Abstract
In this brief review, I have highlighted recent advances in several areas of mitochondrial medicine, including mtDNA-related diseases, mendelian mitochondrial encephalomyopathies, and therapy. The pathogenic mechanisms of mtDNA mutations, especially those affecting mitochondrial protein synthesis, are still largely unknown. The pathogenicity of homoplasmic mtDNA mutations has become evident but has also called attention to modifying nuclear genes, yet another example of impaired intergenomic signaling. The functional significance of the homoplasmic changes associated with mitochondrial haplogroups has been confirmed. Among the mendelian disorders, a new form of “indirect hit” has been described, in which the ultimate pathogenesis is toxic damage to the respiratory chain. Three therapeutic strategies look promising: (i) allogeneic hematopoietic stem cell transplantation in MNGIE (mitochondrial neurogastrointestinal encephalomyopathy); (ii) bezafibrate, an activator of PGC-1α, has proven effective in animal models of mitochondrial myopathy; and (iii) pronucleus transfer into a normal oocyte is effective in eliminating maternal transmission of mtDNA, thus preventing the appearance of mtDNA-related disorders.
Key words: mtDNA-related disorders, mendelian mitochondrial disorders, homoplasmy, pathogenesis, therapy
This paper – as the lecture from which it derives – are dedicated to the memory of Eduardo Bonilla (Fig. 1), a great myologist and a great friend.
Figure 1. Eduardo Bonilla (1937-2010).
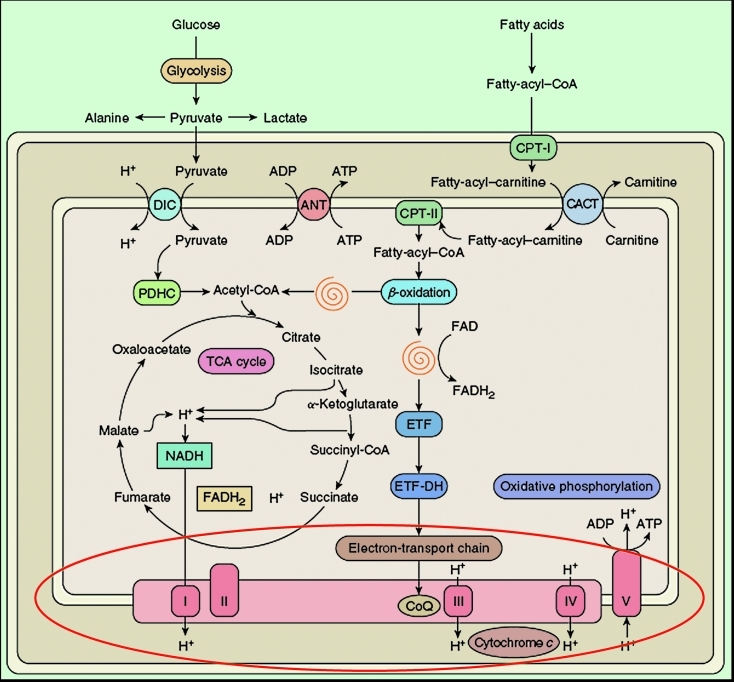
Although mitochondria have multiple functions, it is fair to say that the most important is the generation of energy. In Figure 2, an oversimplified schematic view of mitochondrial metabolism, I have highlighted the respiratory chain, the “business end” of oxidative metabolism, where ATP is actually produced. One “green view” of mitochondria is that they approximate ecologically friendly hydrogen engines: the breakfast that you ate this morning (derived from sunlight) is metabolized through pathways residing mostly outside (glycolysis) or inside (β-oxidation) the mitochondria. Electrons generated in the Krebs cycle from the oxidation of acetyl-CoA are carried along the four multimeric components of the electron transport chain (complexes I-IV) embedded in the inner mitochondrial membrane (IMM) and protons are pumped from the inside of the IMM (mitochondrial matrix) to the intermembrane space (IMS) between the IMM and the outer mitochondrial membrane (OMM). The resulting chemosmotic potential is used to operate the tiniest rotary machine, ATP synthase (complex V), where the influx of protons back into the matrix makes the rotor (F0) turns on the stator (F1) at the respectable speed of 1,000 RPM, bringing together ADP and Pi and releasing ATP (1, 2).
