Abstract
Aims
To confirm and extend to primary care settings prior genome-wide association results that distinguish smokers who successfully quit from individuals who were not able to quit smoking in clinical trials.
Materials & methods
Affymetrix® 6.0 Arrays were used to study DNA from successful quitters and matched individuals who did not quit from the Patch in Practice study of 925 smokers in 26 UK general practices who were provided with 15 mg/16 h nicotine-replacement therapy and varying degrees of behavioral support.
Results
Only a few SNPs provided results near ‘genome-wide’ levels of significance. Nominally significant (p < 0.01) SNP results identify the same chromosomal regions identified by prior genome-wide association studies to a much greater extent than expected by chance.
Conclusion
Ability to change smoking behavior in a general practice setting appears to share substantial underlying genetics with the ability to change this behavior in clinical trials, though the modest sample sizes available for these studies provides some caution to these conclusions.
Keywords: DNA microarray, genetic susceptibility, nicotine dependence, nicotine replacement, smoking cessation
Twin studies document substantial heritability for the ability to successfully abstain from smoking [1,2]. Vulnerability to becoming dependent on addictive substances also displays substantial heritability that is largely shared between dependences on different addictive substances [3–7]. Twin study results suggest that some, but not most, of the genetic influences on the ability to quit smoking are shared with the genetics of developing dependence on an addictive substance [1,2].
Many current analyses of genome-wide association (GWA) data seek individual genomic markers whose allele frequencies display ‘genome-wide’ significant differences between cases and controls in individually genotyped samples. We have recently reported apparent success using different analyses of GWA data for success in quitting smoking in four independent samples of carefully monitored individuals who attempted to quit smoking in clinical trials or in the community [8,9]. No result from any of these studies achieved genome-wide significance. However, the molecular genetic results from these independent samples display substantial convergence with each other in analyses that we have termed ‘nontemplate’. Nominally positive results from each of these samples, and in other work that is under review, cluster in small chromosomal regions to extents far greater than we would expect by chance. These results also display modest convergence with molecular genetic data for vulnerability to dependence on nicotine and other addictive substances [8,10–14]. For example, genes for two cell-adhesion molecules, CDH13 and CSMD1, are identified by clusters of such nominally significant SNPs in at least four of six other samples that compare successful quitters with unsuccessful quitters and at least four of seven other samples that compare substance-dependent with control individuals (Monte Carlo p < 0.0001 for each) [15,16, Rose JE et al., Submitted].
In one of the largest reported trials of smoking cessation in primary care settings, Patch in Practice (PIP) study investigators studied the influences of differing intensities of behavioral support on smoking cessation, aided by 15 mg nicotine patches, in smokers recruited at UK general practices in Buckinghamshire and Oxfordshire [17]. We now report ‘nontemplate’ analyses of GWA data that compares individuals from this trial who were successful versus unsuccessful in achieving biochemically monitored 12-week abstinence. SNP data from Affymetrix® 6.0 Array (Affymetrix, CA, USA) studies of these samples identified many of the same chromosomal regions previously identified in other samples based on data for smoking cessation and vulnerability of smokers to physiological dependence on nicotine. We discuss the significant limitations of this dataset, including substantial limitations based on the modest number of successful quitters in this sample. We also discuss the ways in which the results provide support for previous GWA results for the molecular genetics of smoking cessation from clinical trial and community-based samples employing a variety of behavioral and pharmacological strategies.
Materials & methods
Samples
The PIP study identified individuals who were over 18 years old, smoked more than ten cigarettes per day and were recruited in general practices in Buckinghamshire and Oxfordshire in the UK by referral from their general practitioners, advertisements and direct mail contacts. Smokers were treated with 15 mg/16 h nicotine patches and followed with different levels of behavioral support. Abstinence was monitored at 1, 4, 12 and 26 weeks from quit dates using exhaled carbon monoxide (CO) levels and/or salivary cotinine assessments. The abstinence phenotype used for the current study was based on data from telephone contacts 12 weeks after the quit date. Individuals who reported abstinence were biochemically confirmed by in-person evaluation producing salivary cotinine concentrations of less then 15 ng/ml and/or CO levels of over 10 ppm.
A total of 925 smokers were recruited between July 2002 and March 2005. Approximately 12% were recruited from practices in which smokers were invited by letter.
Samples of DNA extracted from the blood of these subjects was re-quantitated. DNA was available from 108 individuals who remained abstinent at the 12-week time point. These individuals were matched to 216 individuals who were abstinent at no time point and who were similar for gender and degree of baseline dependence assessed using the Fagerström Test for Nicotine Dependence (FTND). These samples provided the basis for six pools (n = 18 in each) that contained DNA from all individuals who self-reported abstinence, with biochemical confirmation, at this 12-week follow-up time point (successful quitters) and 12 pools (n = 18 in each) of individuals who never displayed self-reported abstinence at any follow-up time (unsuccessful quitters), who were selected based on matches to features noted for the successful quitters. Successful and unsuccessful quitters were similar for gender, baseline scores on the FTND and the fraction who reported prior unsuccessful attempts to quit [Uhl G, Unpublished Data].
