Abstract
In the past three decades, scientists have had immense success in identifying genes and their variants that contribute to an array of diseases. While the identification of such genetic variants has informed our knowledge of the etiologic bases of diseases, there continues to be a substantial gap in our understanding of the factors that modify disease severity. Monogenic diseases provide an opportunity to identify modifiers as they have uniform etiology, detailed phenotyping of affected individuals, and familial clustering. Cystic fibrosis (CF) is among the more common life-shortening recessive disorders that displays wide variability in clinical features and survival. Considerable progress has been made in elucidating the contribution of genetic and nongenetic factors to CF. Allelic variation in CFTR, the gene responsible for CF, correlates with some aspects of the disease. However, lung function, neonatal intestinal obstruction, diabetes, and anthropometry display strong genetic control independent of CFTR, and candidate gene studies have revealed genetic modifiers underlying these traits. The application of genome-wide techniques holds great promise for the identification of novel genetic variants responsible for the heritable features and complications of CF. Since the genetic modifiers are known to alter the course of disease, their protein products become immediate targets for therapeutic intervention.
Keywords: genome wide, candidate gene, heritability, variation
Overview
It is generally recognized that genetic, environmental, and stochastic factors contribute to phenotype variation. However, the relative effect of each component is difficult to assess, especially for common diseases where a myriad of environmental factors may play a role. Consider, for instance, the role of drugs (an environmental factor) in the treatment of diseases and the effect they can have in modifying outcome. Parsing patient populations based on multiple environmental exposures can lead to substantial drops in power to detect the responsible genetic variants. Despite the aforementioned challenges, progress has been made in dissecting genetic and nongenetic factors underlying disease variability for several of the more common Mendelian disorders. The “monogenic” diseases provide unique opportunities to dissect components as they each have a single etiology, relatively uniform treatments, and the contribution of the disease-causing gene is known to some degree.1 In many cases, some portion of phenotype variability can be associated with the nature of the mutations in the disease-causing gene. However, disease variability in patients bearing the same combination of mutations emphasizes the role of genetic background and environment, which appears to be the rule rather than the exception.2 Thus, the current challenge for many studying monogenic disorders is to assess the relative contribution of genetic factors distinct from the disease-causing gene and to identify those genes that modify outcome.3
Identification of such modifier genes increases our understanding of the elements that affect disease variability and thereby identifies new targets for therapy. Furthermore, it is possible that genes that modify single-gene disorders contribute to the development and/or progression of common diseases in the general population. These genes may be minimally penetrant in healthy individuals, whereas the altered homeostasis of patients with single-gene disorders may unmask the effect of such modifiers. Thus, the search for genetic modifiers of single-gene disorders could benefit individuals beyond those afflicted with monogenic conditions. Finally, by their very nature, single-gene disorders have one highly penetrant disease-causing gene that could serve as a starting point for modeling of gene–gene and gene–environment interactions.
Rationale for cystic fibrosis
Cystic fibrosis (CF), a single-gene-recessive disorder that affects 60,000 individuals worldwide, is an ideal model for the identification and characterization of factors that cause disease variation.4,5 First, the unfortunate high prevalence of CF provides a large number of accessible patients to perform detailed phenotypic analyses necessary to identify modifier genes in humans. Second, the commonness of CF and its monogenic etiology enable family-based studies to dissect genetic and nongenetic influences upon disease variation. Third, the diagnosis of CF can be confirmed using an objective laboratory measure: the sweat chloride concentration assay. In the vast majority of CF patients, chloride levels in the sweat are raised well beyond the distribution of values observed in the normal and CF carrier population, thereby distinguishing the disease from other conditions with similar features.6
Patients with CF manifest disease in the lungs, pancreas, intestine, liver, male reproductive tract, and sweat gland.6 CFTR, the dysfunctional protein in CF patients, conducts chloride across the apical membranes of polarized epithelia.7 Loss of CFTR function affects the transport of chloride, sodium, and water across epithelial tissues, leading to inadequate hydration of mucous secretions of CF patients. Obstruction of luminal space follows, and recurrent cycles of inflammation and fibrosis ultimately destroys affected organs.6,8 Obstruction of the exocrine pancreas causes intestinal malabsorption and an abnormal nutritional status in almost all CF patients. Obstructive lung disease is the cause of death in almost 90% of patients.9
As noted above, the large number of families with multiple affected offspring facilitated identification of the CF transmembrane conductance regulator (CFTR) gene, an early and remarkable success in positional cloning. The identification of CFTR provided immense insight into the molecular pathophysiology of CF. A network of research labs that joined to form the CF Genetic Analysis Consortium has identified over 1,800 mutations in CFTR (http://www.genet.sickkids.on.ca/cftr/app). One mutation, a deletion of three nucleotides causing the loss of a phenylalanine at codon 508 (p.Phe508del), accounts for approximately 70% of CF alleles in Caucasian patients.10 CF patients homozygous for p.Phe508del constitute the most common CFTR genotype (about 50% of patients, designated as F508del homozygotes from here on), and, as such, have served as a reference population for genotype/phenotype correlations.11 There is also a group of 15–20 less common mutations accounting for 15% of CF alleles in Caucasians.10 A study of the effect of a number of less common mutations upon phenotype demonstrated that allelic variation in CFTR accounts for only a portion of phenotypic variability.11 CFTR genotype is highly correlated with preservation of some function of the exocrine pancreas (termed “pancreatic sufficiency”). Furthermore, sweat chloride levels tend to be less elevated in patients with CFTR genotypes associated with preserved pancreatic function than in those with pancreatic insufficiency.11,12 To date, minimal correlation has been found between CFTR genotype and severity of lung disease, the major cause of mortality for CF patients.13,14 These observations prompted a search for the causes of disease variation independent of CFTR genotype.
