Abstract
Proper simulation of dynamic properties, including molecular diffusion, is an important goal of empirical force fields. However, the widely used TIP3P water model does not reproduce the experimental viscosity of water. Consequently, scaling of simulated diffusion constants of solutes in aqueous solutions is required to effectively compare them with experiment. It is proposed that scaling by the ratio of viscosities of model and real water is appropriate in the regime where the concentration dependence of simulated and experimental solution viscosities is parallel. With this ansatz, viscosity scaling can be carried out for glucose and trehalose up to 20 wt% for simulations carried out with the CHARMM additive carbohydrate force field C35 and TIP3P water; above this value, the concentration dependence of simulated viscosities lags that of experiment, and scaling is not advised. Scaled translational diffusion constants for glucose, and the disaccharides trehalose, maltose, and melibiose at low concentration agree nearly quantitatively with experiment, as do NMR 13C T1’s for glucose, trehalose, and maltose; these results support the use of C35 for simulations of sugar transport properties at low concentration. At high concentrations the scaled diffusion constants for glucose and trehalose underestimate and overestimate experiment, respectively. Hydrodynamic bead model calculations indicate a hydration level of approximately 1 water/hydroxyl for glucose. Patterns for the disaccharides are more complicated, though trehalose binds 0.5 to 1 more water than does maltose depending on the analysis.
1. Introduction
This paper concerns the comparison of diffusion constants obtained from simulation based on empirical force fields and experiment. While many considerations are general, the focus is on validating the recently developed CHARMM additive carbohydrate1,2 force field (FF), C35, using the translational diffusion constant, D, and NMR spin lattice relaxation time, T1, for mono and disaccharides. Both of these properties are sensitive to solute-solvent interactions in that bound water increases the effective hydrodynamic size of the solute, and therefore the translational and rotational friction. T1’s also contain contributions from internal motions such as libration in torsional minima and isomerization, and can thereby provide a sensitive probe of the barriers for the dihedral potentials. In practice, a direct comparison of simulation and experimental D and T1 is confounded by inaccuracies in the viscosity of the water model, and sensitivity to cutoff methods. For example, η20 (the viscosity η at 20 °C) for TIP3P water was shown to equal 0.62 and 0.35 cP when force-switching (12 Å cutoff) and Particle Mesh Ewald (PME), respectively, were used to treat the electrostatic interactions;3 the experimental value is 1.00 cP. More recent evaluations of η20 with PME and 3D-IPS/DFFT to treat the Lennard Jones (LJ) interactions yielded a value of 0.33 cP.4 Hence, a direct comparison of simulated and experimental results is clearly of limited usefulness because the results will be dominated by the errors in the water model.
Rotational and translational diffusion constants for polar molecules in water may be assumed to scale as η−1 under most conditions. Hence, it is reasonable to propose that the viscosity in the simulation system could be scaled to experiment when comparing diffusion constants. This paper considers cases where a scaling approach is appropriate, and when it may lead to fortuitous agreement with experiment. The idea is simple: simulated diffusion constants and rotational relaxation times for solutes in water may be scaled by the ratio of pure experimental and simulated water in the regime where the concentration dependences of viscosities match. The underlying assumption of such a scaling is that differences with experiment arise from deficiencies in the water model, not the solute model.
Following a description of the Methods (Section 2), Section 3.1 reports the simulated viscosities for pure TIP3P water at a range of temperatures, and for glucose and trehalose at a range of concentrations. These provide upper limits for the analysis of simulated and experimental D and T1 presented for glucose, trehalose, maltose and melibiose (Fig 1) in Sections 3.2 and 3.3. Section 3.4 presents the results of hydrodynamic calculations with varying levels of bound water to examine differences in the preceding carbohydrates. The Discussion and Conclusions (Section 4) includes examples where scaling is not advised.
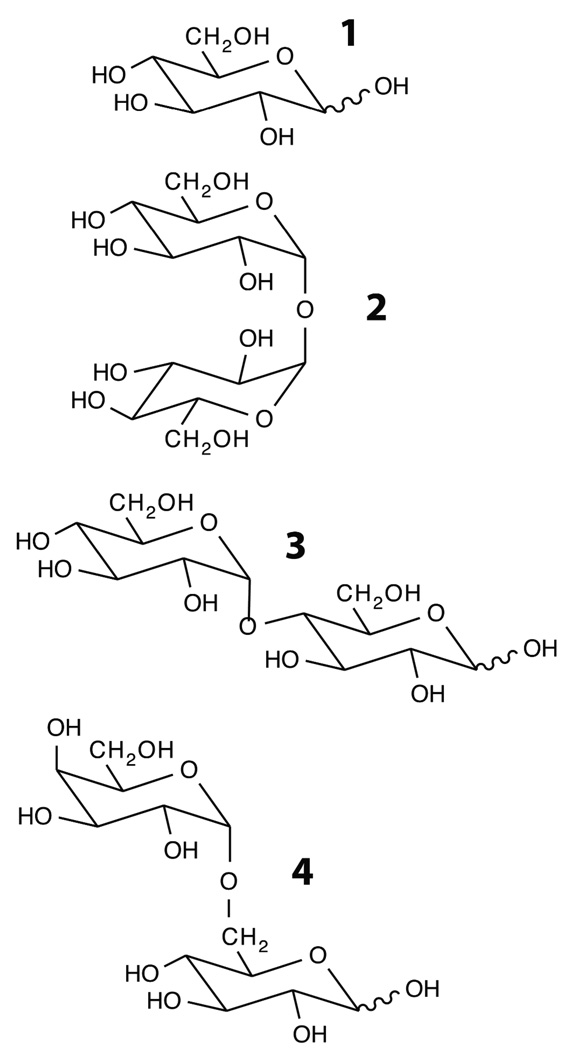
Figure 1.
Molecular structures of D-glucose (1), α,α-trehalose (2), maltose (3), and melibiose (4).