Figure 2. Schematic and simplified view of mitochondrial metabolism. The spirals depict the reactions of the β-oxidation pathway. The red oval highlights the reactions of the respiratory chain.
A recent remarkable achievement in our understanding of energy production by the respiratory chain has been the clarification of the α-helical structure of the membrane domain of complex I of E. coli by Leonid Sazanov’s group (3). The transfer of 2 electrons from NADH to quinone is coupled to the transfer of 4 protons across the IMM, and two mechanisms of coupling had been proposed, a direct, redox-driven mechanism or an indirect, conformation-driven mechanism. When the electrons reach the quinone moiety, conformational changes in complex I subunits (called NuoA/J/K/H) push a long α-helix towards other transmembrane subunits (NuoL/M/N) in a piston-like action [as aptly described by Tomoko Onishi in a News & Views article (4)] thus opening up “trapdoors” through which protons can pass.
Let me consider first recent progress in our understanding of pathogenesis, which unfortunately is still largely “terra incognita” both for mitochondrial DNA (mtDNA)- and for nuclear DNA (nDNA)-related disorders.
Disorders due to mutations in mtDNA
For what concerns mtDNA-related diseases, heteroplasmy and the threshold effect still are the best criteria to explain phenotypic variability. The best example is the NARP (neuropathy, ataxia, retinitis pigmentosa) syndrome, first described by Anita Harding in 1990 in four maternally related relatives: three adults with sensory neuropathy, ataxia, exercise intolerance, retinitis pigmentosa, and dementia; and one child with developmental delay, ataxia, retinitis pigmentosa, and abnormal EEG (5). The relationship between m.8993T > G mutation load and clinical severity was documented by Tatuch et al., who showed that about 70% heteroplasmy in skeletal muscle resulted in an adult-onset syndrome corresponding to the acronymic features of NARP whereas higher degrees of heteroplasmy (around 90%) were accompanied by Leigh syndrome (LS) in infants or children (6).
One would expect that mutations in different mitochondrial tRNA genes, affecting – as they all do – mitochondrial protein synthesis in toto, should cause a “swamp” of largely overlapping symptoms and signs. Contrarywise, clinical experience shows that mutations in individual tRNA genes are often, though not always, associated with specific syndromes. Thus, most patients with MELAS harbor the m.3243A > G mutation in tRNALeu(UUR) whereas most patients with MERRF harbor the m.8344A > G mutation in tRNALys. In addition, single deletions in mtDNA, which also impair mitochondrial protein synthesis globally, almost invariably cause one of three syndromes, the generalized Kearns-Sayre syndrome (KSS), the muscle-specific chronic progressive external ophthalmoplegia (CPEO), or the hematopoietic Pearson syndrome (PS) (7). At this time, the best explanation for this puzzling phenomenon is a spatial selectivity in the distribution of individual mutations, at least in the brain. This concept has been supported by immunohistochemical and in situ hybridization studies showing, for example, a predilection of the MELAS mutation for subpial arterioles (8, 9), of the MERRF mutation for the dentate nucleus of the cerebellum (10), and of single mtDNA deletions for the choroid plexus (11). The obvious but unanswered next question is what “directs” each mutation to a selected area.
The next area of exciting recent development regards homoplasmy. Although the first documented pathogenic point mutation in mtDNA (m.11778G > A in the ND4 gene) was, in fact, homoplasmic and associated with Leber hereditary optic neuropathy (LHON) (12), we have long ignored this lesson, to the point of including heteroplasmy among the canonical criteria of pathogenicity. And this in the face of increasing evidence that homoplasmic mutations were often associated with tissue-selective disorders such as LHON (13), deafness (14), deafness/cardiopathy (15), or tissue-specific disorders such as cardiomyopathy (16).