Evidence for, and quantification of, genetic modifier role for CF traits
Before embarking upon a search for genetic modifiers, it is important to consider the importance of nongenetic contributions to a phenotype. In the case of CF, the impressive increase in mean years of survival from less than 1 year to the upper 30s currently is attributable almost exclusively to the treatments devised for these patients. From that perspective, environmentally mediated factors have a profound effect on disease variability. Specific treatments such as pancreatic enzyme supplementation, nutritional support, and vitamin supplementation have modified CF from a disease of malnutrition and early childhood demise to a disorder marked by respiratory compromise beginning in childhood and progressing into mid-adulthood.6 Extension of longevity has also exposed dysfunction of the endocrine pancreas that manifests as diabetes.15 On the other hand, treatment does not appear to have altered the prevalence of complications that occur in the newborn period (e.g., neonatal intestinal obstruction or meconium ileus) or early to midchildhood (e.g., hepatic cirrhosis).9
Lung disease
A variety of measures have been devised to assess severity of lung disease in CF patients, and the most useful objective gauge is based on airflow. Pulmonary function testing is widely used, and methods for assessing ventilatory capacity and airflow rates are standardized. The forced expiratory volume in 1 sec (FEV1) is an excellent measure of small and large airway obstruction, the major site of disease in CF lungs. Furthermore, FEV1 can track progression of obstruction and is well correlated with survival.16 This lung function measure is highly variable among CF patients with identical CFTR genotypes (e.g., F508del homozygotes).17 With the exception of a few mutations that confer a milder pancreatic phenotype (e.g., p.Arg455Glu18,19), correlation between different CFTR genotypes associated with pancreatic insufficiency and lung function measures is minimal. The small numbers of patients with genotypes other than F508del homozygosity and the high degree of variability among patients with the same CFTR genotype may preclude observation of correlation.11 Indeed, preliminary analysis of nearly 40,000 patients in the CFTR2 database reveals low correlation between CFTR mutations and FEV1 (P. Sosnay, personal communication). Together, these studies indicate that factors other than CFTR genotype determine progression of airway obstruction in CF.
To parse genetic and nongenetic contributions to disease severity and complications, investigators have used family-based studies. Recurrence of complications in affected siblings at rates higher than in unrelated patients suggests genetic effect, although care must be taken to account for the effect of shared environment. A more powerful approach is to compare monozygous (MZ) and dizygous (DZ) twin pairs for concordance for qualitative traits and correlation for quantitative traits. Twin pairs raised together in the same household are controlled, to some degree, for environmental exposures. One then exploits the different rates of overall sharing of genetic variation between MZ pairs (100%) and DZ pairs (50%) to estimate genetic control. When MZ pairs demonstrate higher concordance (or correlation) for a clinical feature than DZ pairs, then genetic factors are assumed to be responsible.20 The aforementioned classic twin study initially proposed by Galton and refined by Siemens has been used to estimate the proportion of phenotype variation attributable to genes (i.e., heritability or h2) for numerous traits in humans.21 This method has been adapted to estimate h2 for variation in lung function and anthropometric measures and the complications of intestinal obstruction and diabetes in CF patients (see below).