Our nontemplate GWA pooled genotyping reduced costs, allowed us to assess high densities of genotypes in these subjects while providing no threat of loss of genetic confidentiality to these individual research volunteers, and allowed us to use methods that provided over 0.98 correlations between individual and pooled genotyping in validating studies [18,19, Drgon T et al., Submitted]. Hybridization probes were prepared with precautions to avoid contamination, as described (Affymetrix Assay 6.0, [18]). For each pool, 250 ng of pooled DNA was processed, labeled and hybridized to Affymetrix 6.0 arrays according to the instructions of the manufacturer (Affymetrix) and [18–20]. Quality controls for assays were performed as recommended (Affymetrix, Online Supplementary Table 1, available at www.futuremedicine.com/doi/suppl/10.2217/pgs.09.156/suppl_file/suppl_table_1.xls).
Identification of nominally positive SNPs
Allele frequencies for each SNP in each DNA pool were assessed based on the intensities of hybridization to the 3–4 ‘perfect match’ cells on each of three arrays, as described [18,19]. We deleted data from SNPs whose chromosomal positions could not be adequately determined and from SNPs on sex chromosomes, allowing us to combine data from male and female subjects and increase overall power. Based on the modest variances obtained, we identified SNPs that display t-values with p-values of less than 0.01 ‘nominally positive’ significance in comparison of data from quit success versus failure pools. Based on the high density of SNPs available on these arrays, we used a criterion of clusters of four nominally positive SNPs (see later).
Identification of genomic regions and genes that contained clustered positive SNP data from these samples
We performed preplanned primary nontemplate GWA analyses as previously described [15]. We identified SNPs that display t-values with p-values of less than 0.01 nominally positive significance in the comparison of data from quit success versus failure pools and cluster in small chromosomal regions, so that at least four of these nominally positive SNPs lay within 10 kb (or 25 kb) of other nominally positive SNPs. A number of these clustered, nominally positive SNPs identify genes; many of them also lie between currently annotated genes.
To seek additional support for the chromosomal regions identified by these clusters of nominally positive SNPs, we sought additional association signals in these same regions from clustered, nominally positive SNPs identified in relevant independent GWA studies: each of the three samples from Uhl et al. (500–600,000 SNP GWA studies of smokers who were successful versus unsuccessful in quitting in clinical trial settings) [8], Drgon et al. (500,000 SNP GWA studies of smokers who quit versus those who continued to smoke in community settings) [9], Bierut et al. (38,000 SNP GWA studies of nondependent [FTND] versus dependent [FTND] smokers) [12] and Drgon et al. (1 million SNP GWA studies of smokers who quit and those who continued to smoke in a clinical trial of denicotinized cigarettes) [16]. To provide insights into some of the genes likely to harbor variants that contribute to individual differences in ability to quit, we identified genes that are identified by overlapping clusters of positive SNPs from the current and at least two other quit success or addiction vulnerability samples.
We compared the observed results to those expected by chance using 10,000 Monte Carlo simulation trials, as described previously [21]. For each trial, a randomly selected set of SNPs from the current dataset was assessed to see if it provided results equal to or greater than the results that we actually observed. The number of trials for which the randomly selected SNPs displayed (at least) the same features as the observed results was then tallied to generate an empirical p-value. These simulations thus correct for the number of repeated comparisons made in these analyses. This is an important consideration in evaluating these GWA datasets. We also performed permutation analyses to provide a secondary assessment of significance. For each trial of the permutation analyses, we labeled results from randomly selected DNA pools as successful or unsuccessful, repeated the analyses used herein, and assessed the extent to which the results demonstrated the extent of chromosomal convergence that was identified by the results of our bona fide analyses.
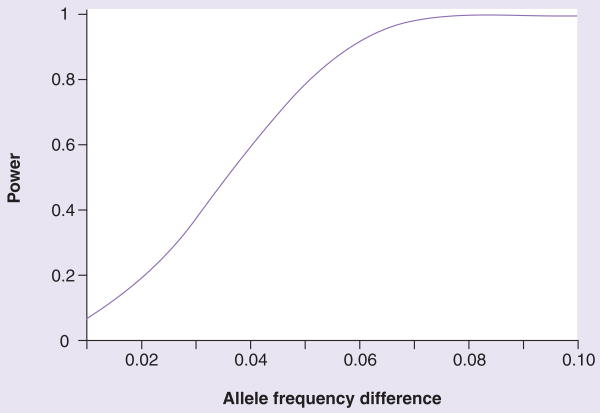
To assess the nominal power of our current approach, we used current sample sizes and standard deviations, the program PS v2.1.31 [22,23] and α = 0.05 (Figure 1). We note that these data provide only nominal power, and do not correct for the large numbers of repeated comparisons provided by the SNPs assessed by these arrays. To provide controls for the possibility that quit success failure differences observed herein were owing to occult ethnic/racial allele frequency differences or noisy assays, we assessed the overlap between the results obtained here and the SNPs that displayed the largest allele frequency differences between African–American versus European–American control individuals evaluated in previous GWA datasets from our laboratory [Drgon T et al., Submitted] and the largest assay ‘noise’.
Figure 1. SNP-wise statistical power of the estimated allele frequency difference between abstinence and nonabstinence pools (t-test).
See ‘Materials & methods’ section; mean standard deviation of the allele frequency estimates between the pools: 0.034, number of abstinence and nonabstinence pools: 6 and 12, respectively, and requirement for ‘nominal’ significance: p = 0.05.
Secondary analyses seeking overall differences between successful quitters & nonquitters using principal component analyses
We performed principal component analyses for the SNP datasets. A first principal component appeared to reflect noise, not relating to phenotype [Uhl G, Unpublished data]. However, a second principal component appeared to distinguish pools of individuals who successfully quit smoking from those who continued to smoke. We performed nominal t-tests to compare the component 2 coefficients for successful versus unsuccessful quitters.
