Abstract
Plants synthesize a great variety of isoprenoid products that are required not only for normal growth and development but also for their adaptive responses to environmental challenges. However, despite the remarkable diversity in the structure and function of plant isoprenoids, they all originate from a single metabolic precursor, mevalonic acid. The synthesis of mevalonic acid is catalysed by the enzyme, 3‐hydroxy‐3‐methylglutaryl coenzyme A reductase (HMG‐ CoA reductase). The analysis of the amino acid sequence of HMG‐CoA reductase from Artemisia annua L. plant showed that it belongs to class I HMG‐CoA reductase family. The three dimensional structure of HMG‐CoA reductase of Artemisia annua has been generated from amino acid sequence using homology modelling with backbone structure of human HMG‐CoA reductase as template. The model was generated using the SWISS MODEL SERVER. The generated 3‐D structure of HMG‐CoA reductase was evaluated at various web interfaced servers to checks the stereo interfaced quality of the structure in terms of bonds, bond angles, dihedral angles and non-bonded atom-atom distances, structural as well as functional domains etc. The generated model was visualized using the RASMOL. Structural analysis of HMG-CoA reductase from Artemisia annua L. plant hypothesize that the N and C‐terminals are positioned in cytosol by the two membrane spanning helices and the C-terminals domain shows similarity to the human HMG‐CoA reductase enzyme indicating that they both had potential catalytic similarities.
Keywords: Mevalonic acid, Hydroxy‐3‐methylglutaryl coenzyme A, Artemisia annua, RASMOL
Background
With over 30,000 isoprenoids being structurally identified, most of them are of plant origin and represent the largest family of natural compounds [10]. They function in respiration, signal transduction, cell division, membrane architecture, photosynthesis, and growth regulation [30]. Furthermore, they also play an important role in the exchange of signals between plants and their environment [1] or in defense against pathogens.These structurally diverse compounds all originate from a branched fivecarbon unit, the so‐called active isoprene, isopentenyl diphosphate (IPP), and its isomer dimethylallyl diphosphate (DMAPP). The key enzyme of the classical mevalonate pathway in plants is 3‐hydroxy‐3‐methylglutarylcoenzyme A (HMG‐CoA) reductase (HMGR, EC 1.1.1.34). It catalyzes the formation of mevalonate by two successive reductions of HMG003200052010CoA, using two molecules of NADPH as cofactor. Genome sequencing has identified hmgr genes in organisms from all three domains of life, and over 150 HMGR sequences are recorded in public databases. Higher animals, archaea, and eubacteria have only a single hmgr gene, where as in plants, which use both HMGR003200052010dependent and HMGR‐independent pathways to synthesize isoprenoids, have multiple HMGR isozymes that appear to have arisen by gene duplication and subsequent sequence divergence [19]. Sequence comparison among HMGRs reveals two distinct classes: eukaryotic HMGRs (class I) and prokaryotic HMGRs (class II) [5]. Artemisinin, a sesquiterpene-lactone, has been isolated from the aerial parts of Artemisia annua L. plants. Besides being currently the best therapeutic agent against both drug003200052010resistant and cerebral malaria causing strains of Plasmodium sp. [20], it is also effective against other infectious diseases such as schistosomiasis, hepatitis B and leishmmaniasis [12, 17,25]. The potent antimalarial sesquiterpene lactone, artemisinin, is produced in low quantities by the plant A. annua L. and hence a serious limitation to the commercialization of the drug [7, 18]. Research on artemisinin metabolic engineering showed that overexpressing a key enzyme in isopeprenoid biosynthesis could elevate the level of the sesquiterpene final product [15]. Also, mevalonate is utilized as precursor in the synthesis of various terpenes including artemisinin hence, we hypothesized that artemisinin biosynthesis and its accumulation in A. annua L. plants can be increased through modulation of HMGR activity and MVA level [18].The present study therefore, has been conducted to understand and elucidate the 3‐D structure of HMG CoA reductase of A. annua by homology modeling. Knowledge gained from its 3‐D structure with functionally important domains and structural features is essential for establishing importance and regulatory mechanism at molecular level as well to target the protein metabolic engineering to enhance biosynthesis of artemisinin.