The first twin-based assessment of gene modifier contribution to CF disease severity used a composite measure of lung function and body mass index (BMI). A higher correlation of this composite measure was observed in 29 MZ versus 12 DZ twins pairs, suggesting genetic control of this trait.22 Analysis of lung function and weight for height as independent measures did not reveal significant differences between the MZ and DZ twin pairs. A subsequent comparison of 38 MZ pairs with six same-sex DZ pairs and 61 same-sex sibling pairs within 3 years of age drawn from the North American CF Twin and Sibling Study estimated heritability of lung function based on FEV1 measurements to range from 0.54 to 1.0.23 The heritability estimate was similar (0.56–0.86) when the analysis was restricted to patients with identical CFTR genotypes (F508del homozygotes). Variance analysis of 231 pairs of affected siblings generated a slightly higher estimate of heritability for the FEV1 measures (0.68–1.0).23 Together, these studies revealed that genetic modifiers play a substantial role in determining FEV1, a key measure of lung function correlated with survival.
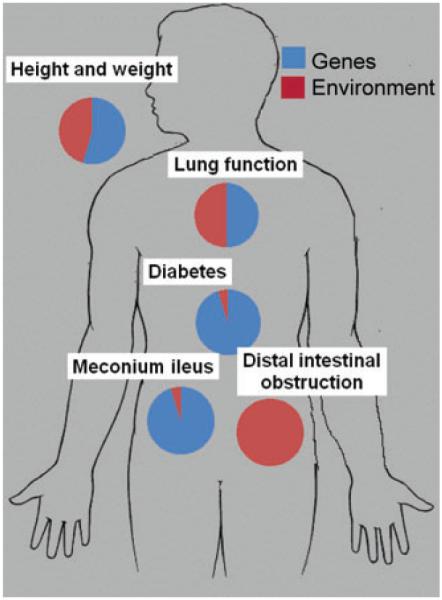
Factors in the environment also contribute heavily to variation in lung function in CF patients. As might be expected from a disorder heavily modified by treatment, socioeconomic factors such as access to care, parental income, and other factors that may compromise optimal treatment have been shown to have significant adverse effects on lung disease and survival.24,25 Pollutants in the air including particulate matter and components of second-hand smoke can adversely affect lung function in CF.26,27 To assess the relative contribution of genetic and environmental factors to variation in FEV1 among CF patients, Collaco et al. evaluated 134 MZ twins and 272 DZ twins and siblings when living together and after moving apart. Differences in lung function between MZ twin pairs while living together in the same household provided an estimate of the contribution of unique environment and stochastic effects. The transition from the home environment to independent living was used to estimate the contribution of shared environment. Genetic contribution was assessed by comparing the similarity in lung function measures in MZ and DZ twin pairs when living together, and then when living apart. These methods revealed that genetic and nongenetic factors had almost equal contribution to variation in lung function (Fig. 1).28 This estimate was unchanged when the analysis was restricted to twins and siblings bearing the same CFTR genotype (F508del homozygotes). Two thirds of the nongenetic effect could be attributed to unique environment and stochastic effects while shared environment was responsible for the remainder (Fig. 1). Analysis of 58 MZ twins and 568 DZ twins and siblings from the European CF Twin and Sibling Study generated similar estimates for the genetic and nongenetic contributions to lung function variance (~60% attributable to genetic factors in all subjects and ~50% in subjects who were F508del homozygotes).29
Figure 1.
Relative contributions of non-CFTR genetic and environmental factors to major manifestations of cystic fibrosis.
Exocrine pancreatic function
Pancreatic exocrine function is abnormal in almost all patients with CF. Most patients have severe dysfunction (pancreatic insufficiency) leading to steatorrhea and malabsorption that manifests in the first year of life.6 As noted above, a fraction of CF patients (~10–20%) do not develop steatorrhea and are termed “pancreatic sufficient.” CFTR genotype is known to be highly predictive of pancreatic exocrine status.30 However, there are patients who appear to have delayed transition to pancreatic insufficiency suggesting a possible role for genetic modifiers. Serum immunoreactive trypsinogen (IRT) is a pancreatic enzyme precursor that is elevated in neonates with CF and, as such, serves as a marker for newborn screening for CF. In young CF patients that develop exocrine pancreatic insufficiency, IRT levels decline rapidly.31 Heritability for variation in IRT levels was estimated in 23 sibling pairs with CFTR genotypes associated with pancreatic insufficiency using variance analysis. Evidence of genetic control was noted in patients at 2 months of age (h2 = 0.51) and at 6 months of age (0.45).32 Genetic effect could not be confirmed in older patients.
