Summary
Circadian (~24 hour) clocks are fundamentally important for coordinated physiology in organisms as diverse as cyanobacteria and humans. All current models of the clockwork in eukaryotic cells are based on transcription-translation feedback loops. Non-transcriptional mechanisms in the clockwork have been difficult to study in mammalian systems. We circumvented these problems by developing novel assays using human red blood cells (RBCs), which have no nucleus (or DNA), and therefore cannot perform transcription. Our results show that transcription is, in fact, not required for circadian oscillations in humans, and that non-transcriptional events appear sufficient to sustain cellular circadian rhythms. Using RBCs, we found that peroxiredoxins, highly conserved antioxidant proteins, undergo ~24 hour redox cycles, which persist for many days under constant conditions (i.e. in the absence of external cues). Moreover, these rhythms are entrainable (i.e. tunable by environmental stimuli), and temperature-compensated, both key features of circadian rhythms. We anticipate our findings will facilitate more sophisticated cellular clock models, highlighting the interdependency of transcriptional and non-transcriptional oscillations in potentially all eukaryotic cells.
Circadian rhythms are a fundamental property of living cells. When held in temporal isolation, organisms from cyanobacteria to humans exhibit behavioural and physiological rhythms that persist with a period of approx. 24 hours1. These rhythms are driven by biological clocks, with two key features. First, their free-running period of ~ 24 hours is temperature-compensated: biological clocks do not run slower at lower temperatures or speed up when hot. Second, they can synchronise to temporally-relevant stimuli such as light, temperature or feeding schedules and thus their definition of internal time becomes predictive of external (solar) time2. Entrained in this way, circadian timing confers selective advantages to organisms by facilitating anticipation of, and thereby adaptation to, the alternating day-night cycle as well as temporally segregating mutually antagonistic processes3. The competitive value of circadian clocks has been demonstrated in prokaryotes and higher plants4,5, whilst disturbance of circadian timing in humans, as seen in rotational shift workers for example, carries significant long-term health costs6.
The molecular clock mechanism is invariably modelled by oscillating transcription-translation feedback, whereby clock proteins feedback to negatively regulate their own transcription, thereby producing rhythmic clock gene expression3. This model has recently been challenged by observations in the simplest organism known to exhibit circadian timing, the cyanobacterium Synechococcus elongatus, in which biochemical oscillations catalysed by several clock proteins occur even in the absence of transcription and translation7,8. Moreover, emerging evidence in multiple eukaryotic systems suggests that assorted cytosolic mechanisms are important to sustain rhythmicity8-13. A critical question that remains, therefore, is whether the nucleus is necessary for circadian rhythms in mammals.
Studying the role of the nucleus in circadian oscillations has been challenging, and in mammals, confined to studies utilising pharmacological inhibitors of transcription14. Such approaches are, however, limited by the pleiotropic effects of pharmacological manipulations (“off-target” effects), and because of their deleterious effects on cell survival14. To circumvent these issues, we developed a unique model system to study circadian rhythmicity, using naturally-occurring anucleate mammalian cells – human red blood cells. Conventional clock gene/protein assays cannot be used in this system because they are not expressed in human red blood cells (see Supplementary Table 1). We therefore developed novel indices of circadian rhythmicity that are not dependent on RNA production.
Peroxiredoxins exhibit circadian rhythms of oxidation and reduction
Peroxiredoxins (PRX) comprise a highly conserved family of anti-oxidant proteins that help to control intracellular peroxide levels15. They share a common catalytic mechanism whereby reducing peroxide, the catalytic active site cysteine residue (CysP) is oxidized to a sulphenic acid (Cys-SOH), which then forms a disulphide bond with a resolving Cys residue (CysR) that is reduced by thioredoxin. In a subclass of PRXs, namely the classical 2-Cys PRXs, a small fraction of the enzyme becomes catalytically inactive through substrate-mediated hyperoxidation of CysP to the sulphinic and sulphonic acid forms (Cys-SO2/3H), and is eventually recycled through ATP-dependent reduction by sulfiredoxin16. Given this mechanism of action, and that circadian post-translational modification of a PRX family member was observed previously17, we hypothesised that PRX oxidation may constitute a novel non-transcriptional rhythmic marker. To test this, we employed human red blood cells (RBCs), which lack nuclei and therefore transcriptional capability, to directly test whether rhythms persist without active transcription.
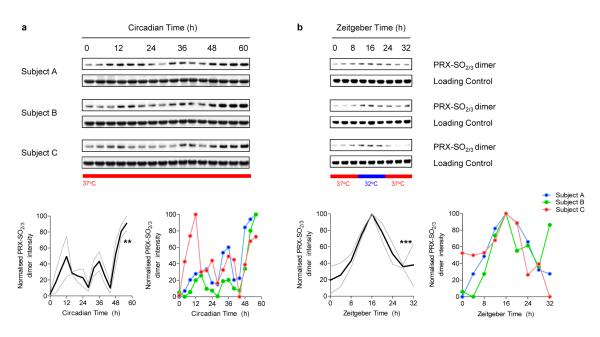
We harvested blood from three aged-matched healthy volunteers (Subjects A, B, and C), and prepared RBC fractions from whole blood. RBCs were then kept in constant conditions of temperature (37°C) and light (complete darkness) and sampled at 4-hourly intervals for 60 hours. Lysates were immunoblotted using an antibody specific to hyperoxidised peroxiredoxin (PRX-SO2/3) to monitor the hyperoxidation status of PRX for each subject (Fig. 1a). For all subjects we observed robust, statistically significant, cycling of PRX-SO2/3 with a period of ~24 hours (Fig. 1a). The clearest and most robust rhythms were exhibited by the dimeric form of PRX (as shown in Fig 1.; a full blot image, illustrating all molecular weight species is shown in Supplementary Fig. 5). Thus, circadian oscillation of post-translational modification of peroxiredoxins occurs in RBCs that are kept in free-running conditions (i.e. in the absence of external time cues), reflecting an endogenous ~ 24 hour (circadian) rhythm.
