Abstract
Endophytic bacteria are harmless in most plant species; and known to boost the growth and development of the host plants probably by secreting growth hormones. The isolation, identification and screening of endophytic bacteria for the plant growth regulators like cytokinin are needed to get the leads for their applications in agriculture sector. We describe the isolation and identification of the bacterial endophytes from the leaves of Sambung Nyawa [Gynura procumbens (Lour.) Merr.] and their screening for cytokinin-like compounds. We isolated three endophytic bacteria from the leaves of G. procumbens collected from the forest research institute of Malaysia (FRIM). They were further identified using amplified 16S rRNA gene sequence based method of bacterial identification. The ethyl acetate extracts of the isolates-broth were analyzed using cucumber cotyledon greening bioassay (CCGB) to determine the presence of cytokinin-like compounds. Consequently, the bacterial putative endophytes were identified as Psuedomonas resinovorans, Paenibacillus polymaxa, and Acenitobacter calcoaceticus. Broth-extracts from two (Psuedomonas resinovorans and Paenibacillus polymaxa) of the three putative bacterial endophytes show the positive results in their screening for cytokinin-like compounds using CCGB. Thus, we hypothesize that the bacterial putative endophytes of G. procumbens that produce cytokinin-like compounds might have a role in the growth and development of G. procumbens.
Abbreviations
CCGB - Cucumber cotyledon greening bioassay, rDNA - Ribosomal DNA, K12, BAP - 6-Benzylaminopurine, Db1, MSA - Multiple sequence alignment. 8081,
Keywords: Bacteria, Cytokinin, Endophytes, Gynura procumbens, Microbial diversity, Plant growth regulators, 16S rRNA gene
Background
Gynura procumbens (Lour.) Merr. is one of the important medicinal plants in Malaysia, Thailand, Indonesia, Korea and Philippines. It is known as ‘ Sambung Nyawa ’ in Malay and as ‘ bai bing ca ’ in Chinese language. It belongs to the Asteraceae (Compositae) plant family. It is distributed in Western and central Africa, and from southern China throughout continental South-East Asia and Papua New Guinea. It is cultivated (at marginal scale) in Malaysia and has limited distribution to the western part of peninsular Malaysia. This plant is used in the traditional treatment of various health aliments [1] such as fever, constipation, kidney diseases, rheumatism, viral diseases of skin, rashes, migraine, and cancer [1]. The leaves of this plant have anti-herpes simplex virus [2], anti-hyperglycemic [3], anti-inflammatory [4], anti hyperlipidemic [5], anti-allergy agent and antihypertensive [6] properties. The leaves of this plant are often consumed in diet; and research shows that leaves contents are not having any toxic effects [1]. The leaves contain flavonoids, unsaturated sterols, terpenoids, polyphenols and essential oil.
The endophytic microbes (bacteria and fungi) live in the tissues of various plant species without causing any disease or damage to their host plants [7]. Endophytes could be used for their various applications in boosting plant growth and development [8] and can make vital contribution in agricultural biotechnology. The endophytic bacteria are known to contribute to the host plants growth and development by producing plant growth regulators [9]. The bacterial endophytes are also known to increase host plants resistance to plant pathogens and parasites [7,10], to promote biological nitrogen fixation [11], and to produce antibiotics [12]. Endophytes are also promising source of pharmaceutically important bioactive compounds; and they are known to produce taxol, the most important and effective antitumor agent developed in the past three decades [13]. In order to be able to exploit their (endophytes) bioactive products in various sectors of biotechnology, the understanding of their diversity in plants, plant-endophyte association and their role in plant growth and development is necessary [14– 16].
The presence of the bacterial endophytes and their interaction and association with agronomic, medicinal [17] and forest plant species is reported for a variety of plant species such as Glycine max, Oryza sativa [18], and Ocimum sanctum [19]. However, relatively very little number of endophytes and plant species for endophytes are being studied so far though endophytes do have several potential applications in agriculture sector and health care sector. Cytokinin is a plant growth regulator which retards the senescence; and cytokinin-like compound producing endophytes can be utilized to prolong the shelf-life of cut-flowers, leafy vegetables and fruits. In addition, endophytes do have potential to produce pharmaceutically important bioactive compounds.