Diabetes
Due to the successful management of CF in childhood, more patients are surviving into adulthood. As an unfortunate result additional complications of the disease are becoming more prevalent. Dysfunction of the endocrine pancreas, generally thought to be a consequence of the exocrine pancreatic disease, leads to loss of beta cell function and decreased insulin secretion. Two percent of children with CF develop diabetes, while 19% of adolescents and over 40% of adults with CF develop this complication.33 Diabetes in CF patients typically occurs in the absence of obesity and is associated with a significantly worse prognosis.34 While CF-related diabetes (CFRD) has been viewed as a condition distinct from diabetes seen in the general population, there are a number of clinical and pathologic similarities with type 1 and type 2 diabetes.35 Both type 1 and type 2 diabetes in the general population show evidence of strong genetic control. Numerous genes and DNA variants have been associated with each form of diabetes.
Twin study has used to assess genetic modifier contribution to CFRD. MZ twins displayed significantly higher rates of concordance than DZ twins and siblings of similar age and same sex generating an estimate of heritability approaching 1.0.36 Pedigree analysis revealed that CFRD and type 2 diabetes may have similar genetic origins. The presence of type 2 diabetes in at least two first-degree relatives was associated with a significantly higher prevalence of diabetes in the CF proband (OR 3.1; P = 0.0009) compared to the absence of a similar family history of diabetes.37 Thus, diabetes in CF patients is quite common, strongly determined by genetic modifiers (Fig. 1) and appears to share some genetic basis with type 2 diabetes in the general population.
Intestinal obstruction
Obstruction of the intestine by abnormal meconium in the neonatal period is a characteristic sign of CF. The condition, termed “meconium ileus” (MI), affects about 15% of newborns with CF.6 While formally lethal, modern treatments consisting of enemas and/or surgery have reduced mortality to less than 10%.38 CFTR genotype appears to make a sizable contribution to risk as patients bearing the missense mutation p.Gly551Asp are at considerably lower risk (6.4%) than p.Phe508del homozygotes (19.5%),39 while patients carrying the nonsense mutation p.Gly542X may be at higher risk.30 However, genetic modifiers make an important contribution as noted by two observations: (1) recurrence risk for MI in affected siblings (25%) is significantly higher than in unrelated CF patients (15%)40 and (2) rates of intestinal obstruction in mouse models of CF differ by strain (i.e., genetic background).41 Intriguingly, neonatal intestinal obstruction that is anatomically and temporally similar to humans occurs with a prevalence of 100% in a porcine model of CF42 and 75% in a ferret model of CF.43 Thus, it appears that humans may have genetic modifiers that protect from this complication.
Twins and sibling analysis has been used to estimate heritability of MI, and a similar phenotype observed in older CF patients termed “distal intestinal obstruction syndrome” (DIOS). Blackman and colleagues noted that concordance for MI in MZ twin pairs (82%) was significantly higher than in DZ and siblings of similar age and sex (22%; P = 0.009) suggesting that heritability for MI approaches 1.040 (Fig. 1). Nongenetic factors must play some role as concordance is not 100% in MZ twins. On the other hand, DIOS showed no differences in concordance rates among MZ and DZ twins and siblings indicating little, if any, genetic modifier effect.40 The latter observation is notable as DIOS was formally termed MI equivalent due to anatomic and pathologic similarities to the neonatal condition.
The intestinal and pancreatic disease in CF leads to substantial disruptions in growth manifesting as short stature and low weight. Aggressive nutritional supplementation has improved both parameters, but chronic illness also affects patients’ entry into puberty. Thus, the use of growth metrics such as BMI or percent of predicted weight for height (WfH%) that have been standardized in healthy individuals presents a challenge for assessing nutritional status in CF. Preliminary analysis of “average” BMI displays moderate heritability (~0.6).44 Genetic control is also noted when BMI measures from select ages are used to avoid confounding by effects of the underlying disease process. Initial analysis of WfH% by the European CF Twin and Sibling Study demonstrated that intrapair discordance for this measure was similar between 29 MZ and 12 DZ twin pairs.22 However, this issue was recently revisited, and a significant trend was noted when comparing intrapair differences in WfH% in 38 MZ, 24 DZ and 396 sibling pairs. Modeling of these three classes of CF siblings estimated that genetic factors account for ~80% of variation in WfH% in 466 related patients (~60% in F508del homozygotes).29 Thus, anthropometric measures also appear to be under genetic control in CF patients. Further study is needed to assess the degree to which height and weight independently contribute to the heritability of these composite measures.