2. Methods
2.1 Simulation details
All simulations and primary data analysis or data extractions were performed with the CHARMM5 program (version c35b3), using the recent C35 carbohydrate FF.1,2 Starting with the c35b1 general release, the program includes a feature to save the pressure tensor at every integration step, essential for the accurate calculation of viscosity. All systems were simulated with a 1 fs time step and the Particle Mesh Ewald (PME) method6 for electrostatics, and a Lennard-Jones (LJ) switching function for the van der Waals interactions;7 the PME real space and LJ cutoff was 12 Å, the parameter set default. The PME calculations used a 5th order polynomial spline lookup, κ = 0.32 Å−1, and grid spacing ≈ 1 Å. Additional water and glucose simulations employed the 3D-IPS/DFFT (Three-Dimensional Isotropic Periodic Sum with the Discrete Fast Fourier Transform extension) non-bond method8 for both electrostatics and LJ interactions, with a real space cutoff of 10 Å. This method, henceforth denoted IPS, obtains both long-range electrostatic and LJ interactions; it is an alternative to PME with a long-range LJ correction. Cubic periodic boundary conditions were used for all simulations, in conjunction with the extended system methods9,10 for the NVT and NPT ensembles (N is particle number, V is volume, T is temperature, and P is pressure).
A box of 1340 TIP3P11,12 water molecules, with a cube edge length of ca. 34 Å, was simulated in triplicate for 12.5 ns at T = 20, 25, 30, 40 and 50 °C, using both PME and IPS non-bond methods (30 simulations total) in the NVT ensemble. By changing the random seed, each of the 3 replicate simulations was assigned different initial velocities. The edge length was set to match the experimental density for water at each T. The Hoover thermostat with a coupling constant of 5000 kcal mol−1 ps−2 was used to regulate the temperature. The last 12 ns were used for data analysis, with standard errors computed over the replicate simulations.
Glucose (D-glucopyranose) solutions were prepared by positioning molecules on a grid based on a perfect cube (27, 64, or 125), with grid locations skipped at random, based on the number of molecules. One third of the molecules were randomly assigned an α anomeric conformation, and the rest were β, resulting in a 2:1 β:α anomeric ratio. In addition, each molecule was given a random rotation, and a small random off-lattice translation. Solutions of ca. 0.3, 0.5, 1, 2, 3, 4 and 5 molal (m) were prepared by first placing 7, 12, 24, 48, 72, 96 and 120 glucose molecules, then combining with a cube of TIP3P water large enough to encompass all of the molecules. Waters overlapping the glucose molecules were then deleted, the water box trimmed if necessary, and finally random water molecules were deleted to obtain a final count of 1320 water molecules. The coordinates and the cubic periodic boundary lattice parameters were then each briefly optimized via minimization. Each solution was then equilibrated via 500 ps of NPT dynamics at 20 °C (30 °C for 0.3 m) using the IPS method, with a pressure piston mass of 500 amu. The average volume was calculated over the last 100 ps, and the stored trajectory frame with a unit cell edge that best matched the average volume was selected for the NVT simulations. Two separate simulations, one using the PME method and one using IPS, were performed at each concentration for at least 16 ns; in order to improve convergence, those at 0.3 m and the PME simulations at 3 m and above were extended to 20 ns. The simulations at 20 °C were used to compute viscosity at the experimental temperature; for translational diffusion, PME simulations were performed via the same protocol at 25 °C for the 0.5 m and higher concentrations. Coordinate sets were saved every ps for all but the 0.3 m simulations; these were used for the rotational relaxation calculations, and coordinate sets were saved every 0.2 ps. Data analysis intervals were 15 and 18 ns, with blocks of 5 or 6 ns used to estimate standard errors.
Three melibiose (D-galactopyranosyl-α-1,6-D-glucopyranoside) solutions of 0.15, 0.20, and 0.25 m were prepared by combining 3, 4, or 5 disaccharides with 1100 waters. The solute molecules were placed on a cubic grid with grid points centered approximately 1/3 the size of the side of the cubic box then TIP3P waters were added to the box. The melibiose molecules were generated with a 2:1 β:α anomeric ratio. Following extensive NPT equilibration, the average volume at each concentration was used to run 37 ns NVT simulations with the PME method, saving coordinate sets every ps. The final 36 ns were used for analysis, with standard errors estimated from four 9 ns blocks.
Five trehalose (D-glucopyranosyl-α,α-1,1-D-glucopyranoside) solutions of ca. 0.10, 0.30, 0.56, 1.11, and 1.26 m were prepared by combining 2, 6, 11, 22, or 25 disaccharides with 1100 waters, analogous to the procedure used for melibiose. A total of seven simulations were run, with three at 25 °C at 0.10, 0.30, and 1.26 m, and four at 30 °C at 0.10, 0.30, 0.56, and 1.11 m. As for melibiose, the average NPT volume at each concentration was used for subsequent NVT simulations, and coordinate sets were saved every ps. Concentrations below 1.0 m were simulated for 25 ns each, while the two high concentrations were simulated for 37 ns each. The first ns of each simulation was used as equilibration, and four blocks of either 6 or 9 ns were used to estimate standard errors.
Solutions of ca. 0.12 m (4 wt%) maltose (D-glucopyranosyl-α-1,4-D-glucopyranoside) and solutions of trehalose at the same concentration were prepared by placing 8 molecules uniformly spaced about the origin, and combining with 3648 TIP3P waters. For each disaccharide, five replicate solutions were made, with random rotations and small random displacements used to generate five unique starting conditions. Each was simulated for 1 ns under NPT conditions; the volume for the NVT simulations was computed as the average over the last 500 ps from all five replicates for each disaccharide. A stored coordinate set which best matched the chosen volume was chosen from the last 500 ps of each replicate, and each was then simulated for 12.5 ns under NVT conditions. For the maltose replicates, 3 of the 8 molecules were assigned α anomeric conformations, and the other 5 were assigned β conformations. Coordinate sets were saved every 0.25 ps for these simulations to improve the accuracy of the short time behavior of the correlation functions used to compute rotational relaxation. The last 12 ns of each simulation were used for analysis, and standard error estimates were calculated over the five replicates. These simulations were also used for hydration shell analysis for these two disaccharides, which counted water O atoms within a shell thickness from any sugar atom (Table 6).
Table 6.
Number of watersa as a function of shell thickness for maltose and trehalose.
| Shell Thickness | maltose | trehalose |
|---|---|---|
| 2.5 | 6.1 | 6.6 |
| 2.6 | 7.5 | 7.8 |
| 2.7 | 9.5 | 9.9 |
| 2.8 | 12.5 | 12.9 |
| 2.9 | 15.6 | 16.0 |
Averages and standard errors (se) computed over 5 replicate 12 ns simulations at 4 wt% and 25 °C; the se = 0.1 for all entries.