Gynura procumbens , an important tropical medicinal plant is studied mainly of its medicinal properties and toxicology of leaves or leaves extracts; and there is no study so far about the endophytes associated with this important plant. The main aim of this study was to isolate and identify the bacterial endophytes from G. procumbens leaves and to screen theputative endophytic bacterial isolates for cytokinin-like compounds using classical CCGB. The isolated and identified bacterial putative endophytes of G. procumbens are being reported in this paper along with the results of their screening for cytokinin-like compounds.
Methodology
Plant material
Gynura procumbens plant twigs along with leaves were collected from the G. procumbens collection available at Forest Research Institute of Malaysia (FRIM), Malaysia. Healthy plant twigs were selected and collected to avoid contamination by plant pathogens. Collected samples were brought to the laboratory within 24 hours after sample collection following guidelines described by Nalini et al . [20] . Plant flowers, fruits, stems, roots and twigs can be used to isolate endophytic bacteria, but leaves were used in this study.
Surface decontamination of collected leaves samples
Leaves were separated from the twigs and washed thoroughly under plenty of running tap water to remove adhering soil particles and the microbes. The surface decontamination method described by Fisher et al . [21] was used with some minor modification. Leaf samples cleaned with detergent (5% Teepol), running tap water and then distilled water were taken into laminar hood and immerged into 70 % ethanol for 30 sec and then into sodium hypochlorite (5%) solution for 3 min. Finally, leaf samples were exposed to absolute alcohol for ˜10 sec and then washed thrice with autoclaved distilled water (ADW). Surface decontaminated leaves were cut into pieces of about 1 cm² using sterile surgical blade. The leaf pieces were aseptically inoculated in Petri dishes containing Luria Bertani (LB) agar medium and then incubated at 37°C for 18 to 20 hours in the dark.
Isolation of bacterial endophytes
Well grown colonies of cultivable bacterial putative-endophytes that are visible on the LB medium (on leaf margins) were randomly chosen for their further analysis. Pure culture of each putative endophyte was cultivated separately in universal bottles containing 10 ml LB medium. The culture-cultivation was carried out at 37°C, 160 rpm for 18 hours [22] . Glycerol stocks were prepared and kept at –80°C to preserve the putative endophytic isolates for the future research work.
Preparation of cell-free broth extracts
The bacterial cells were harvested from overnight grown 10 ml culture of respective isolate. The bacterial cells were used to prepare the lysate and ethyl acetate extract was prepared from 7 ml cell-free broth by mixing it with equal volume of ethyl acetate. The ethyl acetate extract was collected from the vigorously mixed cell-free broth and ethyl acetate. Ethyl acetate was evaporated (70°C) using rotary evaporation and the dried extracts were dissolved in 1 ml analytical grade ethanol. Finally the ethanol was evaporated at 50°C and the dry extracts were dissolved in bioassay solution as described by Fletcher et al . [23] and used in CCGB.
Amplification of the 16S rDNA
The bacterial cells harvested from 10 ml broth were re-suspended in 50 μl sterile distilled water and boiled at 100 ° C for 10 min and centrifuged at 12,000 rpm for 5 min. Five μl lysate was used in polymerase chain reaction (PCR) to amplify 16S rDNA. The 16S rDNA PCR amplification was carried out in a total volume of 50 μl containing 10x PCR buffer, 2.5 mM MgCl 2 , 0.16 mM of dNTPs, 0.75 U of Taq DNA polymerase and 0.2 pmol forward [Bak11W-F; 5 ’ -AGT TTG ATC MTG GCT CAG-3 ’ ], and reverse [Bak-R; 5 ’ -GGA CTA CHA GGG TAT CTA AT-3 ’ ] primer. The PCR conditions consisted of an initial hot start at 95°C for 3 min, 30 amplification cycles of 95°C for 30 sec (denaturation), 52°C for 30 sec (annealing), and 72°C for 30 sec (extension), and finally one cycle of 72°C for 5 min.
