Abstract
Circadian rhythms are ubiquitous in eukaryotes, and co-ordinate numerous aspects of behaviour, physiology and metabolism, from sleep/wake cycles in mammals to growth and photosynthesis in plants1,2. This daily timekeeping is thought to be driven by transcriptional/translational feedback loops, whereby rhythmic expression of clock gene products regulates expression of associated genes in approximately 24-hour cycles. The specific transcriptional components differ between phylogenetic kingdoms3. The unicellular pico-eukaryotic alga, Ostreococcus tauri, possesses a naturally minimised clock, which includes many features that are shared with higher eukaryotes (plants), such as a central negative feedback loop that involves the morning-expressed CCA1 and evening-expressed TOC1 genes4. Given that recent observations in animals and plants have revealed prominent post-translational contributions to timekeeping5, a reappraisal of the transcriptional contribution to oscillator function is overdue. Here we show that non-transcriptional mechanisms are sufficient to sustain circadian timekeeping in the eukaryotic lineage, though they normally function in conjunction with transcriptional components. We identify oxidation of peroxiredoxin proteins as a transcription-independent rhythmic biomarker, which is also rhythmic in mammals6. Moreover we show that pharmacological modulators of the mammalian clockwork have the same effects on rhythms in Ostreococcus. Post-translational mechanisms, and at least one rhythmic marker, appear to be better conserved than transcriptional clock regulators. It is plausible that the oldest oscillator components are non-transcriptional in nature, as in cyanobacteria7, and are conserved across kingdoms.
Over the last two decades, great progress has been made towards delineating the molecular basis of eukaryotic circadian rhythms using model organisms such as Arabidopsis thaliana (plant), Mus musculus (mammal), and Drosophila melanogaster (insect)5,8. In each case mechanistic models of the cellular clock have relied heavily on networks of transcriptional/translational feedback loops (TTFLs) and can successfully account for a wide range of experimental data9. Whilst the identified “clock genes” differ widely across taxa, a growing number of ubiquitous post-translational mechanisms, such as casein kinase II activity5,10,11, have been shown to contribute to timing. Similarly signal transduction pathways, e.g. Ca2+/cAMP, previously viewed as clock inputs have been shown also to be clock outputs, thus becoming indistinguishable from the “core” mechanisms5,12. As a result it is presently unclear whether transcription, per se, is necessary to sustain the eukaryotic cellular clock13,14, especially in light of observations that prokaryotic timekeeping can be reconstituted in vitro using the gene expression products of the cyanobacterial KaiBC/KaiA operons7. Hypothesising that non-transcriptional mechanisms would be competent to sustain cellular rhythms without a transcriptional contribution, we set out to test this using the pico-eukaryote Ostreococcus tauri. This single-celled eukaryote has several advantages. It is readily cultured, possesses a small genome (~12 Mbp), and yet its light-entrainable clock shares the transcriptional architecture of the clock in higher plants, namely a negative feedback loop between the morning-expressed CCA1 and evening-expressed TOC1 genes4.
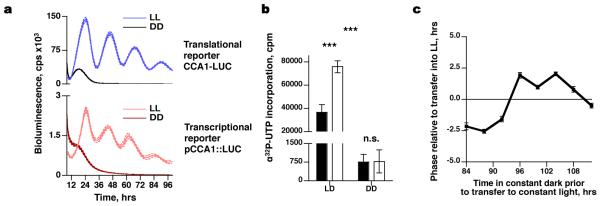
Recently, bioluminescent luciferase (LUC) reporter lines for transcription and translation of O. tauri clock genes were developed to enable non-invasive interrogation of clock mechanisms4. Following entrainment in 12 hour light-12 hour dark cycles, circadian rhythms of bioluminescence from a translational (CCA1-LUC) and transcriptional (pCCA1::LUC) reporter were observed to persist for >4 days in constant light (Fig. 1a), indicating the presence of an underlying circadian clock, able to keep time without reference to any external time cues. Whilst many cellular processes in photosynthetic organisms are light-dependent4,15,16, the cyanobacterial clock was recently shown to persist in darkness7. We therefore determined whether circadian rhythms might similarly persist in O. tauri without light. When placed in constant darkness, bioluminescent traces rapidly dampened to background levels (Fig. 1a). After 96 hours in constant darkness, no incorporation of [α32P]UTP was observed (Fig. 1b), meaning that no nascent RNA was being transcribed. Upon transfer of these transcriptionally incompetent cultures into constant light, circadian rhythms in bioluminescence began at a phase that was not dictated solely by the time of transfer into light (Fig. 1c, Supplementary Fig. 1a,b). If no cellular oscillation had persisted in the dark, we would expect the clock to restart with its phase determined solely by when it was transferred into the light (i.e. complete phase resetting). In contrast, the cultures’ new phase suggested that the response to light was modulated by a pre-existing oscillation, instead of being completely reset by light (Fig. 1c)17. These observations suggest that O. tauri is competent to keep time in the absence of transcription.