The lowest concentration simulations for glucose, maltose, melibiose and trehalose were used to extract heavy atom coordinate sets of a sugar with nearby waters, for a series of calculations of the diffusion constant using a hydrodynamic bead model (described in the next subsection). The nearby waters were defined on the basis of an energy cutoff for the interaction of a water molecule with the sugar molecule; the cutoff was specified as a multiple of kBT, where kB is the Boltzmann constant. For each extraction, the sugar molecule was centered at the origin, and the sugar-water interaction energy computed. The sugar molecule, along with water molecules with an interaction energy more favorable than the cutoff, were written to a formatted coordinate file. Energy cutoffs of -2, -3, -4, and -5 kBT were initially evaluated for each of the four sugars, with a second series of four sugar dependent cutoffs at intervals of 0.1 kBT in the range between -3 and -4 kBT to obtain more exact matches to experimental values. Different sugar molecules were sampled from each simulation trajectory, at intervals of 250 or 500 ps between time points; ca. 200 coordinates sets were extracted for each disaccharide at each multiple of kBT, and ca. 380 coordinates sets were extracted for glucose. The same sugar coordinates were used for the calculation of D and the solvent accessible surface area (SASA) with no hydration.
2.2. Calculation of viscosities, diffusion constants and NMR T1
Viscosities were obtained from
| (1) |
where Pαβ are the off-diagonal elements of the instantaneous pressure tensor. The integrands typically converged between 2 and 5 ps at low concentrations (< 10 wt%), and 10–20 ps at higher concentrations (10–50 wt%). Visual inspection of the integrals was required to confirm the location of the stable plateau region; the viscosity estimate is an average over a window of width 0.5 to 1.5 ps, depending on the width of the stable plateau. The pressure tensor was assumed to be symmetric (Pαβ = Pβα); the CHARMM program was used to compute the correlation function of the 3 unique off-diagonal elements, average them, and then compute the integral.
Diffusion constants evaluated from a simulation in periodic boundary conditions, DPBC, must be corrected for periodic boundary condition artifacts (a correction for viscosity is not required). The formula of Yeh and Hummer 13 is highly accurate:
| (2) |
where L is the cubic box length, and ξ = 2.837297, and η is the viscosity of the solution calculated from the simulation. The continuous (unfolded) center of mass time series for each sugar was used to compute the mean squared displacement <r2>, and DPBC was obtained from a linear fit of <r2> vs. t based on the Einstein relation for diffusion. A FORTRAN program was used to calculate D from the <r2> vs. t data, computing the slope and finite size correction.
Diffusion constants were also computed using a hydrodynamic bead model methodology developed by Garcia de la Torre and Bloomfield,14 and later tailored to included bound water.15–18 For the present application, single sugar molecules with varying amounts of bound water (including none) were prepared as described in the previous subsection. <D> and <Nb> were computed over the results at a specific energy cutoff for bound water, where Nb is the number of bound waters in each coordinate set. Translational and rotational friction tensors were evaluated with the FORTRAN program denoted here HI6 (for Hydrodynamic Interaction, version 6). The original implementation is described in ref. 15. Subsequent improvements include automated recognition of coordinate file type (original free format, PDB, or CHARMM), calculation of vector relaxation in the diffusive frame, and automatic array allocation based on input file size. HI6 is available for download at URL http://www.lobos.nih.gov/mbs/scriptprog.shtml.
Assuming that relaxation is due to dipolar interactions between the 13C nucleus and its N attached protons, the 13C NMR relaxation rates 1/NT1 for each carbon are given by19
| (3) |
where ℏ is Plank’s constant divided by 2π, rC-H is the effective C-H bond length, γH, γC, ωH, and ωC are the gyromagnetic ratios and Larmor frequencies, respectively, of the 13C and 1H nuclei; ωC = γCH and ωH = γHH, where H is the field strength. J(ω) is the spectral density of the second rank reorientational correlation function:
| (4) |
where P2 is the second order Legendre polynomial μ̂(t) and is the unit vector along the C-H bond direction at time t. If tumbling is rapid (all decays of C(t) are much less than 1/ωH), then Eq. (3) reduces to
| (5) |
where τ is the integral of C(t). Setting rC-H=1.117 Å 20 yields the numerical factor in Eq. (5). Time series of individual C-H bond vectors were extracted from the CHARMM trajectories, normalized to the unit vector, and the time correlation function computed; the C(t) were averaged over molecules (and replicate simulations when available), and then integrated numerically to obtain the relaxation time.