Results
We assessed allele frequencies in multiple pools of DNA from smokers who remained abstinent for at least 12 weeks after the quit date versus those who were not abstinent at any time point. For SNPs, there was modest variability among replicate arrays (n = 3) that assessed the same pool; the standard error of the mean value was 0.05. There was also modest variability among the different pools that assessed the same phenotype; the standard error of the mean was 0.018. For the quitter versus nonquitter comparisons, these samples and estimates of variance thus provide 0.26, 0.78, 0.98 and 0.99 nominal power to detect allele frequency differences of 2.5, 5, 7.5 and 10%, respectively (Figure 1).
Unsuccessful versus successful quitters
When we compare data from unsuccessful versus successful quitters, there is significant clustering of nominally positive SNPs in small chromosomal regions. There are 107,455 nominally positive SNPs that display allele frequency differences with nominal p-values of less than 0.01. A total of 41,319 of these nominally positive SNPs fall into 6,743 clusters of at least four nominally positive SNPs separated from each other by 10 kb or less. Monte Carlo p-values for this degree of clustering are less than 0.0001; we would have expected only 21,518 SNPs to cluster in this fashion by chance. Applying a different set of criteria based on distances of 25 kb between nominally positive SNPs leads to the identification of 74,616 clustered, nominally positive SNPs in 8825 clusters (p < 0.0001). In addition, none of the 1000 permutation tests in which six pools were randomly assigned to be ‘pseudoabstinent’ and 12 pools randomly assigned to be ‘nonabstinent’ ever identified as many SNPs that achieved nominally significant results or clustered in small chromosomal regions as found in the actual dataset (thus p < 0.001).
Unsuccessful versus successful quitters: principal components analysis
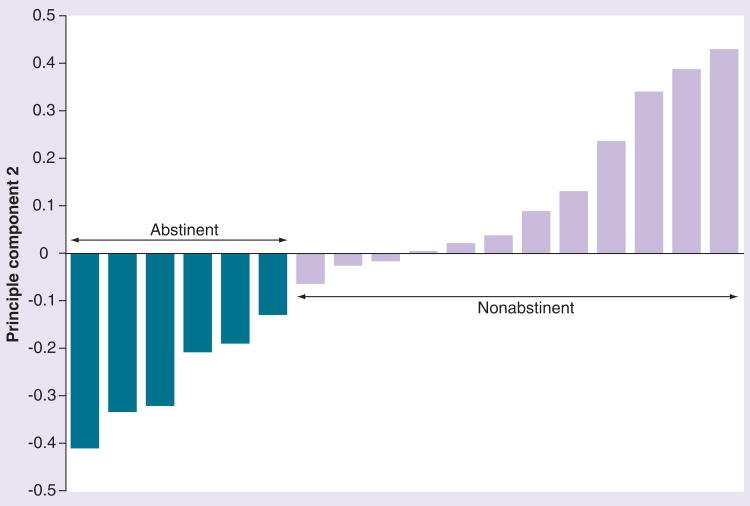
Principal component analyses analyses also provided evidence that helped to confirm the overall assessment of nonrandom differences between genotypes of successful versus unsuccessful quitters. The first principal component from these analyses appeared to reflect noise, and did not appear to separate DNA pools from successful versus unsuccessful quitters [Drgon T, Unpublished Data]. By contrast, the second principal component clearly separated the quitters from nonquitters (Figure 2). Although this component accounted for only 0.5% of the variance in the SNP genotypes, the component 2 coefficient for quitter versus nonquitter pools displayed highly significant differences (p = 0.0001 by t-test; Kolmogorov–Smirnov test D = 1; p = 0.0001). While we cannot exclude the possibility that the second principal component presents a false-positive finding, in conjunction with the primary analysis, it adds to our confidence that there exists a portion of SNPs that distinguish between these two groups of individuals. Monte Carlo models of similar datasets suggest that such an event is highly unlikely [Drgon T, Unpublished Data].
Figure 2. Values for principal component 2 from SNP data for pools of abstinent (darker bars) and nonabstinent (lighter bars) smokers.
The difference, tested by t-test and Kolmogorov–Smirnov test of component 2 coefficients between abstinent and nonabstinent pools, is statistically significant (p = 0.0001).
Overlap with data from previous quit-success samples
These data for clustered, nominally positive SNPs from the current dataset provide significant overlap with genes Table 1 (and intragenic regions, Online Supplementary Table 1, www.future-medicine.com/doi/suppl/10.2217/pgs.09.156/suppl_file/suppl_table_1.xls) that have been identified by other relevant datasets. Thus, they identify the same small chromosomal regions within genes that are identified by nominally positive results in other studies to extents much greater than we would expect by chance. The clustered nominally positive SNPs from the current dataset (e.g., of ≥ four nominally positive SNPs within 25 or 10 kb of each other) provide highly significant overlap with data from each of the three quit success samples reported [8]. Overlaps between the clusters of nominally positive SNPs from this current dataset with previously reported results from the samples from Lerman and coworkers, Niaura and coworkers and Rose and coworkers [8], which identified 100, 217 and 183 clusters of nominally positive SNPs, respectively (p < 0.0001 for each comparison). The four-SNP/10-kb clusters from the current work also displayed significant overlap with the clustered, nominally positive SNPs reported in [9]. Overlaps with these data identified 78 clusters of nominally positive SNPs (p < 0.0001). Finally, these four-SNP/10-kb clusters from the current samples identified 68 clusters from comparisons between successful and unsuccessful quitters studied in clinical trials of denicotinized cigarettes [16]. The overlaps between the clustered, nominally positive SNPs from the 25-k results from the current sample and the clustered, nominally positive SNPs from at least one other sample of successful versus unsuccessful quitters and/or nicotine dependence identified 245 chromosomal regions. A total of 141 of these regions contain 210 annotated genes (Table 1).
