Abstract
Cecropins constitute one of the largest and most potent immune protein families found in insect species with diversified numbers and features. In view of the large number of cecropin proteins existing with much sequence variations among them, an overview of the multigene cecropin family in silkworm Bombyx mori was attempted in this study. Cecropin encodes an inducible 64 residue anti‐bacterial peptide and was clustered into two groups; first group viz. A and second group including B, D, E and Enbocin. Cecropin A consisted of two sub-groups located on chromosome number 6 of B.mori genome. Cecropin B consisted of six sub‐groups, cecropin D and E of one each and Enbocin of two. The second sub‐group formed in tandem array of multigene family locus over a length of 78.62 kb on chromosome number 26 in B.mori genome and was organized in positive as well as opposite orientation. The results indicated that cecropin B genes were organized in a close cluster with the intergenic sequence ranging from 1366 bp to 23526 bp. Interestingly a distantly related cecropin E was also located within the cecropin B multigene locus. Similarly distant members like cecropin D and Enbocin were also located in the 3’ region of cecropin B locus. The maximum intergenic region of 23526 bp observed between Cecropin D and Enbocin indicates that the two genes were distantly evolved. The phylogenetic analysis clearly indicates a positive correlation between the clusters and physical location on the chromosome, as the length of the intergenic region plays a major role to create newer cecropin families. EST database analysis suggests that most of the cecropin A members were expressed in the microbial fat body while, the cecropin B was equally expressed in fat body and other target tissues. The signal peptides were conserved in all the twelve paralogous gene sequences.
Keywords: Bombyx mori, cecropin, genomic organization, multigene, evolution
Background
In animals, efficient innate immunity developed as a first barrier of host defense to kill microbial invades such as bacteria, fungi, and viruses [1]. In contrast, insects have no specific, acquired immunity, but only an innate response that includes so‐called cellular and humoral immune responses [2]. The humoral immune response includes rapid synthesis of a battery of antimicrobial peptides (AMP), such as Cecropin, attacins, defensin, and diptericins in response to bacterial invasion [3] that were important effectors of the innate immune response in insects. The cellular response consists mainly of phagocytosis and encapsulation. In addition to a large number of different AMPs, reactive oxygen species (ROS) and nitric oxide (NO) were known to have a role in humoral defense. Most AMPs were synthesized mainly in the fat body, the major immune‐responsive tissues, and were secreted into the haemolymph [4]. Among the AMPs, Cecropins were well understood and have been investigated in vertebrates as well as insects. Since the discovery of Cecropin in Hyalophora cecropia [5], many Cecropin family genes have been found and isolated in various lepidopteran and dipteran insect [6,. 7,8] Analysis of the Bombyx genomedatabase indicated the presence of A, B, D and E cecropins and these have been cloned [9], except for Cecropin C. Cecropins act against Gram positive as well as Gram negative bacteria. The mature cecropin peptides aggregate on the membranes of infecting bacteria once a threshold concentration has been reached; this aggregation causes disruption of the membrane leading to death of the bacteria. This general immune response in insects stands in contrast to the highly specific response in vertebrates.
Genes that have originated by gene duplication and retained a certain degree of similarity form multigene families. The different members of a multigene family were often arranged in a compact cluster although due to chromosomal rearrangements subsequent to the gene duplications they might be more or less dispersed in the genome. The members of a multigene family can be functional or nonfunctional which were known as pseudogenes. The functional genes can be very similar as the copies might have retained the same function and be redundant. However, one of the copies might have acquired a new function and suffered a certain degree of differentiation. Pseudogenes on the other hand can accumulate substitutions due to the lack of functional constraints [10]. Concerted evolution of the different copies of a gene, facilitated by their compact clustering, can restrict the functional differentiation and loss of function of the copies [11]. On the other hand where concerted evolution is weak or absent the members of that family have a higher probability to become pseudogenes [12].
In Drosophila melanogaster, the Cecropin multigene family consists of both functional and pseudo‐genes and the functional genes code for cecropins [7, 13] . In Drosophila, this response is mediated by at least another eight different kinds of peptides: defensin, attacin, diptericin, drosocin, metnikowin, drosomycin, andropin and lysozyme [7, 13, 14]. Four functional Cecropin genes (Cec A1, Cec A2, Cec B and Cec C) and two pseudogenes (Cec 1 and Cec 2) were detected in a 7-kb region. On bacterial infection, all functional genes were expressed, mainly in the fat body, although at different times during development. The functional genes Cecropin A1 and Cecropin A2 were essentially expressed in larvae and adults, while, Cecropin B and Cecropin C were mainly expressed during the pupal stage [2]. The cecropin cluster has been analyzed thoroughly in D.melanogaster and other lepidopteron insects [10]. In this connection, an attempt has been made to analyze the organization of cecropin multigene cluster in silkworm B.mori.