3. Results
3.1. Viscosity
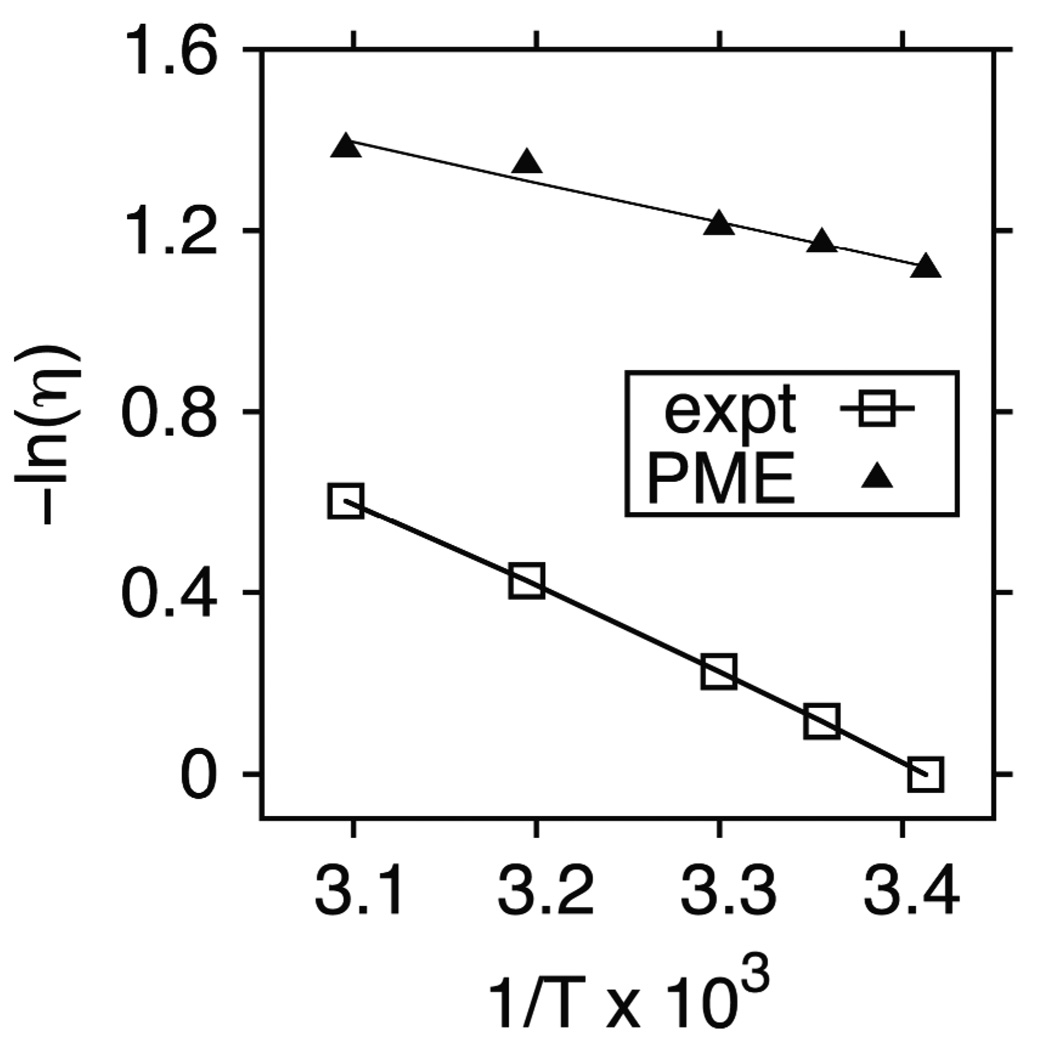
Table 1 compares the simulated viscosities for TIP3P water from 20 to 50 °C. The values at 20 °C, 0.329 cP for PME and 0.332 for IPS, are within statistical error to those obtained previously.4 New results here demonstrate that the temperature dependence of TIP3P also underestimates experiment. The activation energies Ea obtained from a plot of −lnη vs. 1/T (Fig 2) are 1.8 and 3.8 kcal/mol for PME and experimental viscosities, respectively. Interactions between real waters are stronger than those in the TIP3P water model. Consequently, different scalings must be applied at different temperatures to compensate for deficiencies in the model.
Table 1.
Simulated viscosities of TIP3P water for PME and IPS, and experimental values for pure water and D2O (in cP).
Figure 2.
Temperature dependence of water viscosity for experiment and TIP3P (see Table 1). The solid lines are linear fits to all points.
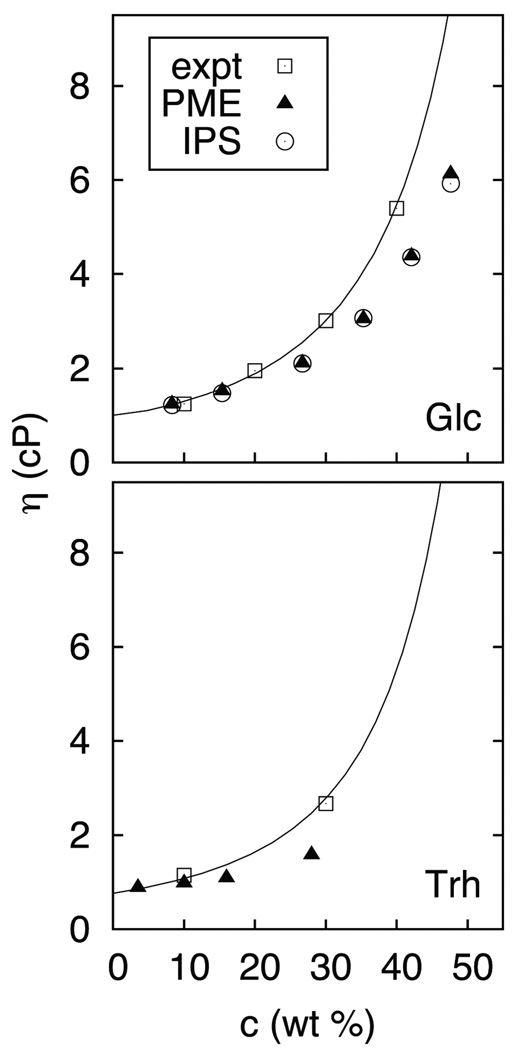
The viscosity of a solution increases with the concentration of solute. Figure 3 plots experimental and simulated viscosities (scaled by the factors in Table 1) for solutions of glucose at 20 °C (top) and trehalose at 30 °C (bottom). Errors for glucose are less than 17% up to 27 wt% (2 m), but increase to 39% for 48 wt% (5 m). Agreement for scaled simulated values and experiment is comparable for trehalose as a function of wt%. Densities of glucose solutions are within 1% of experiment up to 5 m,1 indicating that the structural properties of the FF are more robust than the dynamical ones.
Figure 3.
Experimental and simulated (scaled) viscosities for aqueous solutions of glucose at 20 °C (top) and trehalose39 at 30 °C (bottom); glucose was simulated with both PME and IPS non-bond methods. The empirical equation of Chirife and Buera41 is included with the experimental viscosities. Standard errors of scaled simulated glucose viscosities mostly ranged from 0.02 to 0.08 cP (0.24 cP for ca. 5 m Glc with PME), and were less than 0.03 cP for trehalose.
Based on these results, it is reasonable to propose that simulated diffusion constants and rotational relaxation times for solutes in water may be scaled by the ratio of pure experimental and simulated water in the regime where the concentration dependences of viscosities match; i.e., up to about 20 wt% (2 m for glucose and 1 m for trehalose) for mono and disaccharides. The underlying assumption of such a scaling is that differences with experiment arise from deficiencies in the water model, not the solute model. Scaling at higher concentrations is not advisable because agreement with experiment may be fortuitous. Concentrations used for NMR measurements are typically less than 10 wt%, so the preceding restriction is not severe.