The evolving concept of homoplasmy has resonated with me personally because it has solved a conundrum that has been a thorn in my side for the past 26 years. In 1983, together with my colleagues at Columbia University Medical Center, I reported the puzzling case of an infant who was profoundly floppy at birth and whose initial muscle biopsy showed virtually no staining for cytochrome c oxidase (COX) (17). With vigorous supportive therapy and despite our gloomy expectations, the child improved spontaneously and rather rapidly: his severe lactic acidosis declined, his strength increased, and his muscle biopsy at 7 months of age showed that about 50% of all fibers were now COX-positive. By 3 years of age, the child was neurologically normal and a third muscle biopsy showed, if anything, some excess COX stain. Unfortunately at the time we did not pay enough attention to Eduardo Bonilla’s astute observation that the mother’s muscle biopsy (but not the father’s) showed a few scattered COX-negative fibers. However, it did not escape Rita Horvath’s attention that all 17 patients from 12 unrelated families with virtually identical reversible COX-deficient myopathy harbored a homoplasmic “polymorphism,” m.14674T > C in the tRNAGlu gene of mtDNA (18). This obviously pathogenic change cannot, in and by itself, explain the muscle-specificity of the disease or its reversibility, nor can it explain why some but not all maternal relatives are affected (18).
Pathogenic homoplasmic mtDNA mutations highlight yet another important aspect of the dependence of the mitochondrial on the nuclear genome. If it is true that from the beginning of mitochondrial genetics there has been a lot of handwaving about nuclear factors modulating the phenotypic expression of mtDNA mutations, now this has become a present and immediate question demanding that we identify the putative “nuclear modifiers” and understand their mechanism of action.
In the long course of their migration out of Africa, which started about 150,000 year ago, our ancestors accumulated harmless mtDNA changes (polymorphisms) that differed among different populations and still define ethnic groups (19). It was proposed that these ancient variations are not only harmless but, in fact, adaptive, thus facilitating the settlement of different groups in favorable ecological niches (20). Thus, for example, a mtDNA variation conducive to loose coupling of oxidative phosphorylation (OXPHOS) would enhance the dissipation of energy as heat and be advantageous to people living in frigid climates. Although their effect on OXPHOS would be small, haplogroup-defining mutations might behave as susceptibility factors in multifactorial diseases, in the context of particular environmental or nuclear factors. Such small effect on OXPHOS has been documented by “homogenizing” environmental and nuclear backgrounds with the use of cybrid cell lines, that is, immortalized human cell lines emptied of their own mtDNA and repopulated with haplotype-specific mitochondria (19).
Mendelian mitochondrial disorders
With the term “indirect hits” we refer to mutations in nuclear genes that do not affect respiratory chain subunits directly, but alter proteins needed for the assembly and maintenance of respiratory chain complexes. Numerous such indirect hits have been associated with defects in all five complexes of the respiratory chain (21), but Valeria Tiranti and Massimo Zeviani in Milan, Italy, have discovered a novel type of indirect hit, where the second whammy is toxic instead of structural. First, using integrative genomics, they found that ethylmalonic encephalomyopathy (EE), a devastating early-onset disorder with encephalopathy, microangiopathy, chronic diarrhea, and massively increased levels of ethylmalonic acid and short-chain acylcarnitines in body fluids, was due to mutations in the ETHE1 gene (22). They then documented that ETHE1 is a mitochondrial matrix thioesterase (23) and created an Ethe1-null mouse, which led them to discover that thiosulfate and sulfide accumulate excessively both in the animal model and in affected children due to the lack of sulfur dioxygenase activity (24). As sulfide is a powerful COX inhibitor, what they described was an indirect hit of a toxic kind and likely the prototype of other similar pathogenic mechanisms.