Methodology
Datasets
The HMG Co A reductase protein sequences of A. annua; AHM1, AHM4 and HMGR1 (NCBI accession numbers and AAA68965, AAA68966, AAD47596; UniProt acc. Nos. Q43318, Q43319 and Q9SWQ3) and other sequences examined in this study were retrieved from the public databases [33, 34].
Physicochemical analysis and Homology Modeling
Theoretical pI and molecular weight were determined using the ProtParam at Expasy server [35]. The amino acid sequence of AHM1 protein wassubmitted to TMHMM server for the presence of hydropathic transmembranous regions [36, 2]. BLAST‐p against PDB database at www.ncbi.nlm.nih.gov/BLAST was done to find experimentally determined 3D structural homologue of AHM1 protein. Further, Clustral W analysis showed maximum identity of AHM1 protein with Human HMG‐COA reductase (PDB ID: 1DQ8), (sequence identity 56 %)[28, 29,37]. Therefore, 3‐D structure of AHM1 (566 amino acids) was generatedby using 1DQ8 as template by Swiss Model Server at expasy.org [13, 14,31] and visualized by RASMOL [24, 27]. The functional domains of AHM1 of A. annua were obtained by submitting 3‐D structure to Profunc server at http://www.ebi.ac.uk [34,21 ]. Prediction of secondary structurewas done at Pdbsum http://www.ebi.ac.uk [34,22 ,23].The stereo chemical quality of the structure was analyzed using PROCHECK & WHAT IF serve [23, 26]. The Ramchandran map [8] was generated to check peptide bond planarity, bond lengths, bond angles, hydrogen bond geometry, and side chain conformations of protein structures as a function of atomic resolution.
Results and Discussion
Analysis of protein sequence
The HMG Co A reductase protein sequences of A. annua (Q43318, Q43319 and Q9SWQ3) were retrieved from NCBI. Clustral W results showed the close similarity between all three. The physicochemical analysis of HMGRs of A. annua through Protparm, suggests pI range form 6.52‐8.2 (Table 1, see supplementary material). Sequence comparison among HMGRs reveals two distinct classes: eukaryotic HMGRs (class I) and prokaryotic HMGRs (class II) [5]. The BLAST003200052010p analysis of AHM1 protein against PDB database and Clustral W analysis showed that it has highest sequence identity to HMGR from the human (class I) (56 %; PDB: 1DQ8A) and minimal to HMGR from Pseudomonas mevalonii (class II) suggesting that it belongs to class I HMGRs. The TMHMM plot suggests the presence of two major hydropathic trans‐membranous regions at N terminus (37‐59 and 80‐102) which again supports that it belongs to class I HMGRs as class II HMGRs are soluble proteins and they lack the transmembrane domain. The structure prediction program and intrinsic disorder prediction suggest that HMGR proteins have predominant helical structure (˜18 distinct helical blocks) with coil and extended coil structure (Figure 1a).
Figure 1.
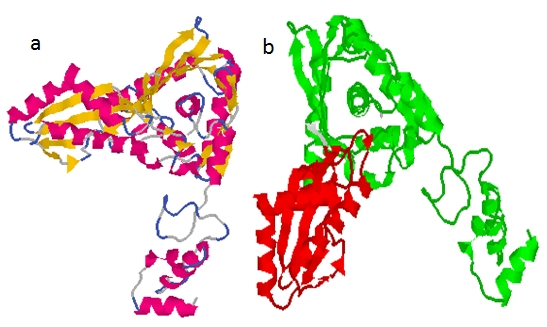
The HMGR1 of Artemisia annua showing major domains. (a)The Substrate binding domain ‐ green (149‐268 and 385‐544) and NADP‐binding domain ‐ red (271‐387). (b) Substrate binding domain resembles prism, with 27‐residue alpha helix forming the central element. Results were obtained from Profunc server and visualized with RASMOL (cartoon).