3.2. Diffusion
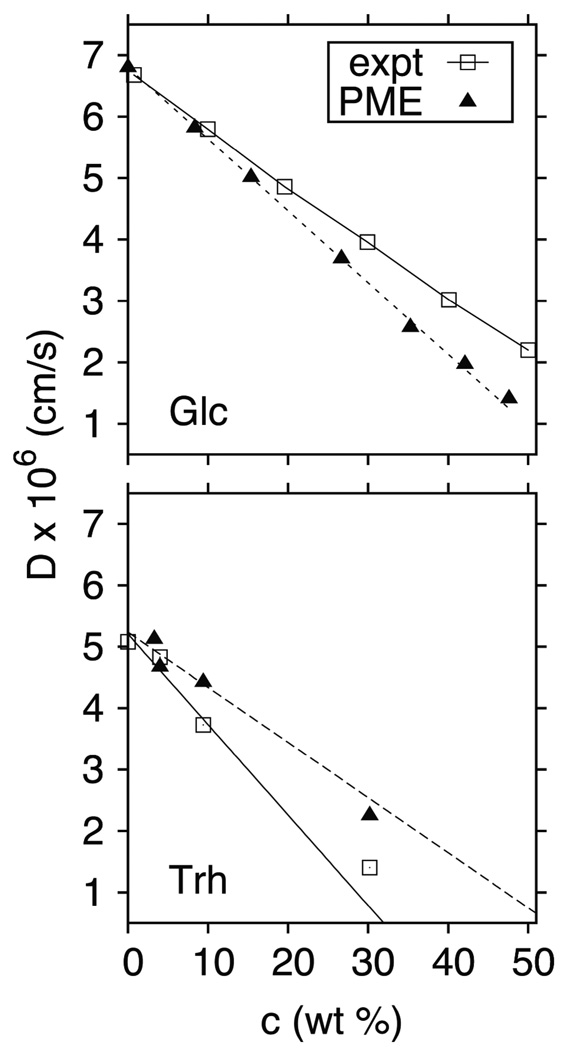
As expected from the preceding discussion of viscosity, simulated diffusion constants for glucose and trehalose evaluated substantially overestimate experiment. However, as evident in Figure 4 and Table 2, the agreement at low concentration is excellent when the simulated results are scaled by the viscosity factors listed in Table 1. In particular, the D0 values (the diffusion constant extrapolated to zero concentration) are within 3% of experiment. Table 2 contains additional results for melibiose and maltose, confirming this good agreement. Simulation and experiment deviate for glucose and trehalose as concentration increases, though in opposite directions (Fig 4). The errors for trehalose appear to be primarily associated with the viscosity; i.e., the (scaled) simulated viscosity at 30 wt% is too low by a factor of 1.5 (Fig 3) and the simulated diffusion constant is too high by a factor of 1.7. In contrast, the simulated viscosities and diffusion constants of glucose are both too low.
Figure 4.
Experimental8,21 and scaled simulated translational diffusion constants for glucose (top) and trehalose (bottom) at 25° C. The solid (experiment) and dashed (simulation) lines are linear fits to the lowest three concentrations for each case. Glucose simulation standard errors are less than 0.1×10−6 cm/s for all but the 1 M (~15 wt%) concentration, which is 0.16×10−6 cm/s; for trehalose, the simulation standard errors are 0.01×10−6 cm/s or less.
Table 2.
Experimental and viscosity scaled (Table 1) simulation translational diffusion constants for glucose and assorted disaccharides at 25 °C; values at zero concentration, D0, are extrapolated.
| c (wt %) | c (m) |
Dsim (10−6 cm2/s) |
Dexp (10−6 cm2/s) |
% error | |
|---|---|---|---|---|---|
| glucose | 0 | 0 | 6.80 | 6.75 a | 0.8 |
| 8.3 | 0.50 | 5.82 | 5.97 | −2.4 | |
| 15.4 | 1.01 | 5.01 | 5.31 | −5.6 | |
| 26.7 | 2.02 | 3.69 | 4.25 | −13.3 | |
| 35.3 | 3.03 | 2.57 | 3.44 | −25.4 | |
| 42.1 | 4.04 | 1.97 | 2.81 | −29.6 | |
| 47.6 | 5.05 | 1.41 | 2.29 | −38.2 | |
| trehalose | 0 | 0 | 5.24 | 5.08 b | 3.1 |
| 3.3 | 0.101 | 5.12 | |||
| 4.0 | 0.122 | 4.67 | 4.83 c | −3.3 | |
| 9.4 | 0.303 | 4.42 | 3.73 d | 18.4 | |
| 30.2 | 1.262 | 2.25 | 1.41 d | 59.6 | |
| melibiose | 0 | 0 | 5.24 | 4.89 b | 7.1 |
| 4.9 | 0.151 | 4.72 | |||
| 6.5 | 0.202 | 4.62 | |||
| 7.9 | 0.252 | 4.42 | |||
| maltose | 4.0 | 0.122 | 4.94 | 4.97 c | −0.6 |
Concentration dependent viscosities and diffusion constants (data not shown) for glucose at 20 °C are nearly identical for PME and IPS. This parallels results obtained earlier,4 and and further supports the notion that that long-range Lennard-Jones forces do not play a significant role in the dynamics of these systems. Long range electrostatics, of course, is critical, as evidenced by the factor of two difference in water viscosities with and without PME.3
3.3. NMR Spin Lattice Relaxation
Eq. (5) has been used directly to evaluate T1’s of alkanes from simulation.21 However, most measurements of carbohydrates are obtained in D2O, so in addition to the scaling applied above for diffusion constants, an additional scaling for the increased viscosity of D2O is required; this is 1.23 near room temperature (Table 1).
Table 3 lists τ for α and β-glucose obtained by integration of the reorientational correlation function, simulated 1/T1 calculated from Eq. (5) with the scaling as noted above, and the experimental 1/T1; the statistical error in the simulated relaxation times, and therefore the 1/T1, is approximately 4%. Results are reasonable for all carbons, with an average difference between simulation and experiment of 12%, not substantially larger than the error of the measurements (usually taken to be approximately 10%). Note that the relaxation times for the two CH vectors of C6 (the exocyclic methyl group) are only slightly less than those of the ring carbons. This implies that on the tumbling time of glucose, there is libration, but no isomerization of the exocyclic torsion. This is consistent with the relatively high barrier leading to isomerizations on the ns, but not ps, time scale.1
Table 3.
