenifar Z, Sciurba FC, Make BJ, Carey V, Estepar RS, Diaz A, et al. CT metrics of airway disease and emphysema in severe COPD. Chest 2009;136:396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel BD, Coxson HO, Pillai SG, Agusti AG, Calverley PM, Donner CF, Make BJ, Muller NL, Rennard SI, Vestbo J, et al. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2008;178:500–505. [DOI] [PubMed] [Google Scholar]
- 11.Maclay JD, Rabinovich RA, MacNee W. Update in chronic obstructive pulmonary disease 2008. Am J Respir Crit Care Med 2009;179:533–541. [DOI] [PubMed] [Google Scholar]
- 12.Boschetto P, Quintavalle S, Zeni E, Leprotti S, Potena A, Ballerin L, Papi A, Palladini G, Luisetti M, Annovazzi L, et al. Association between markers of emphysema and more severe chronic obstructive pulmonary disease. Thorax 2006;61:1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa E, Nakano Y, Ohara T, Muro S, Hirai T, Sato S, Sakai H, Tsukino M, Kinose D, Nishioka M, et al. Body mass index in male patients with COPD: correlation with low attenuation areas on CT. Thorax 2009;64:20–25. [DOI] [PubMed] [Google Scholar]
- 14.Burrows B, Fletcher CM, Heard BE, Jones NL, Wootliff JS. The emphysematous and bronchial types of chronic airways obstruction: a clinicopathological study of patients in London and Chicago. Lancet 1966;287:830–835. [DOI] [PubMed] [Google Scholar]
- 15.McAllister DA, Maclay JD, Mills NL, Mair G, Miller J, Anderson D, Newby DE, Murchison JT, MacNee W. Arterial stiffness is independently associated with emphysema severity in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2007;176:1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson DO, Weissfeld JL, Balkan A, Schragin JG, Fuhrman CR, Fisher SN, Wilson J, Leader JK, Siegfried JM, Shapiro SD, et al. Association of radiographic emphysema and airflow obstruction with lung cancer. Am J Respir Crit Care Med 2008;178:738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan ES, Silverman EK. Genetics of COPD and emphysema. Chest 2009;136:859–866. [DOI] [PubMed] [Google Scholar]
- 18.Kong X, Cho MH, Anderson W, Coxson H, Lomas D, Silverman EK, Bakke PS, Müller N, Pillai S. Genome-wide association study in two independent COPD populations identifies variants in the BICD1 gene associated with emphysema [abstract]. Am J Respir Crit Care Med 2010;181:A3740. [Google Scholar]
- 19.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, Hagan G, Knobil K, Lomas DA, MacNee W, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE). Eur Respir J 2008;31:869–873. [DOI] [PubMed] [Google Scholar]
- 20.Lomas DA, Silverman EK, Edwards LD, Locantore NW, Miller BE, Horstman DH, Tal-Singer R; Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints Study Investigators. Serum surfactant protein D is steroid sensitive and associated with exacerbations of COPD. Eur Respir J 2009;34:95–102. [DOI] [PubMed] [Google Scholar]
- 21.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, Anderson WH, Lomas DA, Silverman EK, Pillai SG; International COPD Genetics Network (ICGN) Investigators. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med 2007;176:167–173. [DOI] [PubMed] [Google Scholar]
- 22.Pillai SG, Zhu G, Gulsvik A, Lomas DA, Silverman EK. SERPINE2 and COPD. Am J Respir Crit Care Med 2007;176:726. [DOI] [PubMed] [Google Scholar]
- 23.Eagan TML, Gulsvik A, Eide GE, Bakke PS. Remission of respiratory symptoms by smoking and occupational exposure in a cohort study. Eur Respir J 2004;23:589–594. [DOI] [PubMed] [Google Scholar]
- 24.Brogger J, Eagan T, Eide G, Bakke P, Gulsvik A. Bias in retrospective studies of trends in asthma incidence. Eur Respir J 2004;23:281–286. [DOI] [PubMed] [Google Scholar]
- 25.National Emphysema Treatment Trial Research Group. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med 2003;348:2059–2073. [DOI] [PubMed] [Google Scholar]
- 26.Crapo RO, Morris AH. Standardized single breath normal values for carbon monoxide diffusing capacity. Am Rev Respir Dis 1981;123:185–189. [DOI] [PubMed] [Google Scholar]
- 27.DeMeo DL, Hersh CP, Hoffman EA, Litonjua AA, Lazarus R, Sparrow D, Benditt JO, Criner G, Make B, Martinez FJ, et al. Genetic determinants of emphysema distribution in the National Emphysema Treatment Trial. Am J Respir Crit Care Med 2007;176:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grydeland TB, Dirksen A, Coxson HO, Pillai SG, Sharma S, Eide GE, Gulsvik A, Bakke PS. Quantitative computed tomography: emphysema and airway wall thickness by sex, age and smoking. Eur Respir J 2009;34:858–865. [DOI] [PubMed] [Google Scholar]
- 29.National Emphysema Treatment Trial Research Group. Patients at high risk of death after lung-volume-reduction surgery. N Engl J Med 2001;345:1075–1083. [DOI] [PubMed] [Google Scholar]
