Abstract
Background
Computer languages can be domain-related, and in the case of multidisciplinary projects, knowledge of several languages will be needed in order to quickly implements ideas. Moreover, each computer language has relative strong points, making some languages better suited than others for a given task to be implemented. The Bioconductor project, based on the R language, has become a reference for the numerical processing and statistical analysis of data coming from high-throughput biological assays, providing a rich selection of methods and algorithms to the research community. At the same time, Python has matured as a rich and reliable language for the agile development of prototypes or final implementations, as well as for handling large data sets.
Results
The data structures and functions from Bioconductor can be exposed to Python as a regular library. This allows a fully transparent and native use of Bioconductor from Python, without one having to know the R language and with only a small community of translators required to know both. To demonstrate this, we have implemented such Python representations for key infrastructure packages in Bioconductor, letting a Python programmer handle annotation data, microarray data, and next-generation sequencing data.
Conclusions
Bioconductor is now not solely reserved to R users. Building a Python application using Bioconductor functionality can be done just like if Bioconductor was a Python package. Moreover, similar principles can be applied to other languages and libraries. Our Python package is available at: http://pypi.python.org/pypi/rpy2-bioconductor-extensions/
Background
The Bioconductor project [1], based on the R language [2], has become a reference for the numerical processing and statistical analysis of data coming from high-throughput biological assays. Starting with microarray data, it became an integrated suite of data structures and functions to perform tasks ranging from reading raw data files to processing algorithms and to data analysis. The project soon expanded to data analysis in bioinformatics at large and to other assays, providing a rich selection of methods and algorithms to the research community.
At the same time, the Python language [3] has matured as a dependable platform for prototype development and data handling. Python is used by many organizations in need of processing or analyzing large volumes of data (Google, NASA, CERN, ILM). Python is a very accessible language and is used in introductory courses to programming for non-computer scientists [4,5]. It is also used by professional programmers in need of increased productivity [6] and agile prototyping.
In the context of bioinformatics, the Biopython project [7] was one of the first Python libraries for bioinformatics, and while a few utilities offered by the Bioconductor project were ported to it, both projects grew independently. A collection of other bioinformatics-related Python libraries has also appeared during the last few years: PyCogent [8], pygr [9], and bx-python [10], to name a few.
We choose the R/Bioconductor-Python duo in the context of bioinformatics to demonstrate how bridging software libraries in different languages can be performed. There exists other bioinformatics libraries in other languages [11-14] with which similar principles could be applied, given the relevant tools for bridging the different languages.
Results and discussion
Communities and translation
Whenever a project spans across several communities, the issue of language arises. Bioinformatics is an example of that: being at the interface between biology, computer science, information technology, and statistics, it requires translating terms when experts in the different fields communicate. Here we are focusing on computer languages but the very same principles apply to disciplines. The analogy is even more appropriate when the practitioners of the different disciplines favor one computer language over another one.
Having a bilingual community is a good way to make cross-language barriers fall, but it has the substantial drawback of being relatively difficult and expensive to achieve. When hiring technical specialists, finding experts in a field can be a difficult task, let alone experts in two fields. Moreover, requiring a bilingual community to operate could cause insidious problems: the imperfect mastery of at least one of the two computer languages can help create issues and keep them unnoticed.
A smaller community of bilingual individuals, we shall call translators or interpreters, is able to bridge two larger communities and is easier to obtain than a bilingual community even when setting high standards of fluency for both languages. Translators can be in charge of exposing written blocks in one language, which are here Bioconductor data structures and functions written in R, into meaningful blocks in another language, here Python. The result is an interface layer that can be used without knowing much of the original language in which the libraries were developed.
The software package presented here demonstrates that a translation layer can provide Python developers access to the Bioconductor project, and allow them to develop applications without knowing R.
Exposing Bioconductor/R structures as native Python structures
The role of translators/interpreters can be restricted to wrapping Bioconductor libraries as Python classes. Here we propose to expose Bioconductor to a Python user, and we rely on the Python-to-R bridge rpy2[15]. This bridge embeds an R interpreter into a Python process and allows seamless access to R objects and functions. This bridge removes the need to deal with the technical issues related to accessing R from Python and lets us focus on presenting Bioconductor libraries to Python programmers.