Reorientational correlation times τ and NMR 1/T1 of α and β-glucose solutions.
| Sim |
Expa | ||||
|---|---|---|---|---|---|
| anomer | carbon | τ (ps) | 1/T1 (s−1)b | 1/T1 (s−1) | Err (%) |
| 1 | 10.3 | 0.624 | 0.735 | −15.1 | |
| 2 | 10.0 | 0.604 | 0.709 | −14.7 | |
| 3 | 10.0 | 0.606 | 0.694 | −12.7 | |
| α | 4 | 10.0 | 0.607 | 0.719 | −15.6 |
| 5 | 10.0 | 0.605 | 0.680 | −11.0 | |
| 6 | 9.4 | 1.139 | 1.190 | −4.3 | |
| 1 | 10.5 | 0.634 | 0.690 | −8.1 | |
| 2 | 10.5 | 0.636 | 0.690 | −7.9 | |
| 3 | 10.5 | 0.639 | 0.725 | −11.9 | |
| β | 4 | 10.5 | 0.637 | 0.735 | −13.3 |
| 5 | 10.5 | 0.638 | 0.699 | −8.8 | |
| 6 | 9.9 | 1.202 | 1.389 | −13.5 | |
The NMR study by Engelsen et al.22 of maltose and trehalose provides both diffusion constants and T1’s, allowing a comparison of both observables obtained under the same conditions. Table 4 lists the T1 results. As for the translational diffusion constants (Table 2), agreement of simulation and experiment is near quantitative. Maltose was also simulated at the experimental conditions of Benesi and Brant23 (the same study reporting the glucose 1/T1). Agreement is comparable to that in Table 1 (data not shown).
Table 4.
Reorientational correlation times τ and NMR 1/T1 of α,α–trehalose and β-maltose solutions.
| Sim |
Expa | ||||
|---|---|---|---|---|---|
| compound | carbon | τ (ps) | 1/T1 (s−1) | 1/T1 (s−1) | Err (%) |
| 1 | 32.1 | 2.10 | 2.07 | 1.4 | |
| 2 | 33.3 | 2.18 | 2.09 | 4.4 | |
| trehalose | 3 | 33.2 | 2.17 | 2.04 | 6.3 |
| 4 | 33.1 | 2.16 | 2.02 | 7.0 | |
| 5 | 33.4 | 2.18 | 2.00 | 8.8 | |
| 6 | 29.3 | 3.83 | 3.40 | 12.5 | |
| 1 | 28.6 | 1.87 | 1.92 | −2.7 | |
| 2 | 31.0 | 2.03 | 1.96 | 3.3 | |
| maltose | 3 | 30.7 | 2.01 | 1.92 | 4.4 |
| 4 | 30.9 | 2.02 | 1.92 | 5.0 | |
| 5 | 30.9 | 2.02 | 1.96 | 3.0 | |
| 6 | 25.0 | 3.27 | 3.13 | 4.6 | |
| 1’ | 26.5 | 1.73 | 1.52 | 14.2 | |
| 2’ | 27.0 | 1.77 | 1.59 | 11.3 | |
| 3’ | 27.2 | 1.78 | 1.61 | 10.4 | |
| 4’ | 27.8 | 1.81 | 1.72 | 5.2 | |
| 5’ | 26.9 | 1.76 | 1.61 | 9.2 | |
| 6’ | 28.1 | 3.67 | 3.45 | 6.4 | |
3.4. Hydrodynamics
The excellent results obtained for translational and rotational diffusion at low concentration for glucose and the disaccharides allow an estimate of the number of bound waters for each carbohydrate. Hydrodynamic calculations of diffusion constants of polar solutes in water based on Stokes’ Law require the addition of bound waters, or a hydration shell, to yield agreement with experiment.24 The same requirement holds for bead model calculations,15,16 as is evident in column 3 of Table 5: the calculated averaged diffusion constants of carbohydrates with no bound water overestimate experiment by approximately 50%. The lower value of <D> for melibiose compared to trehalose and maltose is consistent in that the former has a 1–6 linkage, and is therefore more extended than the others.
Table 5.
Translational diffusion constants (D0 ; 10−6 cm2/s) at 25 °C for glucose and assorted disaccharides from experiment and a hydrodynamic bead model with no bound water, and with bound water (<Nb>) based on energy cutoffs (Ecut), and the solvent accessible surface area (SASA; Å2).