Figure 1. Circadian oscillation of peroxiredoxin (PRX) oxidation in human red blood cells.
a, Red blood cells from three human subjects (A, B, C) were entrained by temperature cycles and then kept under constant conditions (at 37°C, in total darkness) and sampled every 4 hours. b, Red blood cells incubated in alternating 12 hour cycles of high (37°C) and low (32°C) temperature. Representative immunoblots showing oxidised/hyperoxidised peroxiredoxin (PRX-SO2/3) dimer are shown with loading controls. Quantification by densitometry is shown below. Values were normalised to the maximum for each blot. Solid line represents mean normalised intensity, with grey lines indicating s.e.m. boundaries. ** p < 0.01, *** p <0.001 by 1-way ANOVA (effect of time).
To be a useful timing mechanism, or clock, circadian oscillations are tuneable by external cues so that they can be reset when misaligned18. When clocks are stably synchronised to an external cycle, they are said to be “entrained”. Cellular oscillators can be entrained by a variety of stimuli including light, temperature and feeding schedules3,19. We used temperature cycles to entrain cultures of RBCs since this is likely to be a physiologically-relevant time cue as they circulate through the body over a typical day in vivo. Humans experience circadian variation in body temperature, rising to a peak in the late evening (~37.4°C), and falling to reach a minimum (~36.8°C) in the early morning20. We subjected purified RBCs to alternating 12 hour cycles of high (37°C) and low (32°C) temperature for 2 cycles (48 hours) before sampling every 4 hours for a further 32 hours (1½ cycles). We observed PRX-SO2/3 rhythms during temperature entrainment, with low levels seen at high temperatures, and higher levels at low temperatures (Fig. 1b). Importantly, we observed differential entrainment of samples that started in the same phase, but through temperature entrainment, resulted in opposite phases (Supplementary Fig. 1). Together, these results therefore identify PRX oxidation-reduction rhythms as robust, entrainable circadian rhythms in cells lacking the ability to make new RNA.
Rhythmic PRX oxidation is resistant to transcription/translation blockade
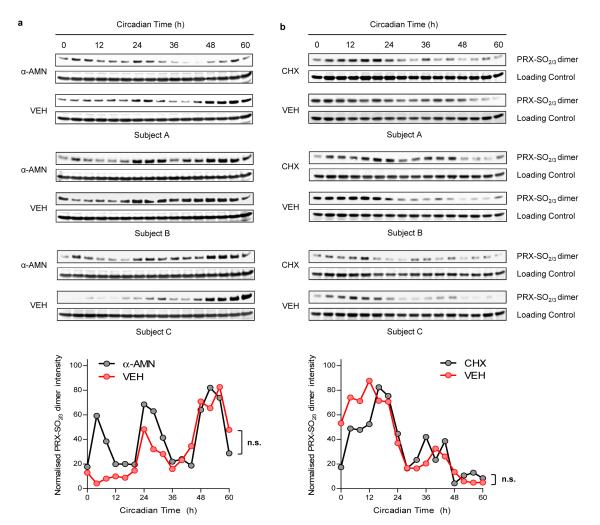
A possible alternative explanation for rhythmicity in our assays could be due to oscillation of contaminating nucleated white blood cells. However, it was clear that at normal ratios, even in whole blood, the maximum number of white cells that could be present cannot contribute significantly to any signal that we observed (Supplementary Fig. 2). To substantiate this beyond reasonable doubt, we performed experiments in the presence and absence of well described transcriptional and translational inhibitors, which should only have effects if nucleated cells contribute significantly to observed PRX oxidation cycles. RBCs were entrained in temperature cycles, as above, and then placed into free-running conditions at 37°C in darkness. Those incubated with the potent cell-permeable transcriptional blocker α-amanitin oleate (α-AMN) had highly reproducible circadian rhythms that, surprisingly, were even more robust than vehicle-treated (VEH) control RBCs (Fig. 2a and Supplementary Fig. 3a). Quantification of PRX-SO2/3 expression revealed no significant difference between the two treatment groups by 2-way ANOVA (Fig. 2a). Similar results were obtained with cycloheximide (CHX), a eukaryotic translation inhibitor (Fig. 2b and Supplementary Fig. 3b). Given that the RBCs were cultured in the presence of antibiotics, we strongly believe that the rhythms we observe in peroxiredoxin oxidation arise only from RBCs and that other cell types, eukaryotic or otherwise, do not contribute to the oscillations we observe in vitro.
Figure 2. Circadian rhythms of peroxiredoxin (PRX) oxidation are not affected by transcriptional and translational inhibition.
RBCs were entrained under temperature cycles and then kept under constant conditions (at 37°C, in total darkness) and sampled every 4 hours. Representative immunoblots showing oxidised/hyperoxidised peroxiredoxin (PRX-SO2/3) dimer are shown for samples incubated with a, α-amanitin (α-AMN), or b, cycloheximide (CHX) for the entirety of the experiments. Quantification by densitometry is shown below. Values were normalised to the maximum for each blot. Each point represents a mean normalised intensity. n.s., not significant. Further details are shown in Supplementary Fig. 3.