Other traits
The secretion of chloride via CFTR and other pathways across epithelial tissues has been studied in twin and sibling pairs. MZ twins were found to have higher rates of concordance (5 of 5 pairs) for the presence of cAMP-mediated chloride conductance in intestinal epithelia than DZ twins (9 of 15 pairs). Furthermore, alternative modes of chloride conductance were also more concordant in MZ versus DZ pairs.45 In a companion study, MZ twins (six pairs) displayed greater concordance for baseline potential difference measurements across nasal epithelia than five sets of DZ twin pairs.46 The latter generally measures the rate of sodium transport across the respiratory epithelia.47 Concordance for other measurements that reflect chloride conductance (e.g., chloride-free solutions and isoproteronol response) across the nasal epithelia did not differ between MZ and DZ twins.46 Abnormal ion transport abnormality in the sweat gland dysfunction is a characteristic finding of CF. Sweat chloride concentration shows amoderate degree of correlation with CFTR mutations grouped by pancreatic status (i.e., PI versus PS48). However, preliminary analysis of twins and siblings has not shown evidence of genetic effect beyond CFTR (Blackman, S. and G.R. Cutting, unpublished observations).
Liver disease is a common problem for CF patients due to obstruction of the common bile duct. A minor fraction (~3–5%) of patients develop severe hepatic cirrhosis and portal hypertension.6 The occurrence of this complication in younger patients suggests that severe liver disease is not merely a complication of chronic liver damage but a discrete phenotype that may be genetically modified.49 Sibling recurrence rates have not been extensively analyzed with the exception of discordance for liver disease in five sets of siblings.50 Hepatic cirrhosis is too uncommon to derive heritability assessments using concordance analysis among the available twins with CF.
Identification of modifier genes
Intense study of the CFTR has uncovered numerous pathways and their components that have high biologic plausibility for modifying CF. Thus, the vast majority of modifier gene studies for CF traits to date have used the candidate gene approach. Testing of variants of known functional effect from carefully selected candidates is attractive for several reasons. The number of variants tested is generally small thereby avoiding severe penalties for multiple comparisons during statistical analysis. Detailed understanding of the candidate gene product and its variants provides mechanistic insight and facilitates experimental studies to evaluate modifier effects. In some cases, compounds that target the protein product of candidate genes may already be available for human trials. On the other hand, use of the candidate approach is fraught with bias due to our very incomplete understanding of CF biology. Since a significant association between a single gene variant and a CF trait can be obtained with very small differences in allele frequencies, it is essential that any association be replicated in a separate population (preferably multiple times). The latter approach is used extensively in genome-wide studies but is equally important in validating candidate gene associations.
In the case of CF, dozens of candidate genes have been evaluated, but replication has been reported for only a fraction of published associations between variants in these genes and the same CF trait. Of these, only a few variants have been tested in more than 1,000 CF patients. At least nine genes can be implicated as modifying some aspect of the CF phenotype based on replication in two independent studies in more than 500 patients in total (Table 1). Since a comprehensive review of all modifier gene studies would be beyond the scope of this review, details will be discussed only for the genes in Table 1. Several recent publications provide detailed lists of all of the genes that have been studied.5,29,51-53
Table 1.
Genetic modifiers of different features of cystic fibrosis53
| Gene | Pulmonary Function (FEV1) |
P. aeruginosa Acquisition/ Colonization |
Intestinal Obstruction |
Diabetes | Liver Disease |
|---|---|---|---|---|---|
| ADIPOR2 | Possible effect87 | ||||
| EDNRA | Probable effect79 | ||||
| IFRD1 | Possible effect74 | ||||
| IL8 | Possible effect75 | ||||
| MBL2 | Probable effect55,62–64,70,94 | Probable effect60,70,94 | |||
| MSRA | Probable effect86 | ||||
| SERPINA1 | No effect57,95–98 | Likely no effect97,99,100 | Possible effect49 | ||
| TCF7L2 | Probable effect37 | ||||
| TGFB1 | Probable effect57,59,69,70,73,78 | No effect70,72,73,78,101 | Likely no effect49,101 |
Probable effect: Association observed in ≥3 independent populations with ≥1000 participants in aggregate.
Possible effect: Association observed in ≥2 independent populations with ≥500 participants in aggregate.
Likely no effect: No association observed in ≥2 independent populations with ≥500 participants in aggregate.
No effect: No association observed in ≥3 more independent populations with ≥1000 participants in aggregate.
N.B.: Some studies include replication populations, which are treated as separate independent populations.
Genome-wide approaches on chromosome 19 met with initial success in the localization of a region modifying risk for neonatal intestinal obstruction (MI).54 Although subsequent studies were unable to replicate linkage to the MI locus (discussed in detail below), the success of genome-wide methods using both linkage and association for common diseases has rekindled enthusiasm for these approaches. The challenge of genome-wide methods for Mendelian diseases is to recruit sufficient patients to attain statistical power to detect variants of relatively small effect. Studies involving tens of thousands of patients as is becoming routine for common conditions are not practical for monogenic disorders, even diseases that are among the most frequent such as CF. However, compelling evidence of substantial genetic effect (i.e., heritability) for lung disease, anthropometry and intestinal obstruction, and diabetes justify the effort required for genome-wide studies. Furthermore, the magnitude of the modifier effect of variants is not known, therefore moderate numbers of CF patients may be sufficient to indentify significant loci. Finally, we do not know if variants conferring modifier effects are common or rare or some combination of both. Candidate gene studies support both concepts as the variants inTGFb1 affecting lung function are common (~30–50% range) while the variant in SERPINA1 that increases risk for hepatic cirrhosis is rare (1.2%).