Homology modeling of AHM1 from A. annua L. plant
Most functional restraints on evolutionary divergence operate at the level of tertiary structure hence 3‐D structures are more conserved in evolution than sequence [16]. The 3-D structure of protein is an important source of information to better understand the function of a protein, its interactions with other compounds (ligands, proteins or DNA) and to understand phenotypical effects of mutations [3]. We have used in present studyhomology modeling method to generate the 3‐D structure. The 3‐D structure generated by SM server and visualized by Rasmol showed the predominance of ‐ helix (18) and β strands (11) arranged in two sheets (Figure 1a). The primary sequence of the initial 146 amino acid residues present at the N-terminus of the AHM1 do not showed any significant sequence homology to the template protein, Human HMGR (1DQ8) sequence deposited at the PDB data bank therefore, this fragment was excluded during the modeling. This initial fragment however, showed the presence of two trans-membranous domains suggesting that they probably anchor protein molecule to the membrane, positioning both N‐ and Ctermini in the cytosol. The Profunc results suggest the presence of two major domains substrate (HMG‐CoA) binding domain and co‐factor (NADPH) binding domain (Figure 1a). The fold of substrate domain is unique to HMGRs. The 3‐D structure of the catalytic portion of AHM1 shows three domains: an N‐terminal ‘N-domain‘, a large ‘L‐domain‘ and a small ‘S-domain‘. The N‐domain is the smallest of the three domains and is α-helical and connects the catalytic portion of HMGR to the membrane domain in full length protein. The fold of the L‐domain (residues 212‐274 and 378‐555) is unique to HMGRs. Its architecture resembles a prism, with 28‐residue helix forming the central core (Figure 1b). The S-domain (residues 276‐351) is inserted into the L‐domain and probably forms the binding site for NADP(H). Detailed analysis show three catalytically active residues E234GC, DK375K, GQD451 (Figure 2a). The solvent accessibility calculation by WESA server showed that E234 and D451 are present in buried form and may be exposed once ligand binds to K375 [9,32]. The HMG‐CoA reductase is phosphorylated and inactivated in vitro by the AMP-activated protein kinase [6]. The presence of S in the conserved sequence NRS555 suggests that AHM1 may also be regulated in the similar manner (Figure 2b).
Figure 2.

Comparison of (a) the catalytically active residues and (b) the phosphorylation site residue in Human HMGR and Artemisia annua HMGR
Homology models validated by PROCHECK essentially satisfy the stereochemical parameters with well‐refined structures at similar resolutions. The distribution of residues in the most favored regions of the Ramachandran plot for AHM1 is 88.3 %. Various studies on structure of catalytic domain have revealed that substrate recognition and catalysis require dimerization of two HMGR molecules through this domain to form the substrate-binding and active site [4]. Thus, it is assumed that in A.annua also, the enzymatically active form of HMGR may be a tetramer. The presence of the HMGR isoforms in A. annua suggests that the isoprenoid biosynthetic pathway is very complex and its metabolic engineering requires the understanding of coordinated activity and specific role of each individual HMGR isoform. Thus, integrating data from metabolomics, proteomics, and transcriptomics will greatly enhance our ability to determine the interactions between components of plant systems and lead to a better understanding of the regulation of plant metabolism networks for enhancing the concentration of desired product.
Conclusions
Although artemisinin is an effective medicine for treating malaria, the application of this medicine is limited by the availability of the source. The artemisinin content in the leaves or florets of A. annua is very low, and the chemical method for the synthesis of this compound is difficult. These factors make the medicine expensive and hardly available on a global scale for patients. Research on artemisinin metabolic engineering showed that overexpressing a key enzyme in isopeprenoid biosynthesis could elevate the level of the sesquiterpene final product.The analysis of structure build through homology modeling shows that AHM1 from A. annua L. plant consists of two major domains: N‐terminal a transmembrane domain that probably anchors the protein molecule to the endoplasmic reticulum membrane and a C‐terminal catalytic domain, which contain the active sites and probably resides in the cytosol where the precursors of sterol biosynthesis are present. Further, the 3‐D structure generated shows the presence of a substrate (HMG‐CoA) binding domain and co‐factor (NADPH) binding domain. The fold of substrate domain is unique and resembles a prism, with 28‐residue helix forming the central core. The homology model, thus generated in this study, could aid in determining the mechanistic function of this important class of proteins.