Yet another indirect mechanism involving sulfur metabolism (the mitochondrial disulfide relay system, DRS) and resulting in multiple respiratory chain enzyme deficiencies (complexes I, II, and IV) was discovered by the group of Giacomo Comi, also in Milan (25). The patients were three siblings born of consanguineous parents. They all had congenital cataracts and various degrees of psychomotor delay, hypotonia, hearing loss, bilateral or unilateral ptosis, sensorineural hearing loss, and lactic acidosis. At age 17 years, the older sibling needed tutorial assistance at school and was hyporeflexic. His brain MRI only showed thinning of the corpus callosum. Muscle histochemistry showed scattered COX-negative, SDH-hyperintense fibers and ultrastructural studies revealed vacuolated mitochondria with thickened cristae. Biochemical analysis showed partial decrease of COX (30%-50% residual activity) and less severe reduction of complexes I and II. Homozygosity mapping led to the identification of a missense mutation in the gene (GFER) whose product belongs to the ERV1/ALR protein family. Yeast Erv1p (and presumably its human counterpart GFER, a sulfhydryl oxidase) oxidizes the disulfide carrier protein Mia40, which, in turn, transfers a disulfide to newly synthesized proteins in the mitochondrial IMS. The reoxidaton of Erv1p is mediated by cytochrome c and COX, thus linking the DRS to the mitochondrial respiratory chain. Comi and coworkers showed that the mutant GFER is unstable and its concentration decreases in mitochondria, thus probably inhibiting the import of DRS substrates, including COX17, TIMM13, and COX6B1 (25). It is noteworthy that a mutation in COX6B1 has been the first example of a “direct hit” in complex IV deficiency (26). It is also noteworthy that defects of the DRS are yet another cause of multiple mtDNA deletions, which were documented in muscle from one of the patients with mutant GFER (25).
As I mentioned in a recent historical review (21) for now at least, the last frontier of research in mitochondrial disorders seems to be the defects of mitochondrial translation. Within the past four years, often through homozygosity mapping, defects have been discovered in genes controlling factors at all levels of the complex translation apparatus, from rRNA base modification, such as MRPS16 (27) to general translation, such as EFG1 (now called GFM1) (28-31) to tRNA processing and base modification, such as PUS1 (32) to tRNA synthetase (33, 34). This subject has been recently and lucidly reviewed by Smits et al. (35).
Therapy
This is very much an area of work in progress, but there are three developments that are worth discussing briefly: stem cell therapy in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE); boosting mitochondrial biogenesis as a therapeutic strategy; and pronuclear transfer as a preventive measure to mtDNA-related disorders.
MNGIE is an autosomal recessive multisystemic disease characterized clinically by progressive external ophthalmoplegia (PEO), ptosis, gastrointestinal dysmotility, extreme cachexia, peripheral neuropathy, leukoencephalopathy, and death in early or mid-adulthood (36). The disease is due to loss-of-function mutations in the TYMP gene that encodes the cytosolic enzyme thymidine phosphorylase (TPase) (37). As a result, there is a dramatic elevation of thymidine and deoxyuridine in blood and tissues (38) and severe deoxynucleotide pool imbalance, which causes multiple mtDNA deletions, depletion, and site-specific point mutations (39). One obvious therapeutic approach is to eliminate the toxic metabolites through hemodialysis, but single treatments had only transient effect in two patients (40) whereas chronic dialysis for over a year failed to slow disease progression in one patient (41). Nor did prolonged peritoneal dialysis fare any better (42). Attempts to replace the missing TPase using erythrocyte-encapsulated TPase or platelet infusion did improve symptoms but paradoxically did not lower plasma nucleotide levels (42).
Michio Hirano took a more radical approach to enzyme replacement therapy by employing allogeneic hematopoietic stem cell transplantation (HSCT), which proved very effective in a first patient (43, 44) and has been successful to date in five of the 11 patients so treated (45). An international therapeutic trial is underway and will hopefully confirm that this approach, though risky, can be a lifesaver in MNGIE.
In recent years, increasing attention has been directed to mitochondrial biogenesis and, more specifically, to the peroxisome proliferator-activated receptor γ coactivator-1α protein (PGC-1α for short), a transcriptional coactivator that binds to several transcription factors and induces gene expression (46). Importantly, PGC-1α is a strong promoter of mitochondrial biogenesis and function (47). This property has been exploited by French clinical scientists, who used bezafibrate (a PGC-1α activator), an approved drug in Europe, to treat patients with imborn errors of fatty acid oxidation (48, 49) and respiratory chain defects (50).
In a series of elegant papers, Tina Wenz and Carlos Moraes in Miami illustrated both the pathogenic role of PGC-1α and its potential therapeutic usefulness. Particularly relevant to the therapy of human mitochondrial myopathies, they used a knock-in mouse model of mitochondrial myopathy with partial COX deficiency due to a mutation in the assembly gene COX10. Promoting mitochondrial biogenesis either by transgenic expression of PG1-α or by administration of bezafibrate resulted in improved respiratory chain function and ATP production, delayed appearance of the myopathy, and prolonged lifespan (51, 52).