Lung disease
One of first candidate modifier studies indicated that functional variants in MBL2, the gene that encodes mannose binding lectin, correlate with lung function measures (FEV1 and FVC).55 At least 12 studies have evaluated the effect of null variants in MBL2 (“O” alleles), and 6 studies involving over 2,049 CF patients have reported that MBL-deficient genotypes (OO or AO) were associated with lower lung function measures (Table 1). Six studies (1,948 patients) did not detect association,56-60 while one study of 109 Danish patients showed the reverse.61 MBL was selected as a candidate due to its role in innate immunity and evidence that MBL deficiency results in predilection to bacterial and viral infection. As infection is a key aspect of CF lung disease, factors that alter resistance are reasonable biologic candidates for CF. Three studies involving 2003 CF patients demonstrated earlier age of infection with Pseudomonas aeruginosa (Pa) associated with MBL deficiency genotypes (Table 1). One study of 112 Swedish patients showed earlier acquisition with sufficient genotypes56 while five studies (943 patients) did not detect association.61-64 Lung disease severity (as measured by FEV1) and infection status are correlated, and both are altered by the age of the patient and by CFTR genotype. Accounting for confounding among these related variables revealed that MBL2 genotype was associated with infection status rather than the other variables.60 Thus, it appears that deficiency in MBL predisposes to early infection with Pa, which, in turn, leads to more severe lung disease than observed in patients of the same age and CFTR genotype who are not MBL deficient.
Transforming growth factor beta 1 (TGFb1) has been investigated numerous times for potential modifier effect on CF. It is a compelling candidate as TGFb1 genotypes have been shown to alter risk for other lung diseases such as asthma and chronic obstructive pulmonary disease.65-67 Furthermore, TGFB1 plays a key role in processes central to CF lung pathophysiology, such as regulation of inflammation and tissue remodeling.68 One of the largest CF genetic modifier studies to date, the Genetic Modifier Study (GMS) analyzed 808 F508del homozygotes drawn from the extreme of lung function (highest 30 percentile and lowest 30 percentile) and reported that alleles in the promoter (−509) and first exon (codon 10) of TGFb1 are associated with worse lung function.57 This finding was replicated in 498 patients with various CFTR genotypes and was independently confirmed when a haplotype composed of the opposite alleles at −509 and codon 10 were associated with improved lung function.69 It also appears that gene and environmental interactions are important to consider in evaluating TGFb1, as variation in CFTR and in MBL2 as well as exposure to second-hand smoke affect association with CF lung function.69-71 Six studies encompassing over 2,500 CF patients report association between TGFb1 and CF lung function (see Table 1) while one study composed of 118 patients did not,72 and another involving 171 patients73 found association with worse lung function and the opposite alleles than reported by Drumm et al.57 and Bremer et al.69 In aggregate, these studies indicate that alleles that increase TGFb1 expression cause worse lung function in CF, although definitive experiments in patients remain to be performed.
Genome-wide methods have been used to identify loci and genes modifying CF lung disease. In a study of 160 mild and 160 severe lung disease F508del homozygous patients drawn from the GMS,57 Karp and colleagues were able to identify 6 candidate loci that associated with lung disease.74 These loci were prioritized using hierarchical clustering for features such as biologic plausibility and regional clustering of associated variants. Three SNPs in the highest ranking gene, the interferon-related developmental regulator 1 gene (IFRD1), showed replication in the entire GMS sample and demonstrated association using transmission-based methods in the family-based CF Twin and Sibling Study (TSS).23 Cell- and mouse-based studies demonstrated that IFRD1 acts via transcriptional mechanisms to modulate neutrophil function in response to bacterial infection.
The concept that modification of CF lung disease may due to altered neutrophil response to infection is supported by evidence that variants in the interleukin-8 (IL-8) gene associate with lung function.75 IL-8 is a mediator of neurophil chemotaxis and is variably and prominently elevated in the airway secretions of CF patients.76,77 Intriguingly, the modifier effect of IL-8 variation may be limited to males (n = 608). A second study of 329 patients failed to detect association between lung function and IL-8 variants, although analysis was not performed using patients separated by sex.78 Taken together, the identification of MBL2, TGFb1, IFRD1, and IL-8 (Table 1) suggests that genetic modifiers may alter lung disease severity by affecting host ability to tolerate infection.