Temperature-compensation of circadian rhythms in RBCs
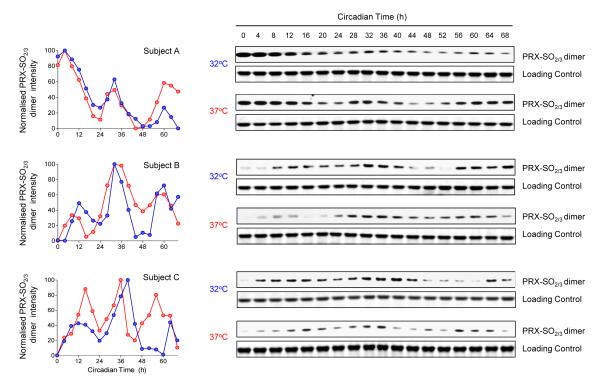
A key property of circadian clocks is that their free-running period length remains ~ 24 hours despite being held at quite different temperatures under steady-state conditions, i.e. they are temperature-compensated. To determine if PRX oxidation rhythms exhibit this property, we entrained RBCs in temperature cycles, and then allowed them to free-run at either 37°C or 32°C (Fig. 3). When released into free-run at the two temperatures, oscillations of PRX-SO2/3 were very similar (Fig. 3). We also attempted to culture the RBCs at a higher temperature (42°C), but found that this was toxic to the cells after 2 days of incubation (they displayed profound haemolysis), precluding any further assessment of PRX rhythms at this temperature in vitro. Together, our findings therefore show that RBCs cultured in vitro exhibit free-running, temperature-compensated, entrainable circadian rhythms of peroxiredoxin oxidation, indicative of the presence of a functioning clock in these non-nucleated cells.
Figure 3. Temperature-compensation of circadian peroxiredoxin oxidation rhythms.
Red blood cells were entrained in temperature cycles (12 h at 32°C, 12 h at 37°C) for two complete cycles and then kept under a constant temperature of either 32°C or 37°C for the rest of the experiment and sampled every 4 hours as before. Immunoblots for oxidised/hyperoxidised peroxiredoxin (PRX-SO2/3) dimer obtained from red blood cells from subjects A, B and C are shown. Loading controls (Coomassie-stained gels showing haemoglobin monomer bands) for each blot are also shown. Quantification of the above immunoblots by densitometry is shown on the left of the figure.
Peroxiredoxin rhythms are complex in their phenotype
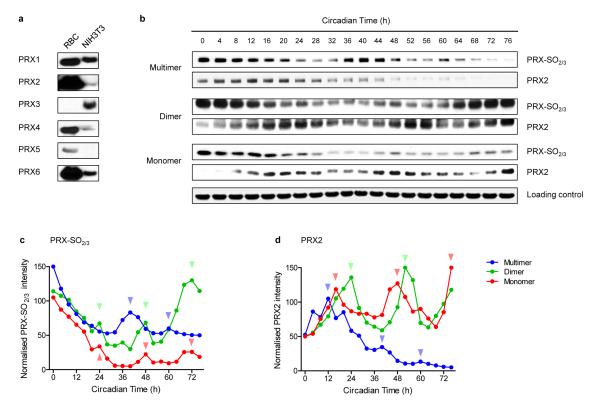
Having established robust circadian oscillations of peroxiredoxin oxidation, we next sought to determine further the nature of these oscillations. Peroxiredoxins are highly conserved across the major phylogenetic kingdoms (eukaryotes, archaea and bacteria)15. In mammals, there are six PRX paralogues, and they differ in subcellular localisation and their anti-oxidant mechanism21. To dissect which peroxiredoxin(s) are relevant to the observed PRX-SO2/3 rhythms in RBCs, we determined the expression of PRX1-6 in human RBCs and nucleated mouse fibroblast (NIH3T3) cells (Fig. 4a). We focused particularly on PRX2 because of its very high expression in RBCs and documented reversible behaviour under oxidising and reducing conditions22 (Fig. 4b). As well as the dimeric form of PRX, other electrophoretic forms exist. Interestingly, although clearly rhythmic, different oligomeric forms of PRX1/PRX2 and PRX-SO2/3 displayed distinct phase relationships, suggesting the possibility of “shuttling” between the forms by reversible oligomerisation (Supplementary Figs. 4c and 5). PRX species thus display complex and likely interlinked time-varying oligomerisation behaviour, some of which is overtly circadian (Fig. 4b-d).
Figure 4. Expression patterns and oligomerisation of peroxiredoxins.
a, Immunoblots showing expression of the human peroxiredoxin paralogues (PRX1-6) in red blood cells (RBC) and in mouse NIH3T3 cells. Loading of each lane was approximately equal. b, Oligomerisation patterns of PRX and PRX-SO2/3 in red blood cells. Following two cycles of temperature entrainment, cells were kept under constant temperature (37°C) for the rest of the experiment, and sampled every 4 hours. Representative immunoblots for PRX2 and PRX-SO2/3 are shown. Whole blot images in Supplementary Fig. 5 illustrate the different oligomeric forms. Immunoblots were quantified by densitometry for c, PRX-SO2/3 and d, PRX2. Arrowheads indicate peaks of abundance.