Table 1.
Genomic regions that identify genes with variants likely to contribute to individual differences in smoking cessation success.
| Chr | Bp start | Bp end | No. cluster SNPs | Convergence samples | Genes | |
|---|---|---|---|---|---|---|
| 1 | 54, 879, 643 | 54, 928, 490 | 10 | B,Bi | ACOT11, C1orf175 | |
| 1 | 58, 227, 024 | 58, 446, 103 | 49 | L, B, Bi | DAB1 | |
| 1 | 63, 929, 785 | 64, 011, 312 | 8 | † | H,L | ROR1 |
| 1 | 76, 162, 374 | 76, 232, 033 | 8 | † | R, Bi | ASB17 |
| 1 | 111, 697, 010 | 111, 792, 259 | 19 | H, L | ATP5F1, C1orf88, OVGP1, WDR77 | |
| 1 | 112, 277, 072 | 112, 430, 579 | 17 | R, B | KCND3 | |
| 1 | 178, 376, 715 | 178, 423, 711 | 8 | H, Bi | QSCN6 | |
| 1 | 199, 507, 386 | 199, 542, 321 | 5 | † | R, B | PKP1 |
| 1 | 204, 786, 441 | 204, 842, 013 | 6 | B, Bi | LGTN, RASSF5 | |
| 1 | 213, 286, 210 | 213, 343, 397 | 10 | † | B, Bi | KCNK2 |
| 1 | 214, 172, 881 | 214, 242, 873 | 14 | R, B | USH2A | |
| 2 | 7, 003, 815 | 7, 232, 079 | 29 | † | B, Bi | RNF144 |
| 2 | 41, 850, 165 | 42, 030, 301 | 32 | L, Bi | FLJ44450 | |
| 2 | 80, 067, 191 | 80, 346, 411 | 37 | † | R, Bi | CTNNA2 |
| 2 | 182, 041, 692 | 182, 133, 792 | 8 | R, B | CERKL, ITGA4 | |
| 2 | 202, 247, 337 | 202, 269, 099 | 6 | L, Bi | ALS2, MPP4 | |
| 2 | 208, 595, 869 | 208, 736, 436 | 18 | R, Bi | CRYGA, CRYGB, CRYGC, CRYGD, CRYGEP1 | |
| 3 | 7, 553, 531 | 7, 659, 290 | 20 | B, Bi | GRM7 | |
| 3 | 8, 476, 282 | 8, 535, 054 | 7 | L, R | LMCD1 | |
| 3 | 8, 950, 156 | 9, 069, 531 | 28 | † | R, Bi | RAD18, SRGAP3 |
| 3 | 29, 464, 039 | 29, 518, 414 | 11 | B, Bi | RBMS3 | |
| 3 | 133, 208, 054 | 133, 423, 529 | 36 | L, R | CPNE4 | |
| 3 | 141, 207, 949 | 141, 227, 150 | 6 | V, B | CLSTN2 | |
| 3 | 189, 622, 323 | 189, 689, 536 | 8 | † | H, Bi | LPP |
| 3 | 191, 188, 236 | 191, 278, 814 | 11 | R, B | LEPREL1 | |
| 4 | 42, 072, 182 | 42, 102, 929 | 4 | H, L | ATP8A1 | |
| 4 | 78, 110, 193 | 78, 165, 841 | 8 | L, Bi | SEPT11 | |
| 5 | 75, 936, 915 | 76, 009, 201 | 15 | B, Bi | F2RL2, IQGAP2 | |
| 5 | 109, 191, 967 | 109, 272, 818 | 11 | H, B | MAN2A1 | |
| 5 | 121, 802, 488 | 121, 935, 316 | 21 | B, Bi | SNCAIP | |
| 5 | 135, 341, 153 | 135, 443, 625 | 21 | † | R, B, Bi | TGFBI |
| 5 | 146, 004, 728 | 146, 050, 103 | 5 | R, B | PPP2R2B | |
| 6 | 13, 369, 609 | 13, 528, 950 | 30 | † | V, H | GFOD1, PHACTR1, TBC1D7 |
| 6 | 37, 590, 901 | 37, 626, 219 | 11 | † | B, Bi | FLJ45825 |
| 6 | 45, 046, 432 | 45, 120, 789 | 6 | R, B | SUPT3H | |
| 6 | 46, 006, 032 | 46, 176, 097 | 25 | † | R, Bi | CLIC5 |
| 6 | 91, 356, 667 | 91, 445, 589 | 17 | B, Bi | MAP3K7 | |
| 7 | 21, 896, 100 | 22, 091, 042 | 23 | † | H, Bi | CDCA7L, DNAH11 |
| 7 | 37, 343, 697 | 37, 418, 238 | 16 | B, Bi | ELMO1 | |
| 7 | 103, 185, 343 | 103, 272, 991 | 10 | R, Bi | RELN | |
| 8 | 3, 423, 347 | 3, 462, 554 | 9 | H, R | CSMD1 | |
| 8 | 3, 724, 342 | 4, 030, 783 | 39 | V.H.R.B | CSMD1 | |
| 8 | 6, 442, 204 | 6, 566, 971 | 14 | † | H, L | AGPAT5, MCPH1 |
| 8 | 8, 805, 453 | 8, 875, 531 | 8 | R, Bi | MRPS18CP2 | |
| 8 | 14, 478, 087 | 14, 676, 266 | 39 | † | R, B | SGCZ |
| 8 | 15, 988, 678 | 16, 012, 684 | 8 | † | H, R | MSR1 |
| 8 | 16, 043, 410 | 16, 157, 007 | 10 | † | H, R | MSR1 |
| 8 | 27, 623, 804 | 27, 649, 069 | 6 | B, Bi | CCDC25 | |
| 8 | 35, 442, 952 | 35, 570, 418 | 22 | † | B, Bi | UNC5D |
| 8 | 36, 722, 386 | 36, 820, 063 | 13 | † | R, Bi | MRPS7P1 |
| 8 | 105, 276, 318 | 105, 402, 479 | 15 | R, Bi | RIMS2 | |