Other mechanisms are likely to contribute to CF lung pathology, as illustrated by evidence that variants in the endothelin receptor type A (EDNRA) gene reproducibly associate with lung disease severity (Table 1). Association between a variant in the 3′ untranslated region of EDNRA was observed in 709 F508del homozygous patients in the GMS study and replicated in three separate samples of CF patients (769 combined).79 Alleles of the EDNRA variant correlated with differences in RNA transcript level, suggesting a possible functional role.79 Given that variation EDNRA has been implicated in vasoconstrictive diseases due to effects on smooth muscle function, it was speculated that this gene may modify CF lung disease by altering smooth muscle tone in the airways and/or vasculature.79
To maximize power to detect novel CF genetic modifiers using genome-wide approaches, the GMS57 and TSS23 have joined forces with a CF modifier study based in Toronto (Canadian Genetics Study; CGS)70 to create the North American CF Gene Modifier Consortium. Three different study designs are incorporated in the consortium: extremes of phenotype (GMS), population based (CGS), and family based (TSS). The former two designs are well-suited to association methods as used in unrelated patients with common diseases. The family-based study can be used for both linkage and association. Using a complementary approach, seven loci meeting criteria for suggestive or significant association at the genome level identified by the two samples of unrelated patients (GMS and CGS) were tested for replication in the TSS. Two suggestive loci showed evidence of association in the TSS but did not meet correction for multiple testing. Sibling analysis revealed a prominent linkage peak on chromosome 20.80-82
Intestinal obstruction
Obstruction of the intestine in mice with a knock-out of the CFTR gene provided an early opportunity to demonstrate the role of genetic modifiers. Strain-specific effects upon the rate of obstruction and survival of CF mice illustrate that genetic background played a significant role in the intestinal trait. By exploiting strain differences, Rozhmahel and colleagues were able to map a locus for obstruction to murine chromosome 7.41 Chromosome 19 contains a region of synteny with the locus on mouse chromosome 7. Using markers from chromosome 19, Zielenski et al. demonstrated association with meconium ileus in 152 sibling pairs (7 concordant affected, 33 discordant, and 112 concordant unaffected).54 Extensive analysis of the chromosome 19 region and the implicated genes failed to identify a causative variant. Two subsequent studies of MI performed in siblings with CF failed to find linkage to chromosome 19. Subsequent mapping studies have identified regions on several mouse chromosomes83,84 as modifying the intestinal phenotype. Other genes that mediate chloride conductance in intestinal tissue have been evaluated in mice and support the conclusion that these genes could modify MI in humans.85
Recent studies using patients collected by the TSS and the CGS have uncovered regions of linkage and association that have been replicated. Blackman and colleagues performed a linkage study of 26 sibling pairs with CF who were both affected with MI. As expected, the CFTR gene was identified with high precision, demonstrating the effectiveness of linkage for genes of moderate to high effect size. Two regions of suggestive linkage (LOD >2.0) were identified on chromosome 8p23.1 and 11q25.40 A subsequent association analysis of the most convincing linkage signal on 8p23.1 was performed using patients from the TSS, GMS, and CGS. Region-wide significance was obtained in the TSS for SNP and haplotype association that implicated the methionine sulfoxide reductase (MSRA) gene. Association between the haplotype (composed of three SNPs with a frequency of 15%) was replicated in a subset of patients from the GMS study and in the entire CGS sample.86 MSRA reduces oxidized methionine residues in proteins that are relevant for intestinal enzymes such as alpha 1 antitrypsin. Variation in MSRA is postulated to alter digestion of intestinal contents contributing to the formation of viscous meconium that obstructs the intestine in CF fetuses and newborns. In a separate study, Dorfman and colleagues used parametric linkage methods to indentify a linkage signal on chromosome 12p13.3 (HLOD 2.935). Association analysis of the linkage region identified a SNP in the adiponectin receptor 2 (ADIPOR2) gene with risk for MI in the CGS population that replicated in the TSS sample.87 Using a similar approach, a broad region on chromosome 4q13 was found to protect from MI. Causative variants have not been found in either MSRA or ADIPOR2, thus further evaluation of these genes may require functional studies. The CF mouse may be ideal for assess how knockout or hypomorphs of these modifier genes affect intestinal development and function given that this animal model develops intestinal obstruction.