16S rDNA sequencing and identification of isolates
The amplified PCR products were purified using the ‘ Wizard ® SV Gel and PCR Clean up system ’ by following guidelines provided by manufacturer (Promega Corporation). Both strands of amplified 16S rDNA weresequenced using Bak11W-F and Bak-R primers. Forward and reverse DNA strand sequence was aligned using BLAST (bl2seq) program available at NCBI [24] . The finalized sequence of amplified 16S rDNA fragment from each isolate was blasted against the collection of non- redundant nucleotide sequence database of NCBI. The isolates were identified based on hits analysis from megablast (highly similar sequences) output. The hits of 16S rDNA sequences were used in phylogenetic analysis of 16S rDNA sequences of isolates to determine 16S rDNA sequence based evolutionary relationship among the isolates and hits.
MSA and tree construction
The MSA of 16S rDNA sequences was carried out using ClustalW (version 1.6) program [25] . The MSA output from ClustalW was used the construction of a rooted dendrogram using NJ (Neighbour Joining) method of Saitou and Nei provided at the http://align.genome.jp/.
Cucumber cotyledon greening bioassay (CCGB)
The cucumber ( Cucumis sativus L.) seeds (F1 Cucumber 303) were purchased from Tai Lung Farm Seed Co Ltd, Taiwan to conduct CCGB. Cucumber seeds were germinated in the plastic tray containing tissue paper towel saturated with autoclaved distilled water. Seeds were incubated at room temperature (RT) in the dark for 6 days following the guidelines reported by Fletcher et al . [23] for cucumber seed germination. Cotyledons were separated from hypocotyls of 6 day old cucumber seedlings. By weighing, cotyledons were added in Petri dishes containing the 10 ml bioassay solution supplemented with broth extracts of respective endophytic bacterial isolate. A negative control and a positive control (BAP) were used in the assay for comparison purpose by following guidelines of Fletcher et al . [23] . The BAP concentration in positive control was 1 ppm.
Extraction and quantitative estimation of chlorophyll content
Cucumber cotyledon samples along with negative and positive control were incubated under fluorescent tube light for 3.5 hours at 27°C ±2. The cotyledons were ground with 80% acetone with mortar and pestle after the incubation. The chlorophyll extracts were collected and centrifuged at 4,000 rpm for ten min. The derived supernatant was analyzed for total amount of chlorophyll using spectrometer (λ 663nm and 645nm) [23] .
Results
The inoculated 18 leaves discs on LB medium produced 24 bacterial colonies in total. Visibly, three different types of bacterial colonies were observed on the leaf discs margins on LB-agar cultures. Three colonies representing three types of colony morphology were selected randomly and inoculated in LB medium for their cultivation. Gp e1, Gp e2, and Gp e3 code names were given to the randomly selected three bacterial putative endophytes. The 16S rDNA fragments were PCR-amplified using the lysate of pure cultures of three (Gp e1, Gp e2, and Gp e3) bacterial putative endophytes. The size of the amplified 16S rDNA fragments was about 794 bp (Figure 1).
Figure 1.
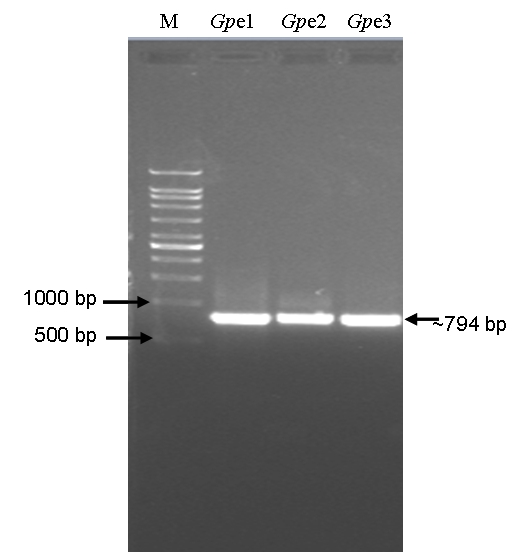
Polymerase chain reaction (PCR) amplified 16S rDNA fragments from three isolates of bacterial putative endophytes isolated from Sambung Nyawa (Gynura procumbens (Lour.) Merr.) leaves. M, 1 kb DNA ladder (DNA marker); Gp e1, Gynura procumbens putative endophyte 1; Gp e2, Gynura procumbens putative endophyte 2; and Gp e3, Gynura procumbens putative endophyte 3.