| 8 | 124, 682, 005 | 124, 795, 511 | 20 | R, Bi | ANXA13, C8ORFK36 | |
| 9 | 3, 971, 734 | 4, 050, 625 | 19 | R, B | GLIS3 | |
| 9 | 4, 566, 451 | 4, 593, 564 | 5 | H, Bi | C9orf68, SLC1A1 | |
| 9 | 8, 234, 384 | 8, 442, 875 | 29 | † | B, Bi | PTPRD |
| 9 | 8, 802, 996 | 8, 884, 013 | 10 | R, B | PTPRD | |
| 9 | 17, 645, 744 | 17, 756, 315 | 32 | R, Bi | SH3GL2 | |
| 9 | 27, 209, 058 | 27, 301, 732 | 9 | L, Bi | C9orf11, TEK | |
| 9 | 36, 112, 297 | 36, 219, 596 | 10 | H, Bi | C9orf19, CCIN, CLTA, GNE, RECK | |
| 9 | 89, 360, 097 | 89, 384, 032 | 6 | V, R, B | DAPK1 | |
| 10 | 18, 415, 180 | 18, 670, 890 | 68 | † | R, B | CACNB2 |
| 10 | 23, 133, 288 | 23, 289, 115 | 21 | † | B, Bi | ARMC3 |
| 10 | 49, 588, 433 | 49, 654, 696 | 12 | † | L, B | C10orf64 |
| 10 | 49, 683, 846 | 49, 715, 477 | 5 | L, B | C10orf64 | |
| 10 | 56, 137, 663 | 56, 173, 483 | 7 | L, B | PCDH15 | |
| 10 | 63, 873, 866 | 64, 005, 316 | 34 | † | R, Bi | ZNF365 |
| 10 | 68, 343, 091 | 68, 547, 563 | 34 | H, Bi | CTNNA3, LRRTM3 | |
| 10 | 69, 603, 138 | 69, 630, 097 | 9 | H, B | MYPN | |
| 10 | 108, 879, 748 | 108, 904, 435 | 7 | V, R | SORCS1 | |
| 10 | 115, 345, 318 | 115, 547, 607 | 28 | † | B, Bi | C10orf81, CASP7, HABP2, NRAP |
| 10 | 118, 951, 893 | 118, 999, 379 | 15 | † | V, B | KCNK18, SLC18A2 |
| 10 | 122, 171, 375 | 122, 234, 298 | 8 | † | L, Bi | PPAPDC1A |
| 11 | 6, 809, 694 | 6, 841, 983 | 4 | R, Bi | OR10A2, OR10A5 | |
| 11 | 34, 384, 953 | 34, 412, 784 | 6 | R, Bi | CAT | |
| 11 | 35, 263, 469 | 35, 339, 791 | 28 | † | R, B | SLC1A2 |
| 11 | 35, 446, 467 | 35, 575, 167 | 12 | R, Bi | DKFZP586H2123 | |
| 11 | 45, 160, 424 | 45, 273, 233 | 12 | L, Bi | PRDM11, SYT13 | |
| 11 | 55, 564, 553 | 55, 652, 591 | 10 | R, B | OR5AQ1P, OR5AS1, OR5BE1P, OR5BN1P, OR5BN2P, OR5J1P, OR8H2, OR8H3, OR8I2, OR8I4P, OR8J3 | |
| 11 | 88, 049, 581 | 88, 130, 053 | 20 | † | B, Bi | GRM5 |
| 11 | 95, 540, 236 | 95, 639, 632 | 9 | H, B | MAML2 | |
| 11 | 109, 803, 757 | 109, 809, 813 | 4 | H, R | FDX1 | |
| 12 | 26, 647, 873 | 26, 814, 162 | 30 | R, B, Bi | ITPR2 | |
| 12 | 66, 793, 406 | 66, 966, 116 | 40 | L, R, Bi | IFNG, IL22, IL26, MDM1 | |
| 13 | 32, 490, 409 | 32, 676, 469 | 29 | † | B, Bi | KL, STARD13 |
| 14 | 20, 275, 863 | 20, 310, 513 | 6 | B, Bi | FAM12A, FAM12B, RNASE6 | |
| 14 | 55, 753, 295 | 55, 873, 368 | 17 | B, Bi | PELI2 | |
| 14 | 57, 337, 787 | 57, 646, 073 | 55 | † | R, B | C14orf37 |
| 14 | 70, 656, 401 | 70, 777, 386 | 18 | R, Bi | PCNX | |
| 14 | 92, 102, 618 | 92, 199, 563 | 13 | R, Bi | RIN3 | |
| 14 | 93, 957, 582 | 94, 007, 781 | 18 | † | H, Bi | SERPINA11, SERPINA9 |
| 15 | 31, 679, 734 | 31, 858, 597 | 31 | H, R | RYR3 | |
| 15 | 51, 677, 685 | 51, 723, 710 | 10 | B, Bi | WDR72 | |
| 15 | 58, 774, 797 | 58, 878, 043 | 18 | L, Bi | RORA | |
| 15 | 69, 460, 239 | 69, 467, 134 | 4 | R, B | THSD4 | |
| 15 | 69, 525, 357 | 69, 604, 738 | 9 | L, B | THSD4 | |
| 15 | 83, 801, 507 | 83, 993, 233 | 32 | † | R, B | AKAP13 |
| 16 | 6, 244, 581 | 6, 413, 198 | 22 | † | H, B, Bi | A2BP1 |
| 16 | 7, 217, 948 | 7, 280, 430 | 13 | L, Bi | A2BP1 | |
| 16 | 7, 526, 679 | 7, 578, 950 | 14 | † | L, B | A2BP1 |
| 16 | 10, 070, 946 | 10, 265, 020 | 37 | R, Bi | GRIN2A | |
| 16 | 26, 865, 197 | 26, 995, 116 | 28 | L, R | TNT | |
| 16 | 54, 316, 342 | 54, 352, 549 | 6 | † | B, Bi | CES4 |
| 16 | 76, 958, 858 | 77, 079, 685 | 14 | † | H, Bi | WWOX |