In B.mori, although 13 cecropin protein genes have been cloned and characterized by different researchers, the genomic organization analyses of cecropin locus to identify the multigene patterns is yet to be attempted. Recently, a complete B.mori genome sequence was obtained through the integration of data from two whole genome sequence projects performed independently by Chinese and Japanese scientists [15]. Utilizing this information, a genome wide screening of cecropin genes B. mori to analyze the organized structure of cecropin multigene clusters in the B.mori genome was adopted. Further, the functional characterization ofdifferent cecropin genes was analyzed by searching both B.mori whole genome sequence as well as EST libraries. We have identified twelve paralogous multigene family sequences organized in two clusters. Further, these paralogous sequences were also characterized based on N‐terminal signal peptide, 5’ region promoter elements and sequence similarity among cecropin paralogous sequences. The phylogenetic analysis of twelve paralogous gene sequences suggested that the duplicated cecropin clusters have evolved independently among insect taxa. Comparison of EST libraries revealed that the multigene cluster group could be correlated to the tissue specificity.
Methodology
Retrieval of cecropin cDNA sequence
The cDNA sequences of cecropin gene were retrieved from the NCBI database and those that expressed exclusively in B.mori were selected. These sequences pertaining to various target tissues were independently deposited by different researchers. The deduced amino acid sequences were obtained through translation of the selected cDNA sequences and were converted in to FASTA format for ClustalX analysis. Then the cDNA sequences were converted into deduced amino acid sequences and the ORF of individual gene was determined using ORF finder (www.ncbi.nlm.nih.gov/projects/gorf/). The conserved domain for cecropin protein was also identified in all translated sequences.
Identification of paralogous gene sequences in B.mori
Identification of paralogous gene sequences in B.mori: The B.mori cDNA sequences were BLAST searched with Silkworm Genome Database (http://silkworm.genomics.org.cn) to identify paralogous multigene family.Using the silkworm database, the functional annotation of genes, paralogous gene sequences, gene products and chromosome mapping were determined. Further, the tools provided in the database were utilized to perform specific genomic BLAST search as well as Map view (a visualization tool that provides a graphical view of selected genes). The organization of paralogous cecropin multigene family on individual scaffold was also analyzed using BLAST search with Gene ID.
Clustal W analysis
Phylogenetic analyses were performed with the multiple sequence alignment using ClustalW through MEGA 4 [16]. The Bootstrap consensus NJ tree for cecropin paralogous gene sequences was constructed with the Bootstrap values. A separate Clustal W alignment was performed with 5’ upstream regions of all paralogous gene sequences. The sequences were manually edited using BIOEDIT programme and the promoter elements were identified using GENERUNNER programme.
Identification of Signal Peptide
Signal peptide cleavage sites were predicted using SignalIP algorithm (www.cbs.dtu.dk/services/SignalIP), based on the Neural Network and Hidden Markov Model.
ESTs of the silkworm B. mori Cecropin
To determine the expression patterns of the individual paralogous genes, a local BLASTN search was performed against the silkworm EST database (http://morus.ab.a.utokyo.ac.jp/). The majority of EST database sequences originated mainly from the tissues of testis, hemocyte, prothoracic gland, brain, antenna, microbial infected fat body, pheromone gland, ovary, midgut and fat body. The specific expressions of the individual paralogous genes were analyzed in EST libraries constructed from different tissues of silkworm B.mori.
Results and discussion
Multigene families often evolve in many ways that violate assumptions necessary for simple and objective gene phylogeny estimation. Some members of the family may evolve at much faster rate and as such are dubbed fast evolving genes. This occurs when one member gene takes on a significantly novel function and thus encounters significantly different selective pressure from other multigene family members. Another usual assumption of molecular tree construction is that each branch of the tree evolves independently from other branches. These families often show coincidental evolution, either indirectly through biased mutational and selective forces or directly by mechanisms such as gene conversions [17].