In essence, Bioconductor packages contain functions, data structure definitions (classes), and data objects (instances). The task of translators is to represent these in Python. This can be done manually, or semi-automatically when relying on the meta-programming tools found in the rpy2 package (See Figure 1). Functions usually do not need much work as they are already automatically exposed by rpy2.
Figure 1.
Implementing a Bioconductor class in Python. Implementing a Bioconductor class can be performed by either declaring all class components explicitly (right), or by relying on meta-programming utilities found in the rpy2 package (left). Meta-programming helps reduce the amount of boilerplate code by automating part of the translation work.
The object system in Python is fairly unified, despite the remaining existence of old and new objects in the Python 2.x series, and is very much central to the language. Most, if not all, Python programmers will be familiar with it. In contrast, the Bioconductor project makes extensive use of the S4 class system for R, a system that remains less known to many R users. The S4 system is related to the one of Common Lisp Object System (CLOS) [16], and offers multiple dispatch for methods. The S4 system is only present in a limited number of languages (beside CLOS, Clojure’s multimethods can be mentioned [17]), and is not available in Python. In this context, the difference in object-oriented programming paradigms have to be resolved by translators/interpreters.
Rpy2 exposes classes and methods from Bioconductor are exposed in such a way that differences in programming languages are alleviated. The resulting overall structure matches the canons of Python programming, which Python programmers refer to as being Pythonic. The translation proposed creates Python classes corresponding to the Bioconductor classes, and creates Python methods for the relevant S4 methods. The class and method names are kept across the translation, with minor exceptions for methods. Suffixes are added to the method name when S4 multiple dispatch results in naming conflicts on the Python side, and in that case, the type of the arguments in the signature are added to the method names. For example, the biostrings class PairwiseAlignedXStringSet has three static methods fromXString_XString(), fromCharacter_Character(), fromCharacter_missing() to represent the three corresponding constructors of PairwiseAlignedXStringSet in Bioconductor. This approach helps keeping a high ressemblance between Python and Bioconductor for the functionalities translated.
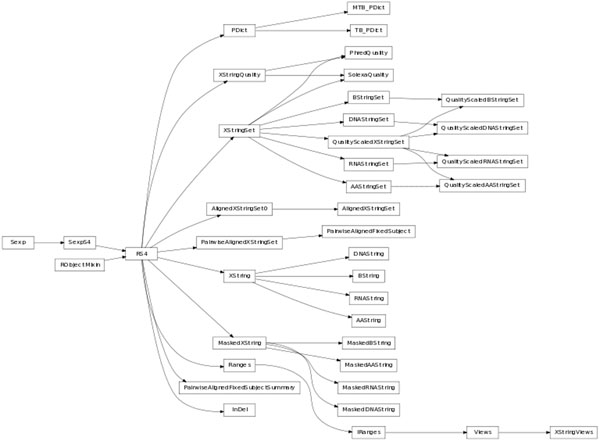
Bioconductor packages can define numerous classes, so it is important that the task of exposing them to Python programmers remains as simple and as short as possible. The Bioconductor package Biostrings alone contains close to 40 classes exposed to Python, as illustrated Figure 2, while the code base for the translation remains of relatively modest size: Biostrings is exposed in less than 600 lines of code, so less than 15 lines of Python code per class exposed on average. In our implementation Python classes are essentially wrappers for R methods of Bioconductor classes, limiting the need for extensive testing.
Figure 2.
Class diagram for the translation of Biostrings. Class from the Bioconductor package Biostrings are exposed to Python as native classes. Parent-child relationships are represented by arrows.
In addition to the above, the task of the translators/interpreters can go beyond exposing the classes. Translating idioms specific to one language into the other language will increase the quality of the translation (for example Python has iterators, not available by default in R and not used in Bioconductor). Translators can also present data structures a different way, and build a new API from the existing Bioconductor libraries. This is of interest in the context of different communities with different views on data structures and methods, as one can quickly rewrap the existing libraries. This can also be helpful for hiding sophisticated options and simplifying the interface, or wrapping sequences of function calls.