| 16 | 79, 581, 073 | 79, 765, 704 | 34 | † | B, Bi | ASCIZ, C16orf46, C16orf61, CENPN, GCSH, PKD1L2 |
| 16 | 81, 666, 609 | 81, 701, 353 | 10 | V, Bi | CDH13 | |
| 16 | 81, 927, 086 | 82, 128, 704 | 46 | L, B, Bi | CDH13 | |
| 16 | 82, 177, 911 | 82, 448, 961 | 66 | † | L, B | CDH13, HSBP1 |
| 16 | 85, 009, 589 | 85, 141, 506 | 31 | R, B | FOXF1, MTHFSD | |
| 18 | 6, 945, 676 | 7, 024, 144 | 14 | R, B | LAMA1 | |
| 18 | 54, 938, 326 | 55, 029, 031 | 17 | V, Bi | GRP, SEC11L3 | |
| 18 | 62, 429, 317 | 62, 520, 134 | 16 | B, Bi | CDH19 | |
| 20 | 4, 653, 718 | 4, 803, 759 | 19 | R, Bi | PRND, PRNT, RASSF2, SLC23A2 | |
| 20 | 10, 117, 643 | 10, 185, 816 | 14 | † | R, B | SNAP25 |
| 20 | 17, 356, 503 | 17, 438, 497 | 20 | B, Bi | BFSP1, PCSK2, RPS27AP2 | |
| 20 | 17, 669, 518 | 17, 756, 526 | 9 | V, B | C20orf179 | |
| 20 | 19, 548, 339 | 19, 678, 620 | 18 | † | B, Bi | SLC24A3 |
| 20 | 40, 278, 482 | 40, 318, 043 | 5 | L, R | PTPRT | |
| 20 | 40, 457, 119 | 40, 515, 646 | 8 | R, B | PTPRT | |
| 20 | 41, 789, 167 | 41, 908, 073 | 15 | L, B | FAM112A | |
| 20 | 49, 854, 068 | 49, 926, 477 | 8 | L, B | SALL4 | |
| 21 | 23, 320, 362 | 23, 443, 841 | 27 | R, Bi | ZNF299P | |
| 22 | 24, 713, 274 | 24, 824, 612 | 20 | † | R, B | MYO18B |
Data from the current study identifies the same small chromosomal regions identified by clustered nominally positive SNPs from at least one other genome-wide association study of ability to quit smoking or of vulnerability to nicotine dependence among smokers. Columns list: chromosome and base pair coordinates for the beginning and end of the genomic region identified by clustered nominally positive SNPs from the current study, number of nominally positive SNPs that lie in clusters within the region in the current sample, code for the confirmatory sample(s) and annotated gene(s) that fall within the chromosomal region. Codes for confirmatory samples: L: Lerman, B: Brown and R: Rose from [8], H: Hamer [9], V: Vector samples [16], Bi: Data from comparison of 38,000 SNPs from [12]. Data from intragenic regions identified in the same fashion in included as Online Supplementary Material (available at www.futuremedicine.com/doi/suppl/10.2217/pgs.09.156/suppl_file/suppl_table_1.xls). Bolding indicates genes that show significant clustering with both 10 kb and 25 kb clustering requirement (see ‘Materials & methods’ section).
Genes identified by clustered SNPs, which display allele frequencies that differ between other African–American versus European–American control samples.
Overlap with data from substance dependence GWA
The clusters of four nominally positive SNPs from the data that compare individuals who are successful versus those who are unsuccessful in quitting smoking in the current trial also overlap at greater-than-chance levels with data from studies that compare nondependent to dependent smokers. Data that overlap with nominally significant results reported by Bierut et al. [12] identifies 1481 SNPs that lie within 50 kb of one of the clusters of nominally positive results from the current data (p < 0.001). The overlaps between the clustered, nominally positive SNPs from the current sample and the clustered, nominally positive SNPs from comparisons of controls to European–American individuals dependent on at least one illegal substance identify 4017 small chromosomal regions (p < 0.0001) [Drgon T et al., Submitted].