Circadian rhythms in reversible haemoglobin oxidation
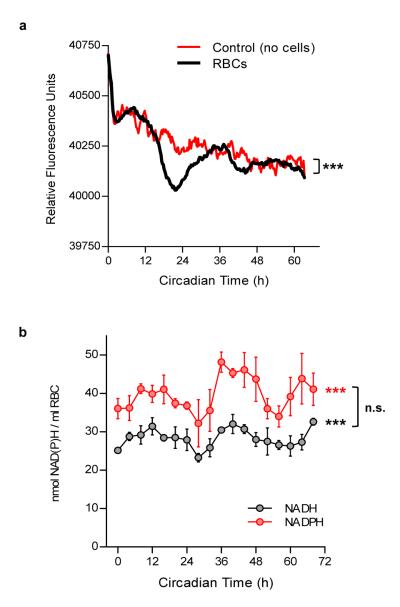
Given the robust circadian rhythms of PRX oxidation, we next explored the possible mechanisms that might underlie them. RBCs transport oxygen in the blood, and haemoglobin (Hb) is essential for this. Hb itself is a source of peroxide intracellularly via auto-oxidation23. Since the dimeric form of Hb displays a ~13 fold higher auto-oxidation rate than the tetramer24, we hypothesised that circadian modulation of Hb tetramer-dimer equilibrium might be linked with rhythms in PRX state. Unlike the normal Hb tetramer, the dimer displays no cooperativity25, and is also far more readily auto-oxidised to methaemoglobin (metHb)24, doubly impairing its ability to transport oxygen. Therefore, if the tetramer:dimer ratio, and thus metHb formation rate were clock-regulated, a circadian variation in oxygen-carrying ability would be expected. Given that blood gas measurements (oxygen concentration) have been shown to be diurnally regulated in vivo26, this line of enquiry seemed promising. To test our theory, we employed intrinsic front-face fluorescence (FFF) as a real-time assay of rhythmicity27. We measured FFF from RBCs for more than 60 hours under free-running conditions and observed highly reproducible circadian oscillations (Fig. 5a and Supplementary Fig. 6a). This strongly indicates reversible low-amplitude oxidation of Hb in RBCs, even when held in vitro.
Figure 5. Circadian rhythms in haemoglobin oxidation and red blood cell (RBC) metabolism.
a, Intrinsic front-face fluorescence (FFF) measurements of RBCs and controls. Experiments performed under constant conditions (at 37°C, in total darkness). Mean values for each time-point are shown (individual traces and further details are in Supplementary Fig. 6a). 2-way ANOVA (group × time) p < 0.001 (***). b, NADH and NADPH concentrations in red blood cells. Mean values (± s.e.m.) for three experimental subjects are shown. 1-way ANOVA (effect of time) for NADH/NADPH data was significant (*** p < 0.001). 2-way ANOVA (metabolite × time) did not reveal a significant difference between NADH and NADPH profiles (n.s., not significant). Individual profiles shown in Supplementary Fig. 6b,c.
Circadian rhythms in red blood cell metabolism
How might rhythms in peroxiredoxins and haemoglobin oxidation interconnect with metabolic pathways in RBCs? The archetypal reducing equivalents NADH and NADPH lie at the core of cellular redox reactions, and thus might be expected to be rhythmic in RBCs27-29. We thus assayed NADH/NADPH in RBCs sampled under free-running conditions, and indeed found ~24 hour rhythms of abundance (Fig. 5b and Supplementary Fig. 6b,c).
Given that red blood cells are dependent on glycolysis for ATP synthesis, and that this contributes significantly to NADH flux, we also assayed the levels of ATP in red blood cells. Although levels damp rapidly over the first 48 hours of sampling, we observed two cycles of circadian oscillation of ATP (Supplementary Fig. 6d), supporting the idea that cycles we observe are metabolic in origin. It is unclear how these low amplitude circadian oscillations of ATP relate to well-described short period (~10 min) glycolytic cycles30-32. A possibility that remains to be tested is whether these rapid oscillations underlie circadian oscillations of ATP, although red blood cells may not be the ideal platform for resolving this.
Peroxiredoxin rhythms in nucleated cells
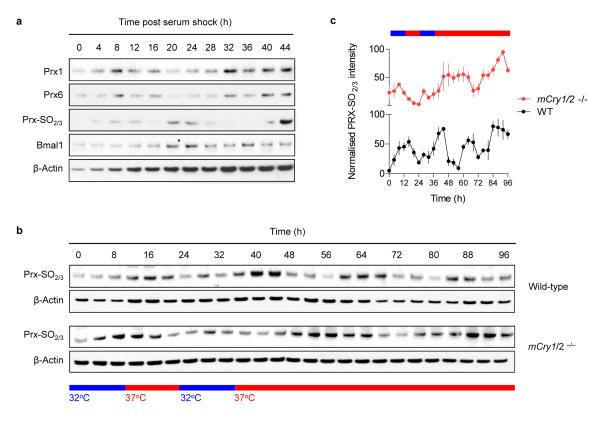
Of particular interest is the circadian oscillation of NADH/NADPH in RBCs because direct modulation of DNA-binding activity of the core circadian transcription factors Clock and Bmal1 by NADH/NADPH could couple “cytoplasmic” and “nuclear” rhythms in nucleated cells33,34 (Supplementary Fig. 8). We therefore mapped the expression of peroxiredoxins in mouse NIH3T3 cells, the best characterised cellular clock model system14,35. Prx1 and Prx6 exhibited high-amplitude circadian cycles, as did oxidised Prx-SO2/3 (Fig. 6a). Thus, anucleate RBCs and nucleated cells share common components displaying circadian rhythms.