Figure 1. Transcriptionally inactive cells show a phase-dependent response to re-illumination.
a, Grouped data showing bioluminescent transcriptional (pCCA1::LUC) and translational (CCA1-LUC) reporter activity in constant darkness (DD) or constant light (LL) (n=16, dotted lines ±SEM). b, After 96 hours in darkness there is no significant incorporation of radiolabelled UTP; 10 minute UTP treatment (black, ±SEM) compared with 30 minute treatment (white, ±SEM) (2-way ANOVA interaction, p<0.001 for time, condition and interaction, n=3; Bonferroni post-test for DD groups, p=0.95). c, Upon transfer from darkness, the phase of CCA1-LUC deviates significantly from the time of transfer into light (2-way ANOVA interaction, p<0.001, n≥16).
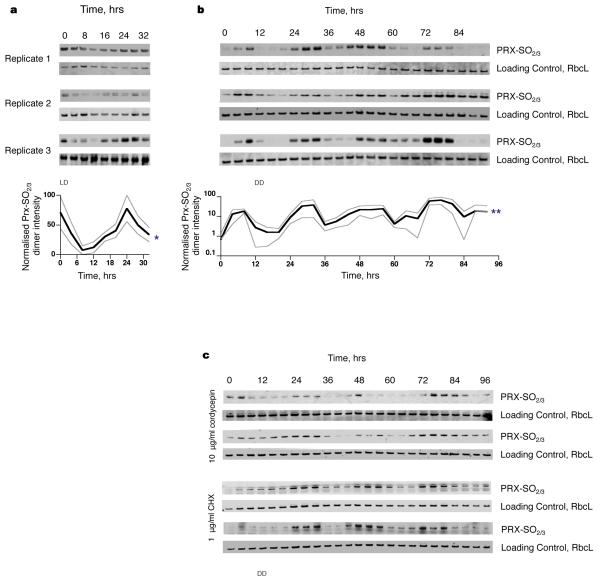
In order to confirm this, we used a novel post-translational biomarker for rhythmicity: peroxiredoxin oxidation. The peroxiredoxins (PRXs) are a ubiquitous family of antioxidant enzymes that scavenge reactive oxygen species, such as hydrogen peroxide, catalysing their own oxidation at a conserved redox-active cysteine (Cys) group to sulphenic acid followed by hyperoxidation to sulphonic acid18. In plants, a subtype of peroxiredoxins (the 2-Cys group) is targeted to chloroplasts where they protect the photosynthetic membrane against photo-oxidative damage19. Oxidation of PRX drives the formation of higher molecular weight multimers with reported chaperone and signalling functions18. Circadian cycles of post-translational modification of PRX have previously been reported in mouse liver6 and recently shown to persist in human red blood cells in vitro20. O. tauri expresses at least one 2-Cys PRX15 (Genbank Accession: CAL55168.1) sharing 61% sequence identity with human PRX2 and 100% sequence identity around the catalytic cysteine residue (Supplementary Fig. 2a,b). Immunoblots using an antibody targeting this highly conserved region20 revealed diurnal regulation of PRX oxidation (sulphonylation) that was highest during subjective day, in advance of RUBISCO large chain expression (RbcL; a highly expressed plant/algal protein) (Fig. 2a). Moreover, in constant darkness, circadian rhythms persisted without transcription (Fig. 2b). PRX oxidation rhythms even persisted in constant darkness in the presence of inhibitors of cellular RNA synthesis (cordycepin) and cytosolic translation (cycloheximide), at concentrations that abolish clock reporter bioluminescence (Fig. 2c and Supplementary Fig. 3c,d), providing strong evidence that new RNA and/or protein synthesis is indeed not required for sustained rhythmicity. RbcL was used as a loading control, because although this protein was rhythmically expressed in a diurnal cycle, its levels were high and stable under constant conditions. In addition, rhythms in PRX oxidation are altered in a long period mutant (TOC1-LUC)4, relative to controls (CCA1-LUC), under constant light; this suggests that post-translational oscillations are coupled with transcriptional/translational cycles under more physiological conditions (Supplementary Fig. 2c,d). Thus, PRX oxidation constitutes the first example, as far as we are aware, of a post-translational circadian biomarker shared between the animal (mouse/human) and green lineages.