Possible alternative explanations for observed results
We would anticipate the observed, highly significant clustering of SNPs that display nominally positive results if many of these reproducibly positive SNPs lay near and were in linkage disequilibrium with functional allelic variants that distinguished subjects who were more able to quit smoking from those who were less able. We would not anticipate this degree of clustering if the results were due to chance. The Monte Carlo p-values noted here are thus likely to receive contributions from both the extent of linkage disequilibrium among the clustered, nominally positive SNPs and the extent of linkage disequilibrium between these SNPs and the functional haplotype(s) that lead to the association with quit success.
Control for occult stratification was based on examining the overlap between the 41,319 clustered positive SNPs from the present quit-success analyses with the 2.5% of the SNPs for which the racial/ethnic differences in control individuals from prior datasets were largest. We identified 1177 SNPs with these properties; 1033 SNPs would have been expected by chance. Controls for noisy SNPs found that 418 of the clustered nominally positive SNPs overlapped with the set of SNPs that provided the largest variance, while 569 would have been expected by chance.
Control of false negatives and false positives arising due to the differences in the local density of available SNPs in different genomic regions was carried out thus: a false-negative region may fail to be detected because it does not contain four SNPs within 10 or 25 kb, while a false-positive region can be detected because of its high local SNP density. Overall genomic SNP density for the SNPs on the arrays is approximately 3.26 SNPs per 10 kb. For the regions detected by the 10-kb clusters, density is 7.85 SNPs per 10 kb; for the 25-kb clusters, density is 5.27 SNPs per 10 kb. However, the fraction of the total genome detected by any single study is quite small. Monte Carlo convergence p-values account for these differences in SNP density (random SNP-dense regions are more likely to be detected in simulation trials, therefore decreasing the Monte Carlo convergence p-values), and show that no two samples would ever converge to the degree shown here by chance.
Discussion
The current results provide few results with genome-wide significance in this dataset when evaluated alone. However, they provide independent support for GWA results from prior studies of smoking cessation success in clinical trial and community settings. The substantial overlaps between these data and those obtained previously support the idea that the ability to quit in primary care settings appears to share genetic influences with the ability to quit in clinical trial or community settings. The current results also appear to support a significant, though more modest, overlap between allelic variants that alter one's vulnerability to develop dependence on an addictive substance versus those that alter one's ability to quit smoking, as we have noted in prior analyses [15].
These observations can be discussed in light of the strengths and limitations of the current dataset. The data display several strengths:
The similar clinical characteristics and the modest pool-to-pool variation provide reassurance that these UK smoking cessation subjects may not differ markedly from the European–American smoking cessation subjects who were previously studied;
The subjects were carefully followed with biochemical monitoring of self-reported abstinence;
The pooled genotyping methods that we use herein have been extensively validated in prior work and demonstrate modest variance in the current studies;
Many more of the positive results from this work (than we would expect to do so by chance) identify the same chromosomal regions that were identified by other studies of smoking cessation and/or vulnerability to develop nicotine dependence in smokers with largely European genetic backgrounds.
There are major and minor limitations of the present work. The major limitation is based on the relatively modest number of successful quitters in this trial. Although we have worked to match these individuals to twice their number of nonquitters, limitations from this modest sample size reduce our confidence in the genes that are identified in this work but not in prior studies, and negative data concerning any gene that has been reproducibly identified in prior studies but not in the current work. Although we have provided extensive validation of the pooled genotyping methods for the SNP probes used here, the modest variance introduced by the pooling approach, combined with the modest sample size, does provide an additional limit to interpretation of data from SNP probes; the chromosome 15 cluster of nicotinic acetylcholine receptor genes that has been reproducibly identified in comparisons of heavy, physiologically dependent smokers with light smokers with less physiological dependence has not been identified in the current work, or in several other comparisons of successful quitters versus unsuccessful quitters [12,14]. The comparisons with results from other studies that help us to validate many of the findings from the current work do not represent conventional meta-analyses that seek to combine data for the same SNPs from multiple samples. Only a modest proportion of the SNPs assessed in the current samples are represented on the arrays used for prior studies. Nevertheless, since virtually all of the SNPs from each study can be accurately positioned on the human genome, identification of overlapping chromosomal regions does provide a reasonable, although less rigorous, means for assessing the extent to which results from different studies provide support for the results from other technically distinct studies of similar phenotypes. Differences in local SNP densities may give rise to both false negatives (in regions where the SNP density is too low for detection of clusters of nominally positive SNPs) and false positives (where the high local SNP density increases the possibility of nominally false-positive SNPs forming a cluster). The latter is addressed by assessing the significance of the cluster convergence with Monte Carlo methods. Last, while we are fairly confident in the significance of the sets of variants, genomic regions and genes presented here, further convergence and fine mapping of the presented candidate genomic regions in independent samples will further increase our confidence in any of the individual regions/loci.
Conclusion
The current results add appreciably to the set of studies that appear to document molecular genetic contributions to the ability to quit smoking in research and community settings. As we work on fine-mapping studies that provide greater specification of the haplotypes that influence quit success, cellular and molecular studies that identify the ways in which these haplotypes alter the brain and other relevant organs and animal model studies that link these alterations to behavioral differences, we can also test if these haplotypes provide genetic determinants of generalized ability to change health-related behaviors. Thus, for both dependent individuals and individuals with other health problems that can be modified through behavior change, these data add to an increasingly rich basis for improved understanding and for the development of personalized treatment strategies.