The purified amplified 16S rDNA PCR products were sequenced using the same forward and reverse (Bak11W-F and Bak-R) primers used in the PCR. The blastn analysis results showed that Gp e1, Gp e2, and Gp e3 isolates are of Psuedomonas resinovorans, Paenibacillus polymaxa , and Acenitobacter calcoaceticus , respectively. The 16S rDNA sequence of Gp e1, Gp e2, and Gp e3 was deposited in the international GenBank/DDBJ/EMBL gene sequence database. The GenBank accession numbers and the length of deposited 16S rDNA sequences are depicted in the (Table 1, see supplementary material )
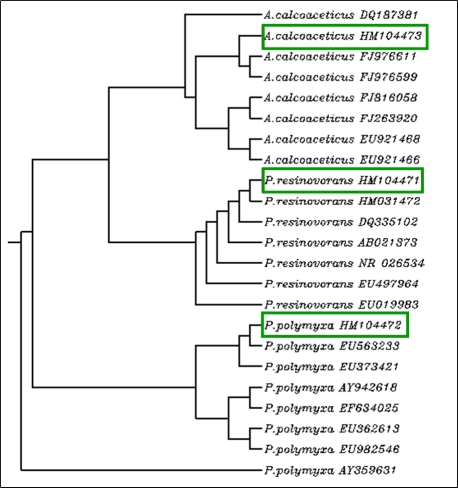
The Gp e1, Gp e2, and Gp e3 16S rDNA sequences and their hits from GenBank were used in phylogenetic analysis to find out phylogenetic relationship between three isolates and the reported other strains of respective species from different countries. The 16S rDNA sequences based dendrogram is shown in Figure 2 . The source and geographical location of different strains of Pseudomonas resinovorans , Paenibacillus polymyxa, and Acinetobacter calcoaceticus used in 16S rDNA sequencesbased dendrogram construction are depicted in (Table 1, see supplementary material.)
Figure 2.
Rooted dendrogram showing clustering of Psuedomonas resinovorans, Paenibacillus polymaxa, and Acenitobacter calcoaceticus isolated from medicinally important Gynura procumbens leaves tissue. Dendrogram was constructed using 16S rDNA sequences of three isolates and other strains of respective species reported from different parts of the world. The 16S rDNA sequences for different strains of respective bacterial species were obtained from NCBI database. The multiple sequence alignment (MSA) of 16S rDNA sequences was carried out using ClustalW (ver. 1.6) by applying default parameters. The MSA output from ClustalW was used for the construction of a rooted dendrogram using NJ (Neighbour Joining) method of Saitou and Nei provided at the http://align.genome.jp/. In the phylogenetic tree, locations of three isolates from G. procumbens are shown in green boxes and source of other strains is given in Table 2 (see supplementary material). The accession number of the respective 16S rDNA sequence is given in front of the respective species (strain) name.
The CCGB results are showed in Figure 3. In positive control, the cucumber cotyledons were treated with 1 ppm authentic cytokinin (BAP; and the total amount of chlorophyll content was 177.37 mg per gram of fresh cucumber cotyledons. The total amount of chlorophyll content in cucumber cotyledons from negative control and cotyledons treated with Gp e1, Gp e2, and Gp e3 isolate broth-extracts is shown in the Figure 3.
Figure 3.
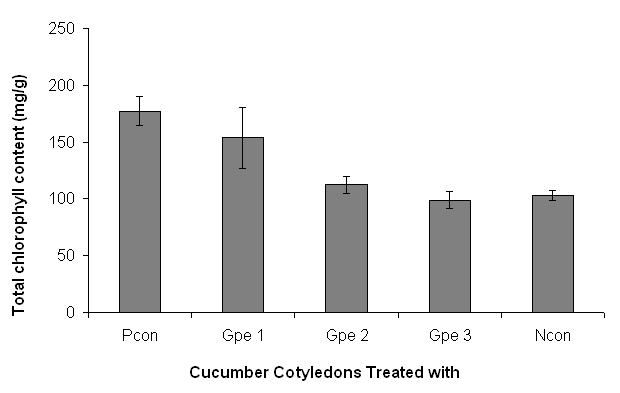
Screening of three endophytic bacterial putative endophytes of G. procumbens for cytokinin-like compounds using cucumber cotyledon greening bioassay (CCGB). The Gp e1, Gp e2, and Gp e3 represent cotyledons treated with broth-extracts of G. procumbens putative endophytes 1, 2 and 3 respectively. Pcon, positive control; Ncon, negative control; positive control is with 1 ppm (BAP), and negative control is without BAP and extract. Bars indicate the standard error.