A total of thirteen B. mori cecropin gene sequences were retrieved from the NCBI database (Table 1 see supplementary material). Among the thirteen sequences, three sequences belonged to cecropin A, seven to cecropin B, one each to cecropin D, E and Enbocin gene sub‐groups. Further, the cecropin cDNA as well as protein sequences were BLAST searched with silkworm genome database (http://silkworm.genomics.org.cn). A total of 12 paralogous gene sequences were retrieved from the genomic database of which two sequences belonged to cecropin A sub‐group, six to cecropin B, one each to the cecropin D as well as E and two to Enbocin. The two cecropin A sequences were located in separate scaffold viz. nscaf 1085 and 2852 of chromosome number 6. However, all the six cecropin B as well as cecropin D, E and two Enbocin sequences were located in the same scaffold (nscaf 1071) of chromosome number 26 (Figure.1). These ten cecropin genes were organized on a single locus and among them six were organized in reverse orientation, while remaining four in the forward orientation. Interestingly, the cecropin E was located in between cecropin B1 and B4. Similarly cecropin D and Enbocin were located in the 3’ region of the scaffold.
Figure 1.
The distribution of Transposable Elements (TEs) in the flanking regions of cecropin family in Bombyx mori. ORFs of cecropin were indicated by black squares and the transcription directions are indicated by arrows. Number below the arrows indicate the intergenic length.
The genomic analysis of Drosophila revealed that, the multigene family originated via retro position and DNA based duplication. The presence of repetitive sequences and TEs in the 5’ and 3’ flanking regions of the multigenes suggested that, gene duplication occurred during the formation of drosomycin multigene family [18]. Earlier reports revealed, only three cecropin families in Drosophila viz. A, B and C. Cecropin A had two sub groups viz. A1 and A2, while, B and C had no sub‐groups indicating that, evolution of paralogous gene sequences was based on the individual requirement and selection pressure in the individual population. The possibility of such gene duplication in the cecropin family was predicted through analysis of the cecropin gene sequences in the B.mori genome which revealed presence of 12 paralogous genes. Presence of the Transposable Elements (TE) in the flanking regions of each paralogous genes confirmed gene duplications giving rise to the cecropin multigene family. The presence of six cecropin B sub‐groups was a unique feature in the silkworm B.mori. Since the cecropin genes B, D, E and Enbocin were located in the same gene locus, it can be inferred that all the four sequences have closely evolved from a single gene.
Comparison of the molecular distance of paralogous gene pairs between different and same species indicates the amount of within‐species coincidental evolution. In B.mori, ten of the twelve cecropin paralogous genes present formed a tight cluster over about 78.62kb length of DNA suggesting that, these original genes probably originated from a common ancestral by gene duplication and later individuals with multiple genes were selected during evolution. If each protein has a slightly different antibacterial spectrum, the presence of multiple proteins should be advantageous for the survival of the insects in various pathogenic environments. Genome analysis of model organisms have shown that over one-third of all protein‐coding genes belong to multigene family originating from the gene duplications [19].
The multigene cluster of cecropin B, D, E and Enbocin was spread over a region of 78.62 kb in length in B. mori genomic DNA (Figure 1). Long intergenic regions were found between cecropin B and Enbocin, and between cecropin B4 and cecropin B5. Each of the cecropin genes invariably contained two exons and one intron. However the lengths of the intron regions varied among the cecropin sub‐groups.
The arrangement of cecropin‐B gene group (cecropin‐B1, B2, B3, B4, B5, B6, D, E and Enbocin) in B.mori within the same locus over an intergenic length ranging from 1336 to 23526 bp indicated tandem gene duplication giving rise to paralogous genes. The findings of the present study reveals that, the major cecropin families probably diverged and formed multigene families in B.mori and did not evolve from a common ancestor of insects.