The implementation presented here covers several Bioconductor infrastructure packages, and is sufficient to handle annotation data, genome sequences, microarray data, and next-generation sequencing.
annotationdbi: infrastructure for handling biological annotations.
biobase: infrastructure for handling data from high-throughput assays.
biostrings: infrastructure for handling biological strings (DNA, RNA, protein sequences)
bsgenome: infrastructure for handling genome sequences
edger: differential digital expression data
geoquery: query data resources from the Gene Expression Omnibus (GEO) repository.
ggbase: infrastructure for genetics of gene expression
ggtools: software and data for genetics of gene expression
goseq: Gene Ontology analysis for RNAseq
gseabase: infrastructure for Gene Set Enrichment Analysis (GSEA) types of methods
iranges: infrastructure for handling interval data
shortread: infrastructure for handling datasets of short reads
Case-study: providing a web-based interface to edgeR
The egdeR method [18] is a popular statistical method for measuring differential abundance in RNA molecule when the measurement technology is based on counts. It is useful for SAGE and RNAseq data. Having the method easily accessible to a community outside the regular Bioconductor user-base expands its reach to the scientific community. In this scenario a simple web application is considered, and the application is written in Python. One strong advantage of Python over R is the presence of many industry-grade solutions for developing web applications, and we choose to demonstrate how to build such a application with edgeR.
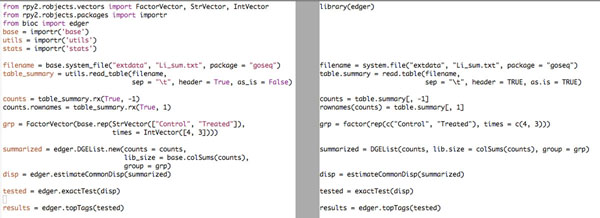
Reproducing R code
The Bioconductor/R edgeR library is exposed to Python in the module bioc. edger, and following the documentation written for R users is straightforward (See Figure 3). As outlined earlier, classes and methods present in the Bioconductor package are represented by matching Python classes and methods, as the translator/interpreter focused on keeping a high resemblance between R and Python code.
Figure 3.
Case study: edgeR with Python. The Python code (left) differs from the R code (right) mostly in the top part, where the Python code is using explicit import procedures.
Building a prototype web server
The code used to perform an edgeR analysis can be wrapped by the Python developer into a Python function, and building a web application that calls this function is trivial (See Figure 4).
Figure 4.
A full webserver for edger. In this scenario Python developers have to develop quickly a prototype website letting users upload a CSV file of gene counts and obtain the table of top differentially counted genes. The main application is minimal (left), relying on three helper functions derived from the earlier example with running edgeR (right). Rendering the result table is not detailed in order to keep the example concise.
A fully functioning self-sufficient prototype, including a web-server, a web-form to upload data, data processing, computation of results from the data uploaded, and an answer returned to a client web browser, can be implemented in less than 100 lines of code.
Having the web server implemented in Python is deemed better because Python has a strong track record of agile web frameworks, the language possesses better error handling mechanisms, and it allows a decoupling of the implementation of data analysis (in R) from the implementation of the application. This separation is important since it allows a programmer specialized in the development of web applications to utilize code developed in R/Bioconductor by data analysts. The translation layer ensures that the code in Bioconductor is exposed in such a way that it can be integrated into the application while retaining all the benefits of the host language.
This example emphasizes the ease with which applications can be built, and relies on a minimal web development framework. There exist more comprehensive and more complex frameworks, such as Django [19] and Plone [20]. Similar implementations have been performed with them. In these cases the development of applications requires highly specialized skills in the corresponding frameworks. In a context where there is specialization of people because of increasingly complex domain-specific knowledge, the availability of a translation layer such as the one proposed is crucial: data analysts can therefore focus on developing algorithms while application developers can focus on the application.
Conclusions
A relatively small community of people fluent in two languages and disciplines can expose data structure definitions and functions from libraries in one language as code directly usable by practitioners of the other language. We demonstrate here how this can be achieved by creating a bridge from the Bioconductor project, a popular set of R libraries for the analysis of bioinformatics data, to the Python language. Work that requires extensive knowledge of both languages can be restricted to a small community of translators/interpreters, and their code be used by Python programmers without the knowledge of R or Bioconductor. The implementation presented here shows that the amount of translation work can be minimal, yet enable the development of Python applications using Bioconductor easily. Our implementation covers key infrastructure packages in the Bioconductor project and can constitute a basis for extending this to more packages in Bioconductor.