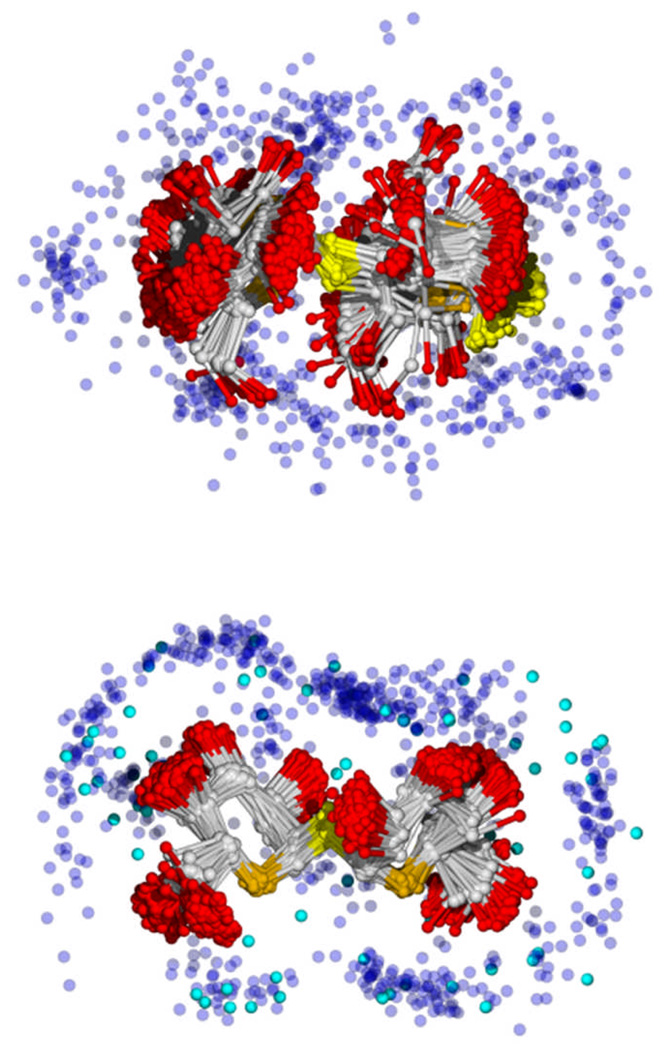
Waters may be placed on a solute in a variety of ways, though an energy cutoff is most natural when using conformations from simulations. In previous estimates, a range of energy cutoffs (Ecut) for carbohydrate-water interaction based on integer multipliers of Ecut/kBT was presented,16,17 with values at 3 and 4 usually bracketing experiment. A slightly different approach is adopted here, in that non-integer multipliers between -3 and -4 kBT with an interval of 0.1 were varied to provide the best agreement (a difference of 0.02×10−6 cm2/s or less) to experiment for each molecule; the averages are denoted <D>E and are listed in Table 5. For glucose, <Nb> =5.1, almost exactly one water per free hydroxyl group. <Nb> varies from 9 to 12 for the disaccharides, though each has only 8 free hydroxyl groups. Trends in <SASA>, Ecut, and <Nb> are readily apparent when these results are listed in order of decreasing D. Melibiose, with the more extended and flexible 1–6 linkage, exhibits the largest value for <Nb>, consistent with the slower diffusion and larger surface area. The noticeable difference in Ecut suggests that the additional bound waters have a weaker interaction with the sugar than those directly interacting with the hydroxyl groups. <Nb> is larger for trehalose than maltose, which is consistent with the lower experimental values for D.25–27 While a range of values is reported in the preceding experimental studies, depending on the method employed, in every case trehalose bound more water than maltose. Fig. 5 compares the hydration shells of these two disaccharides. It is possible that the rigidity of trehalose (with its 1–1 α linkage) allows the formation of a more structured water layer than maltose, allowing the inclusion of more water. This observation is consistent with the average number of waters in different size shells for two disaccharides (Table 6), and the average number of hydrogen bonded waters: 16.2 for maltose and 16.7 for trehalose. It is also consistent with the results from MD simulations listed in Tables 2 and 4 for translational and rotational diffusion, respectively. In each case, trehalose has the larger friction, even though the translational diffusion constant with no bound water is nearly the same as that of maltose (Table 5).
Figure 5.
Aligned overlays of the coordinate sets used for the hydrodynamics calculations for maltose (top) and trehalose (bottom) which best matched the experiment D values. Hydroxyl O atoms are red, ring O5 atoms are orange, anomeric O atoms are yellow, and most water O atoms are transparent blue. For trehalose, the additional water molecules present due to the slightly more permissive value of Ecut are shown in bright blue (cyan). The sugars are oriented on the xy plane; waters with z > 0 (i.e., in front of the sugar) are omitted for clarity.
4. Discussion and Conclusions
The empirical potentials presently used in atomic level simulations are approximate, carbohydrate-water interactions are complex, and transport properties are generally more difficult to model than equilibrium properties. In the present case, the low viscosity of the TIP3P water model (Table 1 and Fig. 2) complicates the validation of the carbohydrate parameter set using experimental results for translational diffusion and rotational relaxation. Simply scaling the viscosity of the solution to experiment at all concentrations is not advisable because discrepancies with experiment may not only arise from the water model: as illustrated in Fig. 3, the viscosities of glucose and trehalose solutions deviate from experiment at concentrations above 20 wt% even after scaling. Given the wide use of TIP3P in the simulation community, it is essential to understand its deficiencies and to develop a procedure for comparing experiment and simulation.
Based on the results in Fig. 3, it is proposed that viscosity scaling is permissible for solutions of mono and disaccharides of less than 20 wt% for simulations using TIP3P water This leads to excellent agreement with experiment for diffusion constants (Table 2) and NMR 13C T1’s (Tables 3 and 4), and thereby further validates the CHARMM parameter set C35. NMR experiments are usually carried at low concentration, so the limit of 20 wt% is not a severe restriction, and C35 can be applied with confidence to analyze relaxation measurements. While the approach presented here can be applied to other water models, the upper range of scaling will need to be determined for each.
The preceding results allow a hydrodynamic analysis of bound waters at low solute concentration (Table 5). While glucose binds one water per free hydroxyl, binding varies for disaccharides. It is proposed that the rigidity of trehalose allows the formation of a more structured hydration shell than maltose (Fig 5), allowing for binding of additional water (Table 5 and 6). Simulations of trehalose solutions at higher concentrations could in principle yield insight into the cryopreservative behavior of this molecule.28 However, the calculated diffusion constants for glucose and trehalose are incorrect at high concentration even after scaling (Fig 4). This result indicates deficiencies in the parametrization, and implies that simulations at higher concentration should be approached with extra caution.
It is possible to avoid viscosity scaling in some circumstances, and still reasonably compare simulation and experiment. For example, the hydrodynamic radius of nearly spherical particles could be evaluated because the different viscosities can be incorporated into Stokes’ Law. However, this is unsatisfactory for nonspherical particles. Likewise, relative relaxation times for intramolecular vectors can be compared without scaling, though reporting calculated T1’s without scaling is potentially confusing. There are circumstances in which scaling should be avoided. In addition to the high concentration regime already discussed, the possibility that the internal motions do not track viscosity should be considered. While there are interesting examples of translation29,30 and rotation31 in some organic solvents with more complicated viscosity dependencies, it is reasonable to assume that the translation and rotational frictions are proportional to water viscosity near room temperatures. However, the situation is less clear for isomerization, where a rapid change in an internal degree of freedom may take place within a cage of solvent, allowing the mode to partially uncouple from viscosity. For example, folding of some α-helical peptides follows a η−0.6 (or “non-Kramers”) dependence in water/sucrose solutions.32 Examples from simulation include spinning of a model methyl group in water,33 and gauche-trans isomerization in alkanes.34
In conclusion, viscosity scaling is reasonable to apply in a defined set of circumstances. It is helpful for validating parameter sets and for obtaining physical insight. It should not be applied where the simulated concentration dependent viscosity departs significantly from experiment, or where internal motions might not track viscosity.