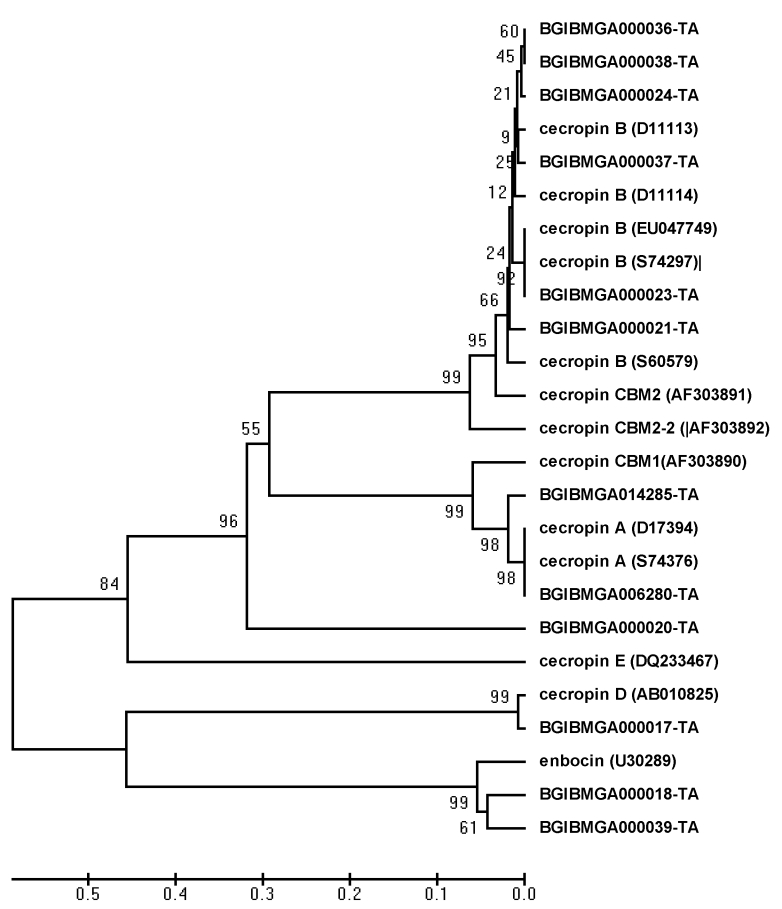
The phylogenetic analysis of different paralogous cecropin gene sequences revealed that, all cecropin B members clustered together (Figure 2). Among the six cecropin B genes, three genes as well as one of the genes formed separate single clusters, while, remaining two genes formed a separate cluster. In contrast, the three cecropin A genes formed a single cluster revealing their unique evolution compared to cecropin B genes. Further, Cecropin E, D and Enbocin formed one out group member in which, Enbocin and cecropin D formed a separate clade. Even though cecropin E formed an out group in the phylogenetic tree, it was located within the cluster of cecropin B sub family. All cecropin B genes, cecropin E, cecropin D and Enbocin were located on the chromosome number 26, while, cecropin A genes were located on the chromosome number 6 (Table 2 see supplementary material).
Figure 2.
Neighbor‐joining tree of 12 paralogous cecropin gene percentage of bootstrap values (based on the 1000 replication) for the main branching nodes shown on the tree. The paralogous gene sequences of silkworm genome database are indicated by Gene ID [BGIBMGA] and the sequences retrieved from NCBI database are indicated by the name and accession number
Analysis of different cecropin sequences with respect to the different target tissues based on the EST library information revealed that, majority of cecropin A genes were expressed in the fat body of bacteria challenged silkworm larvae (Table 3 see supplementary material). However, the cecropin B genes were expressed in different target tissues like testis (12%), hemocyte (3%), prothoracic gland (1%), brain (24%), antenna (2%), microbial fat body (55%) and pheromone gland (1%). Interestingly the cecropin D was expressed less in the fat body (18%), and more in brain tissue (24%), while, Cecropin E was expressed more (46%) in the fat body and less (28%) in brain tissue.
In the above connection, the promoter regions of all the paralogous genes were compared with the help of ClustalW program. The unique promoter elements of cecropin genes were prominently located in the 5’ flanking upstream region such as кβ, GATA, TATAA and CAP site. The promoter elements were also conserved depending upon each sub‐family. The TATAA boxes of all cecropin B paralogous gene sequences were located at exactly the same position.
The hypothesis of Nei’s birth and death model of evolution indicates that, some duplicate genes were maintained in the genome for long‐time but others were deleted or become non‐functional by deleterious mutations [3]. Analyses of expression patterns of individual genes in the cecropin gene family in B.mori comprising of 12 genes is extremely difficult due to high degree of sequence identity. Hence, analysis of different cecropin sequences with respect to the different target tissues based on the EST library was carried out that, indicated that, the cecropin genes of each sub group were selectively expressed in specific target tissue. The location and sequence variations as well as promoter regions were prominent factors in the tissue specificity.
In invertebrates cis‐regulatory motifs (кβ‐like, GAAANN and GATA) as well as the R1 motif are present in the proximal promoter regions of different insect immune genes, the number, order and orientation of which differ. In Drosophila, the two Cecropin A genes present кβ‐like, GATA and R1 motifs; the Cecropin B gene presents both the кβ ‐like and R1 motifs, and the Cecropin C gene presents the B‐like, GATA, and GAAANN motifs. A survey of nucleotide variations in cecropin paralogous genes of B.mori revealed conserved promoter elements like кβ, GATA and TATAA in the 5’ upstream region of the Cecropin gene. However, the positions of the promoter elements are Cecropin gene subgroup specific.