There is a form of gene therapy for mtDNA-related diseases that is ready for experimentation in humans but is stalled by ethical concerns. Pathogenic mtDNA mutations, especially those affecting tRNA genes can be eliminated literally ab ovo by transferring an in vitro-fertilized nucleus from the ooplasm of a woman carrying the mutation to an enucleated oocyte from a normal donor. The resulting embryo, containing normal nuclei from mother and father and normal mitochondrial genomes from the donor woman, can be implanted in the mother’s uterus. The feasibility of cytoplasmic transfer has now been documented by the Newcastle group in the UK (53) and a variant of this approach has been used in the US to produce two healthy “transmitochondrial” rhesus monkeys carrying undetectable mtDNA from their biological mother (54).
I hope that this brief update on the pathogenesis and therapy of mitochondrial diseases conveys the fervor of research in mitochondrial medicine and the exciting realization that effective therapy is finally within our grasp for at least some of these devastating disorders.
Abbreviations
ADP, adenosine diphosphate; ATP, adenosine triphosphate; ANT, adenine nucleotide translocator; CACT, carnitine- acyl-carnitine translocase, CoQ, coenzyme Q; CPT, carnitine palmitoyltransferase; DIC, dicarboxylate carrier; ETF, electron-transfer flavoprotein; ETFDH, ETF dehydrogenase; FAD, flavin adenine dinucleotide; FADH2, reduced FAD; NADH, reduced nicotinamide adenine dinucleotide; PDHC, pyruvate dehydrogenase complex; TCA, tricarboxylic acid; I, complex I; II, complex II; III, complex III; IV, complex IV; V, complex V. Modified from 55
Acknowledgements
Part of the work described here is supported by NIH grant HD32062 and by the Marriott Mitochondrial Disorder Clinical Research Fund (MMDCRF).
References
- 1.Boyer PD. A research journey with ATP synthase. J Biol Chem. 2002;277:39045–39061. doi: 10.1074/jbc.X200001200. [DOI] [PubMed] [Google Scholar]
- 2.Stock D, Gibbons C, Arechaga I, et al. The rotary mechanism of ATP synthase. Curr Opin Struct Biol. 2000;10:672–679. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 3.Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 4.Ohnishi T. Piston drives a proton pump. Nature. 2010;465:428–429. doi: 10.1038/465428a. [DOI] [PubMed] [Google Scholar]
- 5.Holt IJ, Harding AE, Petty RK, et al. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- 6.Tatuch Y, Christodoulou J, Feigenbaum A, et al. Heteroplasmic mtDNA mutation (T > G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am J Hum Genet. 1992;50:852–858. [PMC free article] [PubMed] [Google Scholar]
- 7.DiMauro S, Davidzon G. Mitochondrial DNA and disease. Ann Med. 2005;37:222–232. doi: 10.1080/07853890510007368. [DOI] [PubMed] [Google Scholar]
- 8.Tanji K, Kunimatsu T, Vu TH, et al. Neuropathological features of mitochondrial disorders. Cell Develop Biol. 2001;12:429–439. doi: 10.1006/scdb.2001.0280. [DOI] [PubMed] [Google Scholar]
- 9.Betts J, Jaros E, Perry RH, et al. Molecular neuropathology of MELAS: level of heteroplasmy in individual meurones and evidence of extensive vascular involvement. Neuropathol Appl Neurobiol. 2006;32:359–373. doi: 10.1111/j.1365-2990.2006.00731.x. [DOI] [PubMed] [Google Scholar]
- 10.Sparaco M, Schon EA, DiMauro S, et al. Myoclonic epilepsy with ragged-red fibers (MERRF): an immunohistochemical study of the brain. Brain Pathol. 1995;5:125–133. doi: 10.1111/j.1750-3639.1995.tb00586.x. [DOI] [PubMed] [Google Scholar]
- 11.Tanji K, Schon EA, DiMauro S, et al. Kearns-Sayre syndrome: oncocytic transformation of choroid plexus epithelium. J Neurol Sci. 2000;178:29–36. doi: 10.1016/s0022-510x(00)00354-3. [DOI] [PubMed] [Google Scholar]
- 12.Wallace DC, Singh G, Lott MT, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 13.Carelli V, Barboni P, Sadun AA. Mitochondrial ophthalmology. In: DiMauro S, Hirano M, Schon EA, editors. Mitochondrial Medicine. London: Informa Healthcare; 2006. pp. 105–142. [Google Scholar]