As an example we demonstrated how a complete web application computing differential expression for digital gene expression can be implemented.
List of abbreviations
CERN: Conseil Européen pour la Recherche Nucléaire; CLOS: Common Lisp Object System; DNA: Desoxyribonucleic Acid; GEO: Gene Expression Omnibus; GSEA: Gene Set Enrichment Analysis; ILM: Industrial Light and Magic; NASA: National Aeronautics and Space Administration; RNA: Ribonucleic Acid; RNAseq: Whole Transcriptome Shotgun Sequencing; SAGE: Serial Analysis of Gene Expression
Competing interests
The author declares he has no competing interests
Methods
Software and operating systems
R/ Bioconductor
The principles detailed here were applied to Bioconductor Release 2.6 (April 2010). Bioconductor packages evolve quickly and new versions do not always maintain backward compatibility. Minor adaption might be necessary in order to run what is presented here with other releases. The Bioconductor release 2.6 requires R-2.11, available on the project’s website [21].
Python and libraries
Python 2.6.4 was mainly used for development. Other version in the 2.6 series will work. Python is available with most Linux distribution, and is shipped with OS X Leopard and Snow Leopard (version 2.5 and 2.6 respectively).
A development snapshot of the rpy2[15] package (2.2-dev) was used in this work. Minor adaptations will be required for it to work with the current rpy2 release 2.1.
The lightweight web-framework bottle[22] was used to demonstrate the implementation of a web-based interface.
Operating system
The solution was developed and tested under both Ubuntu Linux 10.04 and 10.10 [23] and Apple OS X Leopard.
Authors contributions
LG designed and implemented the software, and wrote the manuscript.
Acknowledgments
Users, and communities from R, Bioconductor, Python, Biopython. Vincent Davis, Nicolas Rapin, Brad Chapman for discussions. Anonymous reviewers for helping improve the original manuscript. Kam Dahlquist, Editor, for language corrections. LG is funded by an infrastructure grant from the Technical University of Denmark.
This article has been published as part of BMC Bioinformatics Volume 11 Supplement 12, 2010: Proceedings of the 11th Annual Bioinformatics Open Source Conference (BOSC) 2010. The full contents of the supplement are available online at http://www.biomedcentral.com/1471-2105/11?issue=S12.
References
- Gentleman RC, Carey VJ, Bates DM. et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing. Vienna, Austria; 2010. R: A Language and Environment for Statistical Computing.http://www.R-project.org [Google Scholar]
- Python. http://www.python.org
- A Gentle Introduction to Programming Using Python. http://ocw.mit.edu/courses/electrical-engineering-and-computer-science/ 6-189-a-gentle-introduction-to-programming-using-python-january-iap-2010/
- Introduction to Programming using Python. http://www.pasteur.fr/formation/infobio/python/
- Prechelt L. An empirical comparison of seven programming languages. Computer. 2002;33 [Google Scholar]
- Cock PJ, Antao T, Chang JT. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, Maxwell P, Birmingham A. et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- pygr - Scalabable bioinformatics interfaces in Python. http://code.google.com/p/pygr/
- bx-python. http://bitbucket.org/jamestaylor/bx-python/overview
- Stajich JE, Block D, K KB. et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12(10):1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BioRuby - Open source bioinformatics library for Ruby. http://bioruby.org
- Holland RCG, Down T, Pocock M, BioJava: an Open-Source Framework for Bioinformatics. Bioinformatics. 2008. [DOI] [PMC free article] [PubMed]
- McKenna A, Hanna M, Banks E, The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. in press . [DOI] [PMC free article] [PubMed]
- rpy2. http://rpy2.sourceforge.net
- Bobrow DG, Kahn K, Kiczales G, Masinter L, Stefik M, Zdybel F. CommonLoops: merging Lisp and object-oriented programming. SIGPLAN Not. 1986;21(11):17–29. doi: 10.1145/960112.28700. [DOI] [Google Scholar]
- Clojure. http://clojure.org
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:1. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Web framework for perfectionists with deadlines. http://www.djangoproject.com
- Plone CMS: Open Source Content Management. http://www.plone.org
- R project. http://www.r-project.org
- Bottle: Python Web Framework. http://www.bottle.paws.de
- Ubuntu. http://www.ubuntu.org