Supplementary Material
Acknowledgments
We are grateful for the thoughtful advice and discussion from Jed Rose, Dean Hamer, Caryn Lerman, Ray Niaura and Sean David. We would also like to thank all participants in the Patch in Practice trial as well as all clinical and laboratory staff who worked on the trial. We would like to acknowledge excellent technical assistance by Dr Qing Rong Liu.
Footnotes
Financial & competing interests disclosure
Dr George R Uhl is listed as an inventor for a patent application filed by Duke University (NC, USA) based on genomic markers that distinguish successful quitters from unsuccessful quitters in data from other clinical trials. This research was supported by the NIH Intramural Research Program, NIDA, DHSS and a Cancer Research UK Programme Grant. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research: The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
For reprint orders, please contact: reprints@futuremedicine.com
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Broms U, Silventoinen K, Madden PA, Heath AC, Kaprio J. Genetic architecture of smoking behavior: a study of Finnish adult twins. Twin Res Hum Genet. 2006;9(1):64–72. doi: 10.1375/183242706776403046. [DOI] [PubMed] [Google Scholar]; ■■ Defines the heritabilities of smoking behavior.
- 2.Lessov CN, Martin NG, Statham DJ, et al. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol Med. 2004;34(5):865–879. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]; ■■ Defines the heritabilities of smoking behavior.
- 3.Uhl GR, Elmer GI, Labuda MC, Pickens RW. Genetic influences in drug abuse. In: Gloom FE, Kupfer DJ, editors. Psychopharmacology: The Fourth Generation of Progress. Raven Press; NY, USA: 1995. pp. 1793–2783. [Google Scholar]
- 4.Tsuang MT, Lyons MJ, Meyer JM, et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55(11):967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 5.Karkowski LM, Prescott CA, Kendler KS. Multivariate assessment of factors influencing illicit substance use in twins from female–female pairs. Am J Med Genet. 2000;96(5):665–670. [PubMed] [Google Scholar]
- 6.True WR, Heath AC, Scherrer JF, et al. Interrelationship of genetic and environmental influences on conduct disorder and alcohol and marijuana dependence symptoms. Am J Med Genet. 1999;88(4):391–397. doi: 10.1002/(sici)1096-8628(19990820)88:4<391::aid-ajmg17>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Kendler KS, Karkowski LM, Neale MC, Prescott CA. Illicit psychoactive substance use, heavy use, abuse, and dependence in a US population-based sample of male twins. Arch Gen Psychiatry. 2000;57(3):261–269. doi: 10.1001/archpsyc.57.3.261. [DOI] [PubMed] [Google Scholar]
- 8.Uhl GR, Liu QR, Drgon T, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65(6):683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Samples were used for convergence.
- 9.Drgon T, Montoya I, Johnson C, et al. Genome-wide association for nicotine dependence and smoking cessation success in NIH research volunteers. Mol Med. 2009;15(1–2):21–27. doi: 10.2119/molmed.2008.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Samples were used for convergence.
- 10.Uhl GR, Drgon T, Liu QR, et al. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch Gen Psychiatry. 2008;65(3):345–355. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- 11.Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE. Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet. 2007;8:10. doi: 10.1186/1471-2156-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bierut LJ, Madden PA, Breslau N, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Samples were used for convergence.
- 13.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhl GR, Drgon T, Johnson C, et al. Molecular genetics of addiction and related heritable phenotypes: genome-wide association approaches identify “Connectivity Constellation” and drug target genes with pleiotropic effects. Ann NY Acad Sci. 2008;1141:318–381. doi: 10.1196/annals.1441.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drgon T, Johnson C, Walther D, Albino AP, Rose JE, Uhl GR. Genome-wide association for smoking cessation success: participants in a trial with adjunctive denicotinized cigarettes. Mol Med. 2009;15(7–8):268–274. doi: 10.2119/molmed.2009.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Samples were used for convergence.
- 17.Aveyard P, Brown K, Saunders C, et al. Weekly versus basic smoking cessation support in primary care: a randomised controlled trial. Thorax. 2007;62(10):898–903. doi: 10.1136/thx.2006.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson C, Drgon T, Liu QR, et al. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141(8):844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu QR, Drgon T, Walther D, et al. Pooled association genome scanning: validation and use to identify addiction vulnerability loci in two samples. Proc Natl Acad Sci USA. 2005;102(33):11864–11869. doi: 10.1073/pnas.0500329102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhl GR, Liu QR, Walther D, Hess J, Naiman D. Polysubstance abuse-vulnerability genes: Genome scans for association, using 1,004 subjects and 1,494 single-nucleotide polymorphisms. Am J Hum Genet. 2001;69(6):1290–1300. doi: 10.1086/324467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu QR, Drgon T, Johnson C, Walther D, Hess J, Uhl GR. Addiction molecular genetics: 639,401 SNP whole genome association identifies many “cell adhesion” genes. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(8):918–925. doi: 10.1002/ajmg.b.30436. [DOI] [PubMed] [Google Scholar]
- 22.Dupont WD, Plummer WD., Jr Power and sample size calculations. A review and computer program. Control Clin Trials. 1990;11(2):116–128. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 23.Dupont WD, Plummer WD., Jr Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19(6):589–601. doi: 10.1016/s0197-2456(98)00037-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.