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, National Heart, Lung and Blood Institute, by NIH grant GM070855 to ADM Jr., and utilized the high-performance computational capabilities at the National Institutes of Health, Bethesda, MD (NHLBI LoBoS cluster).
References
- 1.Guvench O, Greene SN, Kamath G, Brady JW, Venable R, Pastor RW, MacKerell AD., Jr J. Comp. Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guvench O, Hatcher E, Venable RM, Pastor RW, MacKerell AD. Journal of Chemical Theory and Computation. 2009;5:2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feller SE, Pastor RW, Rojnuckarin A, Bogusz S, Brooks BR. J. Phys. Chem. 1996;100:17011–17020. [Google Scholar]
- 4.Venable RM, Chen LE, Pastor RW. J. Phys. Chem. B. 2009;113:5855–5862. doi: 10.1021/jp900843x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. Journal of Computational Chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 7.Steinbach PJ, Brooks BR. J. Comp. Chem. 1994;15:667–683. [Google Scholar]
- 8.Wu X, Brooks BR. J. Chem. Phys. 2008;129:154115–154115. doi: 10.1063/1.2992601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoover WG. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 10.Nosé S, Klein ML. J. Chem. Phys. 1983;78:6928–6939. [Google Scholar]
- 11.Durell SR, Brooks BR, Bennaim A. J. Phys. Chem. 1994;98:2198–2202. [Google Scholar]
- 12.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 13.Yeh IC, Hummer G. J. Phys. Chem. B. 2004;108:15873–15879. [Google Scholar]
- 14.Garcia de la Torre J, Bloomfield VA. Quarterly Reviews of Biophysics. 1981;14:81–139. doi: 10.1017/s0033583500002080. [DOI] [PubMed] [Google Scholar]
- 15.Venable RM, Pastor RW. Biopolymers. 1988;27:1001–1014. doi: 10.1002/bip.360270609. [DOI] [PubMed] [Google Scholar]
- 16.Rundlof T, Venable RM, Pastor RW, Kowalewski J, Widmalm G. Journal of the American Chemical Society. 1999;121:11847–11854. [Google Scholar]
- 17.Dixon AM, Venable R, Widmalm G, Bull TE, Pastor RW. Biopolymers. 2003;69:448–460. doi: 10.1002/bip.10421. [DOI] [PubMed] [Google Scholar]
- 18.Lee H, Venable RM, MacKerell AD, Pastor RW. Biophysical Journal. 2008;95:1590–1599. doi: 10.1529/biophysj.108.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipari G, Szabo A. Journal of the American Chemical Society. 1982;104:4546–4559. [Google Scholar]
- 20.Ottiger M, Bax A. Journal of the American Chemical Society. 1998;120:12334–12341. doi: 10.1021/ja800854u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klauda JB, Brooks BR, MacKerell AD, Jr, Venable RM, Pastor RW. J. Phys. Chem. B. 2005;109:5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 22.Engelsen SB, Monteiro C, de Penhoat CH, Perez S. Biophysical Chemistry. 2001;93:103–127. doi: 10.1016/s0301-4622(01)00215-0. [DOI] [PubMed] [Google Scholar]
- 23.Benesi AJ, Brant DA. Macromolecules. 1985;18:1109–1116. [Google Scholar]
- 24.Van Holde KE. Physical Biochemistry. Second Edition. Englewood Cliffs, NJ: Prentiss Hall; 1885. [Google Scholar]
- 25.Matsuoka T, Okada T, Murai K, Koda S, Nomura H. Journal of Molecular Liquids. 2002;98-9:317–327. [Google Scholar]
- 26.Iannilli E, Tettamanti E, Galantini L, Magazu S. Journal of Physical Chemistry B. 2001;105:12143–12149. [Google Scholar]
- 27.Branca C, Magazu S, Maisano G, Migliardo F, Migliardo P, Romeo G. Journal of Physical Chemistry B. 2001;105:10140–10145. [Google Scholar]
- 28.Crowe JH, Carpenter JF, Crowe LM. Annual Review of Physiology. 1998;60:73–103. doi: 10.1146/annurev.physiol.60.1.73. [DOI] [PubMed] [Google Scholar]
- 29.Hayduk W, Cheng SC. Chemical Engineering Science. 1971;26:635. [Google Scholar]
- 30.Pollack GL, Kennan RP, Himm JF, Stump DR. Journal of Chemical Physics. 1990;92:625–630. [Google Scholar]
- 31.Bauer DR, Brauman JI, Pecora R. Journal of the American Chemical Society. 1974;96:6840–6843. [Google Scholar]
- 32.Jas GS, Eaton WA, Hofrichter J. Journal of Physical Chemistry B. 2001;105:261–272. [Google Scholar]
- 33.Widmalm G, Pastor RW, Bull TE. Journal of Chemical Physics. 1991;94:4097–4098. [Google Scholar]
- 34.Zhang YH, Venable RM, Pastor RW. J. Chem. Phys. 1996;100:2652–2660. [Google Scholar]
- 35.Korson L, Drosthan W, Millero FJ. Journal of Physical Chemistry. 1969;73:34. [Google Scholar]
- 36.Kestin J, Imaishi N, Nott SH, Nieuwoudt JC, Sengers JV. Physica A. 1985;134:38–58. [Google Scholar]
- 37.Gladden JK, Dole M. Journal of the American Chemical Society. 1953;75:3900–3904. [Google Scholar]
- 38.Uedaira H, Ishimura M, Tsuda S, Uedaira H. Bulletin of the Chemical Society of Japan. 1990;63:3376–3379. [Google Scholar]
- 39.Rampp M, Buttersack C, Ludemann HD. Carbohydrate Research. 2000;328:561–572. doi: 10.1016/s0008-6215(00)00141-5. [DOI] [PubMed] [Google Scholar]
- 40.Ueadaira H, Uedaira H. Bulletin of the Chemical Society of Japan. 1969;42:2140–2142. [Google Scholar]
- 41.Chirife J, Buera MP. Journal of Food Engineering. 1997;33:221–226. [Google Scholar]