- 30.Bankier AA, De Maertelaer V, Keyzer C, Gevenois PA. Pulmonary emphysema: subjective visual grading versus objective quantification with macroscopic morphometry and thin-section CT densitometry. Radiology 1999;211:851–858. [DOI] [PubMed] [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 32.Parr DG, Stoel B, Stolk J, Stockley RA. The influence of BMI on lung CT densitometry in emphysema. Thorax 2002;57:iii58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet 2009;10:387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 2008;32:227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The International HapMap Consortium. A haplotype map of the human genome. Nature 2005;437:1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hersh CP, Washko GR, Jacobson FL, Gill R, Estepar RS, Reilly JJ, Silverman EK. Interobserver variability in the determination of upper lobe–predominant emphysema. Chest 2007;131:424–431. [DOI] [PubMed] [Google Scholar]
- 39.Yuan R, Mayo JR, Hogg JC, Pare PD, McWilliams AM, Lam S, Coxson HO. The effects of radiation dose and CT manufacturer on measurements of lung densitometry. Chest 2007;132:617–623. [DOI] [PubMed] [Google Scholar]
- 40.Madani A, De Maertelaer V, Zanen J, Gevenois PA. Pulmonary emphysema: radiation dose and section thickness at multidetector CT quantification—comparison with macroscopic and microscopic morphometry. Radiology 2007;243:250–257. [DOI] [PubMed] [Google Scholar]
- 41.Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, Willemsen R, Visser P, Grosveld F, Galjart N. Mammalian Golgi–associated Bicaudal-D2 functions in the dynein–dynactin pathway by interacting with these complexes. EMBO J 2001;20:4041–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoogenraad CC, Wulf P, Schiefermeier N, Stepanova T, Galjart N, Small JV, Grosveld F, De Zeeuw CI, Akhmanova A. Bicaudal D induces selective dynein-mediated microtubule minus end–directed transport. EMBO J 2003;22:6004–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matanis T, Akhmanova A, Wulf P, Del Nery E, Weide T, Stepanova T, Galjart N, Grosveld F, Goud B, De Zeeuw CI, et al. Bicaudal-D regulates COPI-independent Golgi–ER transport by recruiting the dynein–dynactin motor complex. Nat Cell Biol 2002;4:986–992. [DOI] [PubMed] [Google Scholar]
- 44.Mangino M, Brouilette S, Braund P, Tirmizi N, Vasa-Nicotera M, Thompson JR, Samani NJ. A regulatory SNP of the BICD1 gene contributes to telomere length variation in humans. Hum Mol Genet 2008;17:2518–2523. [DOI] [PubMed] [Google Scholar]
- 45.Vasa-Nicotera M, Brouilette S, Mangino M, Thompson JR, Braund P, Clemitson JR, Mason A, Bodycote CL, Raleigh SM, Louis E, et al. Mapping of a major locus that determines telomere length in humans. Am J Hum Genet 2005;76:147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savale L, Chaouat A, Bastuji-Garin S, Marcos E, Boyer L, Maitre B, Sarni M, Housset B, Weitzenblum E, Matrat M, et al. Shortened telomeres in circulating leukocytes of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2009;179:566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer LJ, Celedon JC, Chapman HA, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage analysis of bronchodilator responsiveness and post-bronchodilator spirometric phenotypes in chronic obstructive pulmonary disease. Hum Mol Genet 2003;12:1199–1210. [DOI] [PubMed] [Google Scholar]
- 48.Ma C, Quesnelle KM, Sparano A, Rao S, Park MS, Cohen MA, Wang Y, Samanta M, Kumar MS, Aziz MU, et al. Characterization CSMD1 in a large set of primary lung, head and neck, breast and skin cancer tissues. Cancer Biol Ther 2009;8:907–916. [DOI] [PubMed] [Google Scholar]
- 49.Nagayama K, Kohno T, Sato M, Arai Y, Minna JD, Yokota J. Homozygous deletion scanning of the lung cancer genome at a 100-kb resolution. Genes Chromosomes Cancer 2007;46:1000–1010. [DOI] [PubMed] [Google Scholar]
- 50.Yang IA, Seeney SL, Wolter JM, Anders EM, McCormack JG, Tunnicliffe AM, Rabnott GC, Shaw JG, Dent AG, Kim ST, et al. Mannose-binding lectin gene polymorphism predicts hospital admissions for COPD infections. Genes Immun 2003;4:269–274. [DOI] [PubMed] [Google Scholar]
- 51.Dorfman R, Sandford A, Taylor C, Huang B, Frangolias D, Wang Y, Sang R, Pereira L, Sun L, Berthiaume Y, et al. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J Clin Invest 2008;118:1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavarra E, Fardin P, Fineschi S, Ricciardi A, De CG, Sallustio F, Zorzetto M, Luisetti M, Pfeffer U, Lungarella G, et al. Early response of gene clusters is associated with mouse lung resistance or sensitivity to cigarette smoke. Am J Physiol Lung Cell Mol Physiol 2009;296:L418–L429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.