Figure 6. Peroxiredoxin rhythms in nucleated cells.
a, Peroxiredoxin rhythms in mouse NIH3T3 fibroblasts synchronised by a serum-shock. Immunoblots for Prx1, Prx6 and Prx-SO2/3 dimer are shown, in addition to Bmal1 and a β-actin loading control. b,c Peroxiredoxin rhythms in mouse embryonic fibroblasts (MEFs). MEFs from wild-type or mCry1/2 double-knockout mice were entrained in temperature cycles and then kept under constant temperature (37°C) for the rest of the experiment (as shown in the schematic). b, Representative immunoblots of oxidised/hyperoxidised peroxiredoxin (Prx-SO2/3) dimer. c, Quantification of Prx-SO2/3 immunoblots by densitometry. Mean values (± s.e.m.) for n=4 biological replicates are shown.
To examine cross-talk between cytosolic and nuclear rhythms in modulating peroxiredoxin oxidation, we assayed mouse embryonic fibroblasts (MEFs) from mCry1/2 double-knockout mice, which lack cyclical expression of known clock genes/proteins36,37. Cells were sampled under conditions to mimic conditions employed in our red blood cell experiments (Fig. 6b,c). Although present in mCry1/2 knockout cells, rhythms in peroxiredoxin oxidation were altered relative to those seen in wild-type MEFs, with an apparently lengthened period of oscillation (Fig. 6b,c). Therefore, in nucleated cells, peroxiredoxin rhythms are influenced by the transcription-translation feedback loop (Supplementary Fig. 9). This implies direct coupling between nuclear and cytoplasmic rhythms in nucleated cells.
Thus far peroxiredoxins have been treated purely as a rhythmic biomarker, but peroxiredoxins could affect the transcription-translation feedback loop. To assess this, we interrogated a recent genome-wide RNA interference screen which sought to identify genes modifying circadian transcription in human U2OS cells38. Knockdown of PRX2 and PRX4 resulted in a long-period phenotype, whereas siRNAs directed against PRX3 and PRX5 depressed the amplitude of circadian oscillations (Supplementary Fig. 7 and Supplementary Table 2). Therefore, in nucleated cells, there is likely to be an intricate interplay between transcription-dependent processes and non-transcriptional oscillations, since there appears to be reciprocal regulation between these systems (Supplementary Fig. 9).
Discussion
We have shown that human red blood cells display robust, temperature-entrainable and temperature-compensated circadian rhythms, consistent with the presence of a circadian clock within these cells. By demonstrating circadian oscillations in a multitude of cytoplasmic redox parameters (peroxiredoxin oxidation-reduction, haemoglobin tetramer-dimer transitions and NADH/NADPH oscillations), some of which are clearly also present in nucleated cells not involved in oxygen transport, we believe we have uncovered interconnectivity between seemingly distinct cytoplasmic (metabolic) and nuclear circadian processes (Supplementary Fig. 9). Moreover, the interrelationship between post-translational and transcription-dependent oscillations in nucleated cells resembles the emerging model in cyanobacteria39. In this regard, peroxiredoxins are extremely interesting from a phylogenetic perspective, since they are found in virtually all known organisms40. Thus, a testable and important question is whether they (and similar molecules) constitute a circadian marker that transcends phylogenetic kingdoms. This is of great interest since many classical model organisms that are genetically tractable (e.g. yeast and C. elegans), have not been found to express any known “clock genes”, but do exhibit circadian rhythms41,42. In this context, the finding that peroxiredoxin rhythms are exhibited in the alga Ostreococcus tauri43, a novel plant-like clock model organism44, and that they persist in the absence of transcription, echoes this as a potential underlying principle. Furthermore, it has not escaped our notice that rhythmic peroxiredoxin oligomerisation resembles the circadian variation in the formation of hexameric KaiC ring structures in cyanobacteria45, and may therefore be a fundamental part of circadian “logic” in divergent organisms.
It remains to be seen whether peroxiredoxins (and their homologues/orthologues) are essential for circadian rhythmicity, but we anticipate that deleting these fundamental and redundant genes will have generally deleterious cellular consequences. We hope that our findings may shed light on a number of apparent paradoxes concerning circadian clocks, such as how they continue to keep time accurately in the absence of transcription, as occurs for example during cell division cycles. Moreover, as we move towards a deeper understanding of cellular metabolism and redox status in the context of the clockwork, we anticipate that our findings will aid development of current clock models and a reassessment of the role of the nucleus in these.
Methods
Participants
Studies were conducted in accordance with the principles of the Declaration of Helsinki, with ethical approval from the Local Research Ethics Committee (Cambridge, UK). Participants in the study were screened for health (by history, physical examination, and standard biochemistry and haematology), and did not suffer from sleep disorders or excessive daytime sleepiness. All participants provided written, informed consent after having received a detailed explanation of the study procedures.