Supplementary material
Acknowledgments
The authors are thankful to the Jamia Hamdard, New Delhi, India, for providing the infrastructure and financial assistance.
Footnotes
Citation:Abdin et al; Bioinformation 5(4): 146-149 (2010)
References
- 1.Kessler A, et al. Annu Rev Plant Biol. 2002;53:299. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- 2.Krogh A, et al. J Mol Biol. 2001;305:567. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 3.Tramontano A, et al. Methods in Enzymology. 1998;14:293. [Google Scholar]
- 4.Lawrence CM, et al. Science. 1995;268:1758. doi: 10.1126/science.7792601. [DOI] [PubMed] [Google Scholar]
- 5.Bochar DA, et al. Mol Genet Metab. 1999;66:122. doi: 10.1006/mgme.1998.2786. [DOI] [PubMed] [Google Scholar]
- 6.Hardie DG, et al. Trends Biochem Sci. 1989;14:20. [Google Scholar]
- 7.Geldre EV, et al. Plant Mol Biol. 1997;33:199. doi: 10.1023/a:1005716600612. [DOI] [PubMed] [Google Scholar]
- 8.Ramachandran GN, et al. Adv Protein Chem. 1968;23:283. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 9.Chen HL, et al. Nucl Acids Res. 2005;33:3193. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacchettini JC, et al. Science. 1997;277:1788. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 11.Garnier J, et al. Methods Enzymol. 1996;266:540. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 12.Keiser J, et al. J Antimicrob Chemother. 2006a;57:1139. doi: 10.1093/jac/dkl125. [DOI] [PubMed] [Google Scholar]
- 13.Kopp J, et al. Nucleic Acids Res. 2004;32:230. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold K, et al. Bioinformatics. 2006;22:195. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 15.Yan L, et al. J Integr Plant Biol. 2005;47:769. [Google Scholar]
- 16.Bajaj M, et al. Ann Rev Biophys Bioeng. 1984;13:453. doi: 10.1146/annurev.bb.13.060184.002321. [DOI] [PubMed] [Google Scholar]
- 17.Romero MR, et al. Antiviral Res. 2005;68:75. doi: 10.1016/j.antiviral.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Abdin MZ, et al. Planta Med. 2003;69:289. doi: 10.1055/s-2003-38871. [DOI] [PubMed] [Google Scholar]
- 19.Laule, et al. Proc Natl Acad Sci USA. 2003;100:6866. [Google Scholar]
- 20.Newton P, et al. Ann Rev Med. 1999;50:179. doi: 10.1146/annurev.med.50.1.179. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, et al. Nucleic Acids Res. 2005;33:W89. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowski RA, et al. J Mol Biol. 2005;351:614. doi: 10.1016/j.jmb.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski RA, et al. J Appl Crystallogr. 1993;26:283. [Google Scholar]
- 24.Sayle RA, et al. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 25.Sen R, et al. J Med Microbiol. 2007;56:1213. doi: 10.1099/jmm.0.47364-0. [DOI] [PubMed] [Google Scholar]
- 26.Hooft RWW, et al. CABIOS. 1997;13:425. doi: 10.1093/bioinformatics/13.4.425. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez R, et al. Curr Opin Struct Biol. 1997;7:206. doi: 10.1016/s0959-440x(97)80027-9. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, et al. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, et al. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Bach TJ, et al. Lipids. 1995;30:191. doi: 10.1007/BF02537822. [DOI] [PubMed] [Google Scholar]
- 31.Schwede T, et al. Nucleic Acids Res. 2003;31:3381. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan Y, et al. Proteins. 2001;42:23. doi: 10.1002/1097-0134(20010101)42:1<23::aid-prot40>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 33. http://www.ncbi.nlm.nih.gov.
- 34. http://www.ebi.ac.uk.
- 35. http://www.expasy.org/tools.
- 36. http://www.cbs.dtu.dk/services/TMHMM.
- 37. http://www.rcsb.org/pdb.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