In the above connection, the promoter regions of all the paralogous genes were compared with the help of ClustalW program. The unique promoter elements of cecropin genes were prominently located in the 5’ flanking upstream region such as кβ, GATA, TATAA and CAP site. The promoter elements were also conserved depending upon each sub‐family. The кβ, GATA and TATAA elements of all cecropin B paralogous genes were located at exactly the same position indicating functional and structural similarity.
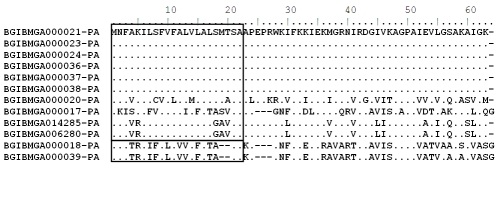
An earlier report indicated that the mature cecropin protein contained sixty four amino acids of which twenty two located in the N- terminal region functioned as signal peptide (Figure 3) [21]. In the present study also, all amino acid signal peptides were also conserved in the cecropin B subfamily and approximately 30% sequence variation was observed in different cecropin genes in B.mori. However the length of each signal peptide did not vary between the cecropin multi gene protein sequences.
Figure 3.
Alignment of the amino acid sequences and phylogenetic relationship of the paralogous gene sequence of Cecropin gene retrieved from Bombyx mori genomic database polypeptide of the Lepidoptera B.mori. The N‐terminal signal peptides of Lepidoptera are indicated by square boxes
Analyses of B.mori genome lead to prediction of 220 putative genes encoding cuticular proteins. More than 80% of these genes were present in gene clusters [22]. Gene clusters of cuticular protein genes have also been reported in Drosophila, Anopheles and Tribolium [15]. In this direction, phylogenetic analysis in B. mori genome reflected distinct organization of cecropin genes wherein Cecropin A and B formed separate clusters. Under the Cecropin B cluster, Cecropin E, D and Enbocin formed one out group member in which, Enbocin and cecropin D formed a separate clade indicating that, these two genes were closely evolved. Even though cecropin E formed an out group member under cecropin B sub group, the clustering of cecropin E within cecropin B sub group is intriguing. The findings revealed that, both cecropin A and remaining cecropin members evolved independently from a common ancestor. Further, the duplication of cecropin B members indicated that, gene duplication occurred exclusively in silkworm B.mori because such phenomenon was not observed in several other insect groups like Drosophila, Anopheles and Tribolium.
In the humoral antimicrobial defense, cecropins show evidence of existence of genes in multi forms. The origin and organization of cecropin gene subfamily in the B.mori has not been studied due to lack of complete information on the silkworm genome. Critically, more studies today focus on evolutionary properties of different immune proteins and their multigene families in Drosophila, Anopheles and Tribolium in view of availability of their genome database.
Conclusion
In the present study the possibility of such gene duplication in the cecropin family was predicted through analysis of the cecropin gene sequences in the B.mori genome which revealed presence of 12 paralogous genes. Presence of the Transposable Elements (TE) in the flanking regions of each paralogous genes confirmed gene duplications giving rise to the cecropin multigene family. The presence of six cecropin B sub‐groups was a unique feature in the silkworm B.mori. Since the cecropin genes B, D, E and Enbocin were located in the same gene locus, it can be inferred that all the four sequences have closely evolved from a single gene.
In B.mori, ten of the twelve cecropin paralogous genes present formed a tight cluster over about 78.62kb length of DNA suggesting that, these original genes probably originated from a common ancestral by gene duplication and later individuals with multiple genes were selected during evolution. The arrangement of cecropin-B gene group (cecropin‐B1, B2, B3, B4, B5, B6, D, E and Enbocin) in B.mori within the same locus over an intergenic length ranging from 1336 to 23526bp indicated tandem gene duplication giving rise to paralogous genes. The findings of the present study reveals that, the major cecropin families probably diverged and formed multigene families in B.mori and did not evolve from a common ancestor of insects.