Red blood cell culture
Approximately 10 ml of blood was collected from each of three subjects for each experiment by using tubes containing sodium citrate anti-coagulant (Sarstedt, UK). Red cell pellets (and white cell fractions) were obtained using gradient centrifugation at 25°C with Accuspin Histopaque-1077 columns (Sigma-Aldrich, UK), following the manufacturer’s protocol. After two washes in approx. 50ml of sterile phosphate-buffered saline (PBS), the pellet (approx. 8 ml volume) was resuspended to a total volume of 30 ml with sterile modified Krebs-Henseleit Buffer (Sigma-Aldrich, UK). Krebs buffer was made up in 500 ml batches with sterile water, and adding 0.1% w/v bovine serum albumin (Sigma-Aldrich, UK), with antibiotics added (100 units/ml penicillin, 100 mg/ml streptomycin) and the osmolarity adjusted to 280 mOsm/L, and pH to 7.4, to match conditions normally found in human plasma. For all experiments, 100 μl of re-suspended RBCs were dispensed into 0.2 ml PCR tubes (Thermo), and then placed in a thermal cycler (Bio-Rad Tetrad) for temperature-entrainment and free-running assessments. When indicated, 100 nM α-amanitin oleate (α-AMN) or 5 μg/ml cycloheximide (CHX), purchased from Sigma-Aldrich, were added to the RBCs prior to aliquoting, and thus the drugs were present for the entirety of the experiment. Drugs were made up in sterile dimethyl sulfoxide (DMSO), which was also used therefore as the vehicle control (VEH) for those experiments. Samples were taken directly from the thermal cycler and either flash-frozen (with liquid nitrogen) or 75 μl immediately lysed in 250 μl 2X LDS buffer (Life Technologies) and heated to 70°C for 10 mins with constant shaking (600 rpm) in a thermomixer (Grant, UK). Samples and lysates were stored at −80°C until use.
NIH3T3 cell culture
NIH3T3 cells were obtained from the American Type Culture Collection (ATCC) and used at passage 6 for these experiments. Cells plated into 35 mm circular standard cell culture dishes, with at least n=3 replicates for each time-point to be assayed. Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Sigma-Aldrich), supplemented with 10% fetal bovine serum (Hyclone), 100 units/ml penicillin and 100 mg/ml streptomycin. Cultures were maintained under standard sterile cell culture conditions, at 37°C and in 5% CO2, in a humidified incubator. Experiments were performed when cultures were fully confluent (100%). Serum starvation was instituted for 24 hours, prior to serum-shock with 50% horse serum for 2 hours, and then incubated in standard supplemented high-glucose DMEM for subsequent sampling. Dishes were taken from the incubator 0-44 hours after the serum-shock, and cells lysed immediately in LDS Buffer (Life Technologies). Lysates were immediately heated to 70°C for 10 mins with constant shaking (600 rpm) in a thermomixer (Grant, UK). Samples and lysates were stored at −80°C until use. For MEF experiments, temperature cycles were imposed on fully confluent cells growing in 12-well plates using an automated temperature-cycling incubator. Sampling at each time-point was performed on four biological replicates, and lysates prepared as above, after initial lysis in non-reducing CHAPS/Urea buffer to facilitate protein loading correction46.
Flow cytometry and cell viability assays
Cells were stained with Vybrant DyeCycle Ruby (Life Technologies), at 1:500 dilution for 60 mins at 25°C, and then assessed by flow cytometry using a FACScalibur system (BD Biosciences, UK). Data were analysed with the CellQuest Pro software. 2 × 104 cells were counted for each of the three experimental subjects. For quantification, a FL3 (red) channel cut-off level of >100 (determined empirically), was used to classify cells as being nucleated. For cell viability assays, either whole blood or purified red cell fractions were subjected to haemolysis by resuspending respective pellets (approx. 200 μl volume) in 800 μl hypotonic solution (50% PBS, pH 7.2) for 1 min at 25°C. Following incubation with hypotonic solution (which lyses red cells but not white cells), the tonicity is brought back to 100% PBS by adding 110μl of 10X PBS concentrate (Life Technologies). Aliquots of the haemolysed samples from each subject were then assayed, using 0.4% trypan blue, with the automated Countess system (Life Technologies) following the manufacturer’s instructions.
Real-time intrinsic front-face fluorescence (FFF) measurements
Red blood cells were prepared as above and then diluted 1:100 for FFF. Fluorescence measurements were performed in 96-well, black, clear-bottom sterile culture plates, using an Enspire plate reader (PerkinElmer, UK) held at 37°C continuously, in its light-tight enclosure. Emission was recorded at 325 nm, using an excitation wavelength of 280 nm, every 15 mins for 64 hours in total. The excitation/emission wavelength is optimal for detecting maximal fluorescence in red blood cells47.
NADH and NADPH assays
Colourimetric assays were performed using commercial kits from Abcam, UK (ab65348, ab65349). Cell pellets (100μl per time-point, per biological replicate) were processed following the instructions in the kit. NADH/NADPH were extracted in the recommended extraction buffer, and all samples were processed in a single run to aid quantification and comparability. Colourimetric measurements were made at 25°C using OD450 measurements on a Packard Fusion plate-reader. We performed n=3 technical replicates for each of n=3 biological replicates (Subjects A, B, C) at each time-point. OD450 measurements were converted to nmol/ml red blood cells (RBC) using a standard curve for both NADH and NADPH.
ATP assays
We used the Promega ENLITEN ATP assay system, following the manufacturer’s protocol. ATP was extracted from cell pellets (100 μl per time-point, per biological replicate) with an equal volume of 0.38N (6%) trichloroacetic acid (Sigma). Samples were then diluted/neutralised 1:50 with Tris-Acetate buffer to final pH 7.75. This step brought the samples into an appropriate pH range for the bioluminescence assay, and also decreased the trichloroacetic acid concentration to (0.1%), both of which are necessary to avoid inhibition of the luciferase reaction. Luminescence measurements were performed in triplicate for each biological replicate using a Berthold Centro LB 960 high-sensitivity system, integrating photon counts over 2 secs per well.