Figure 2. Circadian cycles of PRX sulphonylation are detected during light/dark cycles and in constant darkness, and persist during drug inhibition of gene expression.
a, Individual blots and grouped mean intensities for three O. tauri time series sampled under 12:12 light:dark cycles (*Friedman test, p=0.04 for time effect). b, Individual blots and grouped mean intensities for three O. tauri time series sampled under constant darkness (**Friedman test, p=0.005 for time effect). c, PRX-SO2/3 immunoblots of O. tauri time series sampled under constant darkness in the presence of inhibitors of transcription (cordycepin) and translation (CHX).
Although experimentally useful for dissecting the algal clockwork, constant darkness potentially represents an exotic environmental challenge to O. tauri, and hence we sought to also examine non-transcriptional rhythms in constant light using real-time bioluminescence reporter assays. CCA1-LUC and pCCA1::LUC reporter lines were incubated with a range of concentrations of cordycepin and cycloheximide (CHX) during bioluminescent recordings, in order to assay the effects of inhibiting cellular RNA synthesis and cytosolic translation, respectively. At lower concentrations we observed dose dependent dampening with both drugs, and a robust increase in circadian period with increasing cordycepin concentration (Supplementary Fig. 3a,b), in agreement with observations in the marine mollusc Bulla21. At saturating doses, both drugs resulted in immediate dampening and arrhythmia in the transcriptional reporter lines (Supplementary Fig. 3c). Intriguingly, the translational reporter exhibited an additional cycle of CCA1-LUC synthesis in the presence of saturating transcriptional inhibitor (Supplementary Fig. 3d), which was not observed in the transcriptional reporter line. We interpret this to mean that when CCA1-LUC mRNA is present, post-transcriptional mechanisms are sufficient to drive an additional cycle of correctly timed protein accumulation.
Clearly in the context of a living cell, transcription is ultimately required for any biological process, including circadian rhythms, since the mRNAs have limited half-lives and can only be replaced through transcription. In a biological clock context, it seems natural that some mRNAs are cyclically expressed in anticipation of cellular need. Microarray studies in several organisms have shown >10% of the transcriptome is regulated on a daily basis6,15,22. This implies that circadian cycles in transcription factor activity are a normal feature of cell physiology. Some of this transcriptional activity will contribute to timekeeping, directly or indirectly. If the natural state of a eukaryotic cellular clock revolves around reciprocal interplay between post-translational oscillations and established transcriptional feedback loops, it becomes of great interest to identify at what phases this interconnection is regulated.
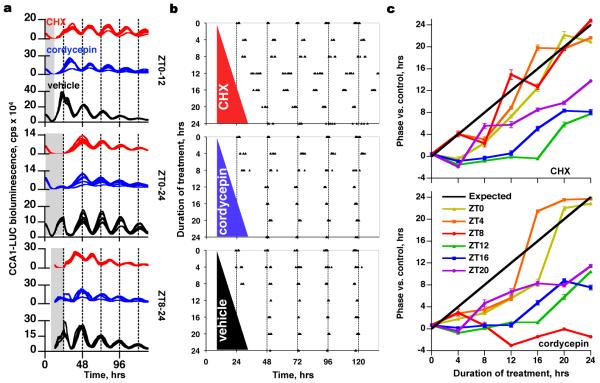
Ostreococcus cultures are amenable to drug treatment that can be reversed by wash-off, because O. tauri grown in liquid culture forms aggregates at the bottom of microplate wells (Fig. 3a). To ascertain at what phases of the circadian cycle gene expression exerts priority over non-transcriptional mechanisms, we performed a ‘wedge’ experiment21, in which transcription or translation was reversibly inhibited starting at 4 hour intervals across the circadian cycle, for increasing durations, in constant light. Resultant phases were determined by the timing of CCA1-LUC expression peaks over the interval following removal of the drug (Fig. 3b). As with the earlier experiments using constant dark, our null hypothesis was that if the clock was immediately arrested by drug treatment then phase would be set by the time of drug wash-off. Phase is described relative to the zeitgeber, or time giver, during entrainment where ZT0 is dawn and ZT12 is dusk (ZT denotes zeitgeber time).