Discussion
In the natural habitat, plants live in a complex ecosystem where microbial (bacteria and fungi) communities interact continuously with plants. The endophytic microbes compete for nutrients and water in the host tissues regardless of plant category. The knowledge of the diversity of endophytic bacteria in plants in general and medicinal plants in particular is important to explore their capabilities in various sectors of biotechnology.
Different plant organs can be used to isolate the cultivable endophytic bacteria. However, in this study we have used leaves of G. procumbens plant to isolate the cultivable bacterial endophytes. This plant bears lots leaves and leaves are easily available in plenty and hence used in the isolation of bacterial endophytes. But, other parts of the plant such as roots [26–27] , stems, seeds [27–28] , petioles, tubers tissue, and flowers [29] can also be used in isolation of endophytes.
The 16S rDNA coded 16S rRNA is a component of the 30S subunit of bacterial (prokaryotic) ribosome. The 16S rDNA is 1,542 bp in length [30] . The prokaryotic 16S rDNA sequence provides bacterial species-specific signature sequences and can be used in bacterial identification. Hence, we amplified 16S rRNA encoding gene sequence for the rapid and accurate bacterial identification [31] instead of using traditional phenotypic and biochemical tests based bacterial identification methods. We have used the forward and reverse primers that are commonly used for the 16S rDNA amplification in sequence based identification of bacteria [32– 33] . The amplified 16S rDNA fragments are of ˜ 794 bp as expected based on location of primers. However, only clean sequence was used in 16S rDNA based identification of the isolates.
The Gp e1, Gp e2, and Gp e3 cultivable bacterial putative endophytic isolates (Table 1) were identified as Psuedomonas resinovorans, Paenibacillus polymaxa , and Acenitobacter calcoaceticus , respectively. There are only 61, 338 and 534 nucleotide entries for various strains of Psuedomonas resinovorans, Paenibacillus polymaxa , and Acenitobacter calcoaceticus , respectively (September 10, 2010, GenBank Search). However, there are very few or no reports on endophytic roles of Psuedomonas resinovorans, Paenibacillus polymaxa , and Acenitobacter calcoaceticus . The P. resinovorans strain P-1, and P. resinovorans strain LAM 9 reported from Taiwan and Italy (Table 2) are known to have phenol degrading and heavy metal resistance properties [34– 35] . The P. polymyxa strain YRL13 (accession No: EU373421) and P. polymyxa strain GBR-602 (accession No: AY359631) both reported from Korea are known to have antimicrobial and plant growth promoting properties that are not verified. The A. calcoaceticus strain LCR102, A. calcoaceticus strain HIRFA36 and A. calcoaceticus strain HIRFA33 (Table 2) are believed to have Phosphate solublizing and plant growth promoting properties but claims are not verified.
The phylogenetic analysis indicates that G. procumbens endophyte 1 (Gp e1) is closely related to the Psuedomonas resinovorans strain mol11 reported from Korea; the G. procumbens endophyte 2 (Gp e2) is closely related to the Paenibacillus polymyxa strain 12170504 reported from China; and the G. procumbens endophyte 3 (Gp e3) is closely related to the Acenitobacter calcoaceticus strain LCR102 reported from India (Table 2). It is of interest to note that we have used 16S rDNA sequences of Pseudomonas resinovorans reported from China, Japan, Italy, Korea, Taiwan, and USA; but interestingly P. resinovorans reported from Korea shows close evolutionary relationship with Gp e1 (see Figure 2 and Table 2). The A. calcoaceticus strain BA60 (accession No: FJ263920) similar to Gp e3 is isolated from human blood culture. However, there is no more information about role of this bacterium in human body or in the plant host like G. procumbens .