Gel electrophoresis and immunoblotting
We used NuPAGE Novex 4-12% Bis-Tris gradient gels (Life Technologies), and ran them using the manufacturer’s protocol with a non-reducing MES SDS buffer system, allowing characterisation of proteins between 10-260 kDa. Protein transfer to nitrocellulose for blotting was performed using the iBlot system (Life Technologies), with a standard (P3, 7 min) protocol. Nitrocellulose was then washed briefly, and then blocked for 30 mins in 0.5% w/w BSA/non-fat dried milk (Marvel, UK) in Tris Buffered Saline/0.05% Tween-20 (TBST). After three brief washes in TBST, membranes were incubated in antibody diluted in blocking buffer (0.5% milk/BSA) overnight at 4°C. The following day, membranes were washed for 5 mins three times (in TBST) and then incubated with 1:10,000 HRP-conjugated secondary antibody (Sigma-Aldrich) for 30 mins. Four more 10 mins washes were then performed before performing chemiluminescence detection using ECL Plus reagent (GE Healthcare). To check protein loading was even in the gels, they were stained with Coomassie SimplyBlue (Life Technologies). Antisera against peroxiredoxins were obtained from Abcam (Cambridge, UK) and used at the recommended dilutions (PRX-SO2/3: ab16830, PRX1: ab59538, PRX2: ab15572, PRX3: ab16751, PRX4: ab16943, PRX5: ab16944, PRX6: ab16947, 2-Cys PRX: ab16765). Rabbit anti-Bmal1 antiserum was used at 1:2000 in 0.5% BSA46. A mouse β-actin antibody (Santa Cruz Antibodies, USA) was used at 1:5000 in 0.5% milk/BSA.
Image and statistical analysis
Coomassie-stained gel images were obtained using a Licor Odyssey system, and immunoblot films were scanned using a back-illuminated flat-bed scanner. Densitometric quantification of images was performed using NIH ImageJ software. Parametric statistics (1-way and 2-way ANOVA) were performed using Graphpad Prism v5 software.
Methods Summary.
For red cell experiments, ~10 ml of blood was collected from each of three subjects for each experiment by using tubes containing sodium citrate anti-coagulant. Red cell pellets (and white cell fractions) were obtained using gradient centrifugation at 25°C. After two washes in sterile PBS, the pellet was resuspended in sterile modified Krebs-Henseleit Buffer (pH 7.4, 280 mOsm/L). For all experiments, 100 μl of re-suspended RBCs were dispensed into 0.2 ml PCR tubes, and then placed in a thermal cycler for temperature-entrainment and free-running assessments. When indicated, 100 nM α-amanitin oleate (α-AMN) or 5μg/ml cycloheximide (CHX), were added to the RBCs. DMSO was used as the vehicle control (VEH) for those experiments. Samples were lysed in LDS buffer and heated to 70°C for 10 mins with constant shaking (600 rpm) in a thermomixer. Electrophoresis was performed using pre-fabricated 4-12% Bis-Tris gradient gels, using a non-reducing MES SDS buffer system, allowing characterisation of proteins between 10-260 kDa. Immunoblotting was performed after protein transfer to nitrocellulose membranes. After blocking, membranes were incubated in antibody diluted in blocking buffer (0.5% milk/BSA) overnight at 4°C. The following day, membranes were washed and bands visualised with chemiluminescence detection. To check protein loading was even in the gels, they were stained with Coomassie blue. Front Face Fluorescence measurements were performed in 96-well clear-bottom sterile culture plates, using a PerkinElmer Enspire plate reader held at 37°C continuously, in its light-tight enclosure. Emission was recorded at 325 nm, using an excitation wavelength of 280 nm, every 15 mins for 64 hours in total. Colourimetric NADH/NADPH assays were performed using kits purchased from Abcam, following the manufacturer’s protocol.