Figure 3. Circadian timing can survive the inhibition of cellular transcription, or cytosolic translation.
a, Treatment (from top, at ZT0-12, ZT0-24, ZT8-24: shaded) with inhibitors of transcription (10 μg/ml cordycepin, blue lines) or translation (1 μg/ml CHX, red lines) may shift the phase of reporter expression, compared with vehicle (0.04% DMSO, black lines), depending on the treatment phase and duration. b, peak times of CCA1-LUC expression from individual wells (n>5) are plotted, after treatments of different durations, starting at ZT8. c, Summary of phase shifts (±SEM, n>5) relative to vehicle-treated controls, for all treatment durations (x-axis) and starting times (see legend in lower panel). Black line represents the expected result, assuming total resetting by wash-off.
A general trend towards the anticipated wedge shape was observed (Supplementary Fig. 4a,b). However, there were significant exceptions to this pattern, summarised as follows: (1) the clock was insensitive to transcriptional inhibition for up to 24 hours in treatments starting from ZT8 (Fig. 3b, cordycepin treatment); (2) transcriptional inhibition outside ZT0-8 did not affect phase, and (3) after treatments spanning this window, the clock resumed at dusk if treatment began during the subjective night, or at dawn if treatment began during the subjective day (Fig. 3c, Supplementary Fig. 4a,c,d); (4) Translational inhibition (CHX treatment) outside ZT4-12 did not affect phase (Fig. 3c, Supplementary Fig. 4b-d).
The simplest interpretation is that transcription of mechanistically relevant clock genes is licensed by post-translational mechanisms, and occurs around the first half of the subjective day. These transcripts are translated around the second half of the subjective day and non-transcriptional mechanisms keep time during the subjective night. Presumably when inhibition of transcription occurs at midday, for example, the resumption of stalled gene expression following wash-off overrides the phase of the non-transcriptional oscillations and the clock resumes from the nearest expected light/dark transition to when inhibition began. This runs contrary to current understanding of clocks in eukaryotes, in which transcription of key clock genes is active almost continuously around the circadian cycle23. Even in O. tauri, transcription of TOC1 and CCA1 would span the full cycle except for the interval ~ZT2-8, after the peak of CCA1 mRNA and before the rise in TOC14.
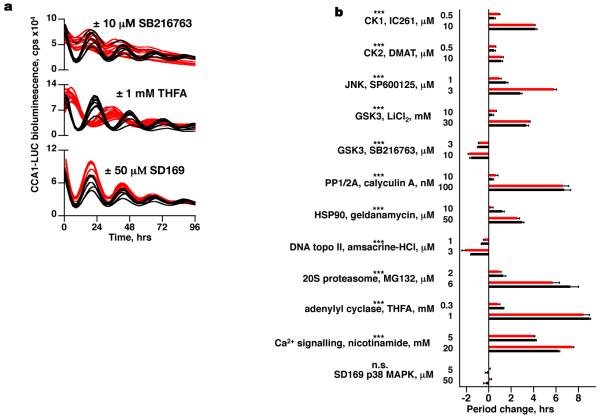
The final question of importance is: what non-transcriptional mechanisms are involved in sustaining the clock? We hypothesised that the components that sustain these post-translational rhythms are likely to be ubiquitous and highly conserved. Certainly the O. tauri genome encodes close homologues of enzymes such as casein kinase II that tend to exhibit greater sequence conservation across kingdoms than canonical transcriptional clock genes (Supplementary Table 1). In the last two years a number of high-throughput chemical biology screens on mammalian cellular rhythms in culture have been published, identifying a number of potent modulators of free-running period12,24-27. Since many such inhibitors target an enzyme’s active site, it seemed plausible that drug action might be similarly conserved. The effects of specific pharmacological inhibitors that have been demonstrated to significantly affect free-running period in mammalian cells, and/or other model organisms (Supplementary Table 2), were therefore tested in O. tauri. In all cases, dose-dependent effects were observed on circadian period that correlated with their action in other species such as mouse or Neurospora (Fig. 4a,b and Supplementary Fig. 5a). Critically, where tested, such pharmacological perturbations also delayed the timing of transcriptionally incompetent cells, firstly with respect to the additional cycle of CCA1-LUC expression observed during transcriptional inhibition in constant light (Supplementary Fig. 6a). Secondly, the period of rhythmic PRX oxidation in constant darkness was also lengthened by the treatments tested (Supplementary Fig. 6b). Although drugs can have pleiotropic effects on cell biology, the drug effects on the clock are conserved across taxa. The parsimonious interpretation is that, both with and without transcription, conserved post-translational mechanisms are necessary to keep biological time.