- 14.Li X, Zhang LS, Fischel-Ghodsian N, et al. Biochemical characterization of the deafness-associated mitochondrial tRNASer(UCN) A7445G mutation in osteosarcoma cell cybrids. Biochem Biophys Res Comm. 2005;328:491–498. doi: 10.1016/j.bbrc.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Santorelli FM, Tanji K, Manta P, et al. Maternally inherited cardiomyopathy: an atypical presentation of the 12S rRNA A1555G mutation. Am J Hum Genet. 1999;64:295–300. doi: 10.1086/302188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor RW, Giordano C, Davidson MM, et al. A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited cardiomyopathy. J Am Coll Cardiol. 2003;41:1786–1796. doi: 10.1016/s0735-1097(03)00300-0. [DOI] [PubMed] [Google Scholar]
- 17.DiMauro S, Nicholson JF, Hays AP, et al. Benign infantile mitochondrial myopathy due to reversible cytochrome c oxidase deficiency. Ann Neurol. 1983;14:226–234. doi: 10.1002/ana.410140209. [DOI] [PubMed] [Google Scholar]
- 18.Horvath R, Kemp JP, Tuppen HA, et al. Molecular basis of infantile reversible cytochrome c oxidase deficiency. Brain. 2009;132:3165–3174. doi: 10.1093/brain/awp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Duran A, Pacheu-Grau D, López-Gallardo E, et al. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet. 2010;19:3343–3353. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- 20.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–180. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 21.DiMauro S. A history of mitochondrial diseases. J Inher Metab Dis. 2010 doi: 10.1007/s10545-010-9082-x. [DOI] [PubMed] [Google Scholar]
- 22.Tiranti V, D’Adamo P, Briem E, et al. Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein. Am J Hum Genet. 2004;74:239–252. doi: 10.1086/381653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiranti V, Briem E, Lamantea E, et al. ETHE1 mutations are specific to ethylmalonic encephalopathy. J Med Genet. 2006;43:340–346. doi: 10.1136/jmg.2005.036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiranti V, Viscomi C, Hildebrandt T, et al. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nature Med. 2009;15:200–205. doi: 10.1038/nm.1907. [DOI] [PubMed] [Google Scholar]
- 25.Fonzo A, Ronchi D, Lodi T, et al. The mitochondrial disulfide relay system protein GFERis mutated in autosomal-recessive myopathy with cataract and combined respiratory-chain deficiency. Am J Hum Genet. 2009;84:594–604. doi: 10.1016/j.ajhg.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massa V, Fernandez-Vizarra E, Alshahwan S, et al. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus- encoded subunit of cytochrome c oxidase. Am J Hum Genet. 2008;82:1281–1289. doi: 10.1016/j.ajhg.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller C, Saada A, Shaul N, et al. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol. 2004;56:734–738. doi: 10.1002/ana.20282. [DOI] [PubMed] [Google Scholar]
- 28.Antonicka H, Sasarman F, Kennaway NG, et al. The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum Mol Genet. 2006;15:1835–1846. doi: 10.1093/hmg/ddl106. [DOI] [PubMed] [Google Scholar]
- 29.Coenen MJH, Antonicka H, Ugalde C, et al. Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N Engl J Med. 2004;351:2080–2086. doi: 10.1056/NEJMoa041878. [DOI] [PubMed] [Google Scholar]
- 30.Smeitink JAM, Elpeleg O, Antonicka H, et al. Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am J Hum Genet. 2006;79:869–877. doi: 10.1086/508434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valente L, Tiranti V, Marsano RM, et al. Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EGF1 and EFTu. Am J Hum Genet. 2007;80:44–58. doi: 10.1086/510559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bykhovskaya Y, Casas KA, Mengesha E, et al. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am J Hum Genet. 2004;74:1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheper GC, Klok T, Andel RJ, et al. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nature Genet. 2007;39:534–538. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- 34.Edvardson S, Shaag A, Kolesnikova O, et al. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am J Hum Genet. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smits P, Smeitink JAM, Heuvel B. Mitochondrial translation and beyond; Processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotechnol. 2010 doi: 10.1155/2010/737385. doi:1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirano M, Nishigaki Y, Marti R. Mitochondrial Neurogastrintestinal Encephalomyopathy (MNGIE): a disease of two genomes. Neurologist. 2004;10:8–17. doi: 10.1097/01.nrl.0000106919.06469.04. [DOI] [PubMed] [Google Scholar]