The isolates screening for cytokinin-like compounds using CCGB suggest that Gp e1 and Gp e2 isolate extracts do contain compounds that acts like cytokinin. The cucumber cotyledons treated with Gp e1 and Gp e2 extracts showed 49.5% and 9.3% extra amount of chlorophyll in cucumber cotyledons in comparison to negative control used in CCGB. Our previous study showed that the cell-free crude broth of endophytes is not suitable in CCGB [36] . Therefore, extracts were prepared from the cell-free broth using ethyl acetate and used in the CCGB. Though, Gp e1 and Gp e2 broth extracts are showing positive results in CCGB; these results need to be verified using other methods such as liquid chromatography-tandem mass spectrometry (LC-TMS) [37–38] and or by using other methods described by Ge et al . [39].
The G. procumbens is an important medicinal plant and in this study only three types of bacterial endophytes were found in their leaves. Any individual plant species could contain several types of endophytes [28] The leaves samples used in the isolation of cultivable bacterial putative endophytes in this study are only from one place; and leaves samples are from plants that were grown in restricted conditions (Figure 4 ). It could be a possible reason for limited number of putative endophytes in the leaf samples used. The endophytic population in the other organs of the plant such as stem could contain high number of endophytes than that of the leaves. The results reported by Magnani et al . (2010) also shows that the plant stem could harbors a larger number of different endophytes [28] compared to leaves, through it could be seasonal.
Gao et al . (2005) has reported that in medicinal plants a seasonal fluctuation of the endophytes occurs [40] . Therefore, we hypothesize that various types of endophytes might be colonizing the G. procumbens in addition to the three bacterial putative endophytes reported in this study. Furthermore, we have isolated only bacterial endophytes. However, more comprehensive study needs to be done in order to get a complete understanding of the association of various (bacterial and fungal)endophytes with the G. procumbens and their benefits to the G. procumbens . This type of comprehensive study might shade the light on the possible roles of endophytes in the synthesis of the medicinally important compounds found in G. procumbens .
Conclusion
The results data show that G. procumbens leaves contains cultivable bacterial endophytes (putative) namely, Psuedomonas resinovorans, Paenibacillus polymaxa, and Acenitobacter calcoaceticus . Furthermore, this study also show that two (Gp e1 and Gp e2) out of three isolated bacterial putative endophytes of G. procumbens do produce cytokinin-like compounds. However, these cultivable bacterial putative endophytes need to be studied further to determine their beneficial roles to the host plant, G. procumbens .
Supplementary material
Figure 4.

The plantlets collection of Sambung Nyawa [ Gynura procumbens (Lour.) Merr.] at Forest Research Institute of Malaysia (FRIM), Malaysia. The plant material used in isolation of endophytic bacteria was collected from FRIM (Photography credit: Preveena J., AIMST University).
Acknowledgments
Authors are grateful to the Malaysia ' s Ministry of Agriculture and Agro Based Industry (MoA) for financial support (Grant Code Number: 05-02- 16-SF1001), and to the Forest Research Institute of Malaysia (FRIM) for providing plant material for this study.