Diabetes
Candidate gene studies for diabetes were informed by the observation that diabetes in CF may share genetic origins with type 2 diabetes.37 SNPs near the transcription factor 7-like 2 (TCF7L2) gene88 demonstrate the most consistent association with type 2 diabetes with an estimated odds ratio of ~1.5 per allele.89 Transmission testing of the most highly associated SNP (rs7903146) in 539 families (998 patients) in the TSS revealed significant association with risk for diabetes (P = 0.004). The association was replicated in 802 unrelated patients from the GMS (P = 0.02; combined P = 0.0002).37 Treatment with systemic glucocorticoids, an independent environmental risk factor for diabetes in CF,90 obscured the association with TCF7L2. However, risk for diabetes associated with TCF7L2 was substantially increased (HR 2.9 per allele; P = 0.00011) when patients who had not been treated with glucocorticoids in the past year were analyzed.37 Together, these observations indicate that TCF7L2 modifies risk for diabetes in CF patients who have not had recent or prolonged exposure to systemic steroids. TCF7L2 is postulated to play a role in proliferation and function of the beta cells of pancreatic islets.91,92 Thus, implication of TCF7L2 as a modifier of risk in CF-related diabetes suggests that factors intrinsic to the beta cells of the pancreas contribute to the development of this common complication of CF.
Future directions and implications for other monogenic disorders
The candidate gene approach has been of moderate success in finding genetic modifiers of CF, and several associations have been replicated numerous times. These replicated modifiers account for a small fraction of the heritable contribution to each trait (<1%). The identification of loci and genes using genome-wide methods appears highly promising. The sample sizes accrued by the North American CF Modifier Consortium (~3,500 pts) have been sufficient to find several loci/genes responsible for variation in lung function and for risk of MI and diabetes. Initial screens found at least seven loci meeting or exceeding suggestive evidence of association. Thus, it is reasonable to predict that additional modifier loci and genes, each contributing small effect sizes, exist, and should be discoverable with larger sample sizes. Furthermore, environmental factors are likely to work in concert with genetic modifiers to amplify effect, as in the case of TGFb1 and second-hand smoke, or diminish effect, as noted for steroid exposure and TCF7L2. Longitudinal studies of patients with single-gene disorders are likely to be needed to identify key exposures that contribute to gene–environment interactions. The unfortunate chronicity of monogenic conditions requires frequent medical evaluation, facilitating collection of detailed exposure histories. Furthermore, the near-universal screening for CF in newborns provides an ideal opportunity to recruit patients for longitudinal studies.
A primary reason to identify disease modifiers is to inform the development of therapies and to provide accurate prediction of disease course. Achieving these goals will require characterization of the mechanism of disease modification. One needs to know if improvement in function is afforded by increased activity of a modifier protein, or the reverse. The availability of various mouse strains carrying CFTR mutations associated with mild and severe disease provides an excellent foundation for functional studies and investigation of interactions between CFTR and specific modifier genes. The recent development of pig and ferret models of CF that more completely reflect human CF pathology should be useful for therapy evaluation.43,93 Prognostication will be useful when the effect of genetic and nongenetic factors can be combined to accurately estimate a meaningful change in outcome. The latter may likely require prospective studies in sizable patient cohorts.
The experience in CF modifier research holds some useful insight for other single gene disorders. The importance of consistent phenotype measures and complementary study designs cannot be overemphasized. Family studies can help estimate the contribution of genetic and nongenetic factors. Sibling analysis should be considered if sufficient twins are not available for statistically robust concordance/correlation analyses. By their very nature, Mendelian studies lend themselves to recruitment of families that can be employed in linkage and association studies. Genotyping of parents provides accurate assessment of descent in linkage analysis and enables transmission disequilibrium methods. Ascertaining unrelated patients from the across the severity spectrum is essential for identification and replication studies using by association tools. Finally, recruitment of singleton patients is facilitated by their Mendelian condition as they can be verified to have disease due to the same etiologic factor.
Footnotes
Conflicts of interest
The author declares no conflicts of interest.
References
- 1.Antonarakis SE, Beckmann JS. Mendelian disorders deserve more attention. Nat. Rev. Genet. 2006;7:277–282. doi: 10.1038/nrg1826. [DOI] [PubMed] [Google Scholar]
- 2.Scriver CR, Waters PJ. Monogenic traits are not simple: lessons from phenylketonuria. Trends Genet. 1999;15:267–272. doi: 10.1016/s0168-9525(99)01761-8. [DOI] [PubMed] [Google Scholar]
- 3.Nadeau JH. Modifier genes in mice and humans. Nat. Rev. Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- 4.Drumm M. Modifier genes and variation in cystic fibrosis. Respir. Res. 2001;2:125–128. doi: 10.1186/