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust (083643/Z/07/Z), the MRC Centre for Obesity and Related metabolic Disorders (MRC CORD) and the NIHR Cambridge Biomedical Research Centre. We thank M. Jain and R. Edgar for helpful discussion about the manuscript, A. Coles and J. Jones for providing access to samples, and G. van der Horst and F. Tamanini for providing access to mCry1/2 knockout (and wild-type) mouse embryonic fibroblasts.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author information Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9(10):764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2009 doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks; an experimental assessment in cyanobacteria. Curr Biol. 2004;14(16):1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 5.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309(5734):630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 6.Barger LK, Lockley SW, Rajaratnam SM, Landrigan CP. Neurobehavioral, health, and safety consequences associated with shift work in safety-sensitive professions. Curr Neurol Neurosci Rep. 2009;9(2):155–164. doi: 10.1007/s11910-009-0024-7. [DOI] [PubMed] [Google Scholar]
- 7.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307(5707):251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 8.Lakin-Thomas PL. Transcriptional feedback oscillators: maybe, maybe not. J Biol Rhythms. 2006;21(2):83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- 9.Dodd AN, et al. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science. 2007;318(5857):1789–1792. doi: 10.1126/science.1146757. [DOI] [PubMed] [Google Scholar]
- 10.Johnson CH, et al. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science. 1995;269(5232):1863–1865. doi: 10.1126/science.7569925. [DOI] [PubMed] [Google Scholar]
- 11.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320(5878):949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN. Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J Neurosci. 2007;27(46):12489–12499. doi: 10.1523/JNEUROSCI.3680-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woolum JC. A re-examination of the role of the nucleus in generating the circadian rhythm in Acetabularia. J Biol Rhythms. 1991;6(2):129–136. doi: 10.1177/074873049100600203. [DOI] [PubMed] [Google Scholar]
- 14.Dibner C, et al. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 2009;28(2):123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins--structures, mechanisms and functions. FEBS J. 2009;276(9):2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhee SG, Jeong W, Chang TS, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int Suppl. 2007;(106):S3–8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- 17.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 18.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4(8):649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 19.Hastings MH, Maywood ES, Reddy AB. Two decades of circadian time. J Neuroendocrinol. 2008;20(6):812–819. doi: 10.1111/j.1365-2826.2008.01715.x. [DOI] [PubMed] [Google Scholar]
- 20.Wright KP, Jr., Hull JT, Czeisler CA. Relationship between alertness, performance, and body temperature in humans. Am J Physiol Regul Integr Comp Physiol. 2002;283(6):R1370–1377. doi: 10.1152/ajpregu.00205.2002. [DOI] [PubMed] [Google Scholar]
- 21.Barranco-Medina S, Lazaro JJ, Dietz KJ. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 2009;583(12):1809–1816. doi: 10.1016/j.febslet.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 22.Woo HA, et al. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J Biol Chem. 2003;278(48):47361–47364. doi: 10.1074/jbc.C300428200. [DOI] [PubMed] [Google Scholar]
- 23.Cho CS, et al. Irreversible inactivation of glutathione peroxidase 1 and reversible inactivation of peroxiredoxin II by H2O2 in red blood cells. Antioxid Redox Signal. 12(11):1235–1246. doi: 10.1089/ars.2009.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffon N, et al. Tetramer-dimer equilibrium of oxyhemoglobin mutants determined from auto-oxidation rates. Protein Sci. 1998;7(3):673–680. doi: 10.1002/pro.5560070316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hewitt JA, Kilmartin JV, Eyck LF, Perutz MF. Noncooperativity of the dimer in the reaction of hemoglobin with oxygen (human-dissociation-equilibrium-sulfhydryl-absorption-x-ray analysis) Proc Natl Acad Sci U S A. 1972;69(1):203–207. doi: 10.1073/pnas.69.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Latenkov VP. Diurnal rhythm of acid-base equilibrium and blood gas composition. Biull Eksp Biol Med. 1986;101(5):614–616. [PubMed] [Google Scholar]
- 27.Kennett EC, et al. Investigation of methaemoglobin reduction by extracellular NADH in mammalian erythrocytes. Int J Biochem Cell Biol. 2005;37(7):1438–1445. doi: 10.1016/j.biocel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Ogo S, Focesi A, Jr., Cashon R, Bonaventura J, Bonaventura C. Interactions of nicotinamide adenine dinucleotides with varied states and forms of hemoglobin. J Biol Chem. 1989;264(19):11302–11306. [PubMed] [Google Scholar]
- 29.Jacobsen MP, Winzor DJ. Characterization of the interactions of NADH with the dimeric and tetrameric states of methaemoglobin. Biochim Biophys Acta. 1995;1246(1):17–23. doi: 10.1016/0167-4838(94)00174-f. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh AK, Chance B, Pye EK. Metabolic coupling and synchronization of NADH oscillations in yeast cell populations. Arch Biochem Biophys. 1971;145(1):319–331. doi: 10.1016/0003-9861(71)90042-7. [DOI] [PubMed] [Google Scholar]
- 31.Hess B, Brand K, Pye K. Continuous oscillations in a cell-free extract of S. carlsbergensis. Biochem Biophys Res Commun. 1966;23(1):102–108. doi: 10.1016/0006-291x(66)90276-2. [DOI] [PubMed] [Google Scholar]
- 32.Betz A, Chance B. Phase Relationship of Glycolytic Intermediates in Yeast Cells with Oscillatory Metabolic Control. Arch Biochem Biophys. 1965;109:585–594. doi: 10.1016/0003-9861(65)90404-2. [DOI] [PubMed] [Google Scholar]
- 33.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 34.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293(5529):510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 35.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119(5):693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Yagita K, Tamanini F, van der Horst GTJ, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 37.van der Horst GTJ, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 38.Zhang EE, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139(1):199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin X, Byrne M, Xu Y, Mori T, Johnson CH. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 2010;8(6):e1000394. doi: 10.1371/journal.pbio.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood ZA, Schroder E, Harris J. Robin, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28(1):32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 41.Eelderink-Chen Z, et al. A circadian clock in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 107(5):2043–2047. doi: 10.1073/pnas.0907902107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kippert F, Saunders DS, Blaxter ML. Caenorhabditis elegans has a circadian clock. Curr Biol. 2002;12(2):R47–49. doi: 10.1016/s0960-9822(01)00670-4. [DOI] [PubMed] [Google Scholar]
- 43.O’Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2010 doi: 10.1038/nature09654. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corellou F, et al. Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell. 2009;21(11):3436–3449. doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson CH, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol. 2008;18(17):R816–R825. doi: 10.1016/j.cub.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 47.Hu T, et al. PEGylation of Val-1(alpha) destabilizes the tetrameric structure of hemoglobin. Biochemistry. 2009;48(3):608–616. doi: 10.1021/bi801880y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.