- 37.Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 38.Valentino ML, Martí R, Tadesse S, et al. Thymidine and deoxyuridine accumulate in tissues of patients with mitochondrial neurogastrintestinal encephalomyopathy (MNGIE) FEBS Lett. 2007;581:3410–3414. doi: 10.1016/j.febslet.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez LC, Akman HO, García-Cazorla A, et al. Umbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum Mol Genet. 2009;18:714–722. doi: 10.1093/hmg/ddn401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spinazzola A, Marti R, Nishino I, et al. Altered thymidine metabolism due to defects of thymidine phosphorylase. J Biol Chem. 2002;277:4128–4132. doi: 10.1074/jbc.M111028200. [DOI] [PubMed] [Google Scholar]
- 41.Marca G, Malvagia S, Casetta B, et al. Pre- and post-dialysis quantitative dosage of thymidine in urine and plasma of a MNGIE patient by using HPLC-ESI-MS/MS. J Mass Spectrom. 2006;41:586–592. doi: 10.1002/jms.1013. [DOI] [PubMed] [Google Scholar]
- 42.Yavuz H, Ozel A, Christensen M, et al. Treatment of mitochondrial neurogastrointestinal encephalomyopathy with dialysis. Arch Neurol. 2007;64:435–438. doi: 10.1001/archneur.64.3.435. [DOI] [PubMed] [Google Scholar]
- 43.Hirano M, Martí R, Casali C, et al. Allogeneic stem cell transplantation corrects biochemical derangements in MNGIE. Neurology. 2006;67:1458–1460. doi: 10.1212/01.wnl.0000240853.97716.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schon EA, DiMauro S, Hirano M, et al. Therapeutic prospects for mitochondrial disease. Trends Mol Med. 2010;16:268–276. doi: 10.1016/j.molmed.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schupbach M, Benoist J-F, Casali C, et al. Allogeneic hematopoietic stem cell transplantation (HSCT) for mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) Neurology. 2009;73:332–332. [Google Scholar]
- 46.Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1- alpha drives the formation of slow-twitch muscle fibers. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 47.Rohas LM, St-Pierre J, Uldry M, et al. A fundamental system of cellular energy homeostasis regulated by PGC-1alpha. Proc Natl Acad Sci USA. 2007;104:7933–7938. doi: 10.1073/pnas.0702683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Djouadi F, Bastin J. PPARs as therapeutic targets for correction of inborn mitochondrial fatty acid oxidation disorders. J Inher Metab Dis. 2008;31:217–225. doi: 10.1007/s10545-008-0844-7. [DOI] [PubMed] [Google Scholar]
- 49.Gobin-Limballe S, Djouadi F, Aubey F, et al. Genetic basis for correction of very-long-chain acyl-coenzyme A dehydrogenase by bezafibrate in patients fibroblasts: towards a genotype-based therapy. Am J Hum Genet. 2007;81:1133–1143. doi: 10.1086/522375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bastin J, Aubey F, Rotig A, et al. Activation of peroxisome proliferator- activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients’ cells lacking its components. J Clin Endocrinol Metab. 2008;93:1433–1441. doi: 10.1210/jc.2007-1701. [DOI] [PubMed] [Google Scholar]
- 51.Wenz T, Diaz F, Spiegelman BM, et al. C.T. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–255. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Wenz T. PGC-1alpha activation asa therapeutic approach in mitochondrial disease. IUBMB Life. 2009;61:1051–1062. doi: 10.1002/iub.261. [DOI] [PubMed] [Google Scholar]
- 53.Craven L, Tuppen HA, Greggains GD, et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010;465:82–85. doi: 10.1038/nature08958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tachibana M, Sparman M, Sritanaudomchai H, et al. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]