Figure 4. Circadian period in O. tauri can be modulated pharmacologically in a dose-dependent manner by the application of inhibitors that have been previously validated in other taxa.
a, Examples of drug effects (red) on CCA1-LUC bioluminescence compared with vehicle controls (black): SB216763 shortens period, THFA increases period, SD-169 has no effect. b, Summary showing grouped dose-responses on circadian period of transcriptional (red) and translational (black) reporter lines for selected drugs with previously demonstrated action in other taxa (±SEM, n≥8, *** indicates p<0.0001, 2-way ANOVA for concentration; n.s. for SD169 negative control, n=8, p=0.90).
Whilst the importance of transcription to circadian rhythms is self-evident, our observation that eukaryotic rhythms persist in the absence of transcription challenges the general paradigm for eukaryotic clocks, suggesting a functional equivalent to cyanobacterial timekeeping28, though undoubtedly more complex. This is supported by increasing numbers of observations in diverse organisms5,13,20,29. Most prominently, the observation of a rhythmic post-translational marker that persists in the absence of transcription in species as diverse as a unicellular green alga and humans20 raises exciting prospects for our understanding of how circadian clocks evolved. We note that both PRX and cyanobacterial KaiB are clock-relevant members of the thioredoxin-like superfamily28, that associate into higher molecular weight complexes with catalytic function. We speculate this may reflect conserved remnants of a proto-clock in the last common ancestor of eukaryotes and prokaryotes.
Methods Summary
All materials were purchased from Sigma-Aldrich (UK) unless otherwise stated. Transgenic Ostreococcus tauri lines4 were cultured in Keller media-supplemented artificial sea water (Km) under 12:12 h blue (Ocean Blue, Lee lighting filter 724) light:dark cycles (17.5 μE/m2). For imaging, cultures were transferred to 96-well microplates (Lumitrac, Greiner Bio-one) at a density of ~15×106 cells/ml and entrained for 7-10 days. No density effects on clock output were observed under relevant density ranges, and cell division in microplates was found to be close to zero. One day prior to recording, 150 μl Km was replaced with 150 μl Km containing 333 μM luciferin (Km+). Drugs were made up in DMSO or Km+, diluted in Km+ and added to replicates of 8 or 16 wells immediately prior to recording. For incubations in constant darkness, Km+ was supplemented with 200 mM sorbitol and 0.4% glycerol in order to increase cell viability. Bioluminescent recordings were performed on a TopCount (Packard) under constant darkness or constant red+blue LED light (5-12 μE/m2). For wash-off of reversible inhibitors, cell aggregates formed in the bottom of the wells were quickly and gently washed twice with Km+, using multi-channel pipettes, and returned to recording conditions. Analysis of period was performed with FFT-NLLS (BRASS 330) using time windows ≥3 days; mFourfit (BRASS 3) was used to assess phase and confirmed manually. Statistical analysis was performed using GraphPad Prism. For de novo RNA synthesis analysis by [α32P]UTP uptake, 1 ml cell aliquots were either incubated in darkness or light/dark cycles for 4 days. 0.2 MBq of [α32P]UTP was added, and after incubation cells were collected and washed twice with Km. Incorporation was measured using scintillation counting. Immunoblots were performed as described elsewhere20. Sequence alignments were performed using EBI Jalview, BLAST searches were performed using NCBI BLASTp under the default BLOSSUM62 settings.