Analysis of different cecropin sequences with respect to the different target tissues based on the EST library was carried out indicating that, the cecropin genes of each sub group were selectively expressed in specific target tissue. The location and sequence variations as well as promoter regions were prominent factors in the tissue specificity. The unique promoter elements of cecropin genes were prominently located in the 5’ flanking upstream region such as кβ, GATA, TATAA and CAP site. The promoter elements were also conserved depending upon each sub‐family. The кβ, GATA and TATAA elements of all cecropin B paralogous genes were located at exactly the same position indicating functional and structural similarity.
Phylogenetic analysis in B. mori genome reflected distinct organization of cecropin genes wherein Cecropin A and B formed separate clusters. Under the Cecropin B cluster, Cecropin E, D and Enbocin formed one out group member in which, Enbocin and Cecropin D formed a separate clade indicating that, these two genes were closely evolved. Even though cecropin E formed an out group member under Cecropin B sub group, the clustering of Cecropin E within Cecropin B sub group is intriguing. The findings revealed that, cecropin A and remaining cecropin members evolved independently from a common ancestor. Further, the duplication of cecropin B members indicated that, gene duplication occurred exclusively in silkworm B.mori because such phenomenon was not observed in several other insect groups like Drosophila, Anopheles and Tribolium.
The information retrieved from the recently created silkworm genome database allowed us to draw comprehensive conclusions regarding adaptive evolution as well as functional significance of cecropin multigene family in B.mori. This forms a very vital basis to understand evolution of the immune system genes of B.mori with respect to interaction with the natural diversified pathogens in the ever changing environment.
Supplementary material
Acknowledgments
This research work was supported by grants from Department of Biotechnology, Ministry of Science and Technology, Government of India (BT/PR11397/PBD/19/192/2008 dated 30th June 2009).
Footnotes
Citation:Ponnuveletal; Bioinformation 5(3): 97-103 (2010)
References
- 1.Vilmos P, et al. Immunology Letters. 1998;62:59. doi: 10.1016/s0165-2478(98)00023-6. [DOI] [PubMed] [Google Scholar]
- 2.Hultmark D. Trends in Genetics. 1993;9:178. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- 3.Boman HG, et al. Annual Review of Microbiology. 1987;41:103. doi: 10.1146/annurev.mi.41.100187.000535. [DOI] [PubMed] [Google Scholar]
- 4.Uvell H, et al. Trends in Genetics. 2007;23:342. doi: 10.1016/j.tig.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Steiner H, et al. Nature. 1981;292:246. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 6.Dickinson L, et al. Journal of Biological Chemistry. 1988;263:19424. [PubMed] [Google Scholar]
- 7.Kylsten P, et al. EMBO Journal. 1990;9:217. doi: 10.1002/j.1460-2075.1990.tb08098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniai K, et al. Biochemistry Biophysics Acta. 1992;1132:203. doi: 10.1016/0167-4781(92)90013-p. [DOI] [PubMed] [Google Scholar]
- 9.Cheng T, et al. Genomics. 2006;87:356. doi: 10.1016/j.ygeno.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 10.Ramos-Onsins S, et al. Genetics. 1998;150:157. doi: 10.1093/genetics/150.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh JB. Genetics. 1987;117:543. doi: 10.1093/genetics/117.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh JB. Genetics. 1995;139:421. doi: 10.1093/genetics/139.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tryselius Y, et al. European Journal of Biochemistry. 1992;204:395. doi: 10.1111/j.1432-1033.1992.tb16648.x. [DOI] [PubMed] [Google Scholar]
- 14.Meister M, et al. BioEssays. 1997;19:1019. doi: 10.1002/bies.950191112. [DOI] [PubMed] [Google Scholar]
- 15.Xia Q, et al. Insect Biochemistry and Molecular Biology. 2008;38(12):1036. doi: 10.1016/j.ibmb.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S, et al. Briefings in Bioinformatics. 2004;5:150. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 17.Roach CJ, et al. Proceedings of National Academy of Sciences. 2005;102:9577. [Google Scholar]
- 18.Deng J, et al. Journal of Biomedicine and Biotechnology. 2009;1 [Google Scholar]
- 19.Meisel PR. Gene. 2009;438:1. doi: 10.1016/j.gene.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Nei M, et al. Proceedings of National Academy of Sciences. 1997;94(15):7799. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong MS, et al. Bioscience, Biotechnology, Biochemistry. 2008;72(8):1992. doi: 10.1271/bbb.80082. [DOI] [PubMed] [Google Scholar]
- 22.Futahash K, et al. Insect Biochemistry and Molecular Biology. 2008;38:1138. doi: 10.1016/j.ibmb.2008.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.