Footnotes
Citation:Bhore et al; Bioinformation 5(5): 191- 197 (2010)
References
- 1.Rosidah, et al. J Ethnopharmacol. 2009;123:244. doi: 10.1016/j.jep.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Nawawi A, et al. Phytother Res. 1999;13:37. doi: 10.1002/(SICI)1099-1573(199902)13:1<37::AID-PTR382>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 3.Akowuah GA, et al. Akowuah. 2002;40:405. [Google Scholar]
- 4.Iskander MN, et al. Plant Foods Hum Nutr. 2002;57:233. doi: 10.1023/a:1021851230890. [DOI] [PubMed] [Google Scholar]
- 5.Zhang XF, Tan BK. Singapore Med J. 2000;41:9. [PubMed] [Google Scholar]
- 6.Kim MJ, et al. J Med Food. 2006;9:587. doi: 10.1089/jmf.2006.9.587. [DOI] [PubMed] [Google Scholar]
- 7.Hallmann J, et al. Can J Microbiol. 1997;43:895. [Google Scholar]
- 8.Mengoni A, et al. Microbiol Res. 2003;158:363. doi: 10.1078/0944-5013-00216. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, et al. J Appl Microbiol. 2006;100:938. doi: 10.1111/j.1365-2672.2006.02843.x. [DOI] [PubMed] [Google Scholar]
- 10.Reiter B, et al. Appl Environ Microbiol. 2002;68:2261. doi: 10.1128/AEM.68.5.2261-2268.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldani JI, et al. Int J Syst Bacteriol. 1986;36:86. [Google Scholar]
- 12.Strobel G, Daisy B. Microbiol Mol Biol Rev. 2003;67:491. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gangadevi V, et al. Sheng Wu Gong Cheng Xue Bao. 2008;24:1433. doi: 10.1016/s1872-2075(08)60065-5. [DOI] [PubMed] [Google Scholar]
- 14.Germida JJ, et al. FEMS Microbiol Ecol. 1998;26:43. [Google Scholar]
- 15.Sturz AV, et al. Plant Pathol. 1999;48:360. [Google Scholar]
- 16.Strobel GA, et al. Microbes Infect. 2003;5:535. doi: 10.1016/s1286-4579(03)00073-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhao k, et al. Curr Microbiol. 2010 [Google Scholar]
- 18.Ikeda s, et al. Plant Cell Physiol. 2010 [Google Scholar]
- 19.Tiwari R, et al. Curr Microbiol. 2010;60:167. doi: 10.1007/s00284-009-9520-x. [DOI] [PubMed] [Google Scholar]
- 20.Nalini MS, et al. Mycopathologia. 2005;159:245-249. doi: 10.1007/s11046-004-5497-y. [DOI] [PubMed] [Google Scholar]
- 21.Fisher PJ, et al. New Phytol. 1992;122:299. doi: 10.1111/j.1469-8137.1992.tb04234.x. [DOI] [PubMed] [Google Scholar]
- 22.Reinhold HB, et al. Crit Rev Plant Sci. 1998;17:29. [Google Scholar]
- 23.Fletcher RA, et al. Plant Physiol. 1982;59:675. doi: 10.1104/pp.69.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. http://www.ncbi.nlm.nih.gov/
- 25. http://align.genome.jp/
- 26.Asraful ISM, et al. Curr Microbiol. 2010 [Google Scholar]
- 27.Zhang X, et al. Wei Sheng Wu Xue Bao. 2010;50:530. [PubMed] [Google Scholar]
- 28.Magnani GS, et al. Genet Mol Res. 2010;9:250. doi: 10.4238/vol9-1gmr703. [DOI] [PubMed] [Google Scholar]
- 29.Reiter B, Sessitsch A. Can J Microbiol. 2006;52:140. doi: 10.1139/w05-109. [DOI] [PubMed] [Google Scholar]
- 30.Case RJ, et al. Environ Microbiol. 2007;73:278. doi: 10.1128/AEM.01177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarridge JE, et al. III Clin Microbiol Rev. 2004;17:840. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosshard PP, et al. Int J Syst Evol Microbiol. 2002;52:1263. doi: 10.1099/00207713-52-4-1263. [DOI] [PubMed] [Google Scholar]
- 33.Dunbar J, et al. Appl Environ Microbiol. 2002;68:3035. doi: 10.1128/AEM.68.6.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F, Lee CM. Int Biodeterior Biodegradation. 2006;59:206. [Google Scholar]
- 35.Alisi C, et al. Sci Total Environ. 2009;407:3024. doi: 10.1016/j.scitotenv.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Bhore SJ, Sathish G. World J Agric Sci. 2010;6:345. [Google Scholar]
- 37.Kojima M, et al. Plant Cell Physiol. 2009;50:1201-14. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussain A, et al. Curr Microbiol. 2010 [Google Scholar]
- 39.Ge L, et al. Electrophoresis. 2006;27:4779. doi: 10.1002/elps.200600195. [DOI] [PubMed] [Google Scholar]
- 40.Gao XX, et al. FEMS Microbiol Lett. 2005;249:255. doi: 10.1016/j.femsle.2005.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.