One-line summary.
Circadian rhythms in a eukaryote can be sustained solely by non-transcriptional mechanisms, which are conserved across taxa.
Supplementary Material
Acknowledgements
CSBE is a Centre for Integrative Systems Biology funded by BBSRC and EPSRC award D019621. CT is supported by a BBSRC/ANR joint project F005466 awarded to FYB and AJM and by the HFSP. ABR is supported by the Wellcome Trust (083643/Z/07/Z) and the MRC Centre for Obesity and Related metabolic Disorders (MRC CORD).
Footnotes
Supplementary information accompanies the paper on www.nature.com/nature
Competing interests: The authors declare no competing financial interests.
References
- 1.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 2.Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2009 doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakin-Thomas PL. Transcriptional feedback oscillators: maybe, maybe not. J Biol Rhythms. 2006;21:83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- 4.Corellou F, et al. Clocks in the Green Lineage: Comparative Functional Analysis of the Circadian Architecture of the Picoeukaryote Ostreococcus. Plant Cell. 2009 doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 6.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 7.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 8.Roenneberg T, Merrow M. Circadian clocks - the fall and rise of physiology. Nat Rev Mol Cell Biol. 2005;6:965–971. doi: 10.1038/nrm1766. [DOI] [PubMed] [Google Scholar]
- 9.Ueda HR. Systems biology flowering in the plant clock field. Mol Syst Biol. 2006;2:60. doi: 10.1038/msb4100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merrow M, Mazzotta G, Chen Z, Roenneberg T. The right place at the right time: regulation of daily timing by phosphorylation. Genes Dev. 2006;20:2629–2623. doi: 10.1101/gad.1479706. [DOI] [PubMed] [Google Scholar]
- 12.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woolum JC. A re-examination of the role of the nucleus in generating the circadian rhythm in Acetabularia. J Biol Rhythms. 1991;6:129–136. doi: 10.1177/074873049100600203. [DOI] [PubMed] [Google Scholar]
- 14.Morse DS, Fritz L, Hastings JW. What is the clock? Translational regulation of circadian bioluminescence. Trends Biochem Sci. 1990;15:262–265. doi: 10.1016/0968-0004(90)90050-l. [DOI] [PubMed] [Google Scholar]
- 15.Monnier A, et al. Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles. BMC Genomics. 11:192. doi: 10.1186/1471-2164-11-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moulager M, et al. Light-dependent regulation of cell division in Ostreococcus: evidence for a major transcriptional input. Plant Physiol. 2007;144:1360–1369. doi: 10.1104/pp.107.096149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konopka RJ. Genetic dissection of the Drosophila circadian system. Fed Proc. 1979;38:2602–2605. [PubMed] [Google Scholar]
- 18.Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins--structures, mechanisms and functions. Febs J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baier M, Dietz KJ. The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 1997;12:179–190. doi: 10.1046/j.1365-313x.1997.12010179.x. [DOI] [PubMed] [Google Scholar]
- 20.O’Neill JS, Reddy AB. Circadian Clocks in Human Red Blood Cells. Under review at Nature. 2010 doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalsa SB, Whitmore D, Bogart B, Block GD. Evidence for a central role of transcription in the timing mechanism of a circadian clock. Am J Physiol. 1996;271:C1646–1651. doi: 10.1152/ajpcell.1996.271.5.C1646. [DOI] [PubMed] [Google Scholar]
- 22.Edwards KD, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dodd AN, et al. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science. 2007;318:1789–1792. doi: 10.1126/science.1146757. [DOI] [PubMed] [Google Scholar]
- 25.Eide EJ, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirota T, et al. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci U S A. 2008;105:20746–20751. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isojima Y, et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson CH, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol. 2008;18:R816–R825. doi: 10.1016/j.cub.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eelderink-Chen Z, et al. A circadian clock in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2010;107:2043–2047. doi: 10.1073/pnas.0907902107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwards KD, et al. Quantitative analysis of regulatory flexibility under changing environmental conditions. Mol Syst Biol. 2010;6:424. doi: 10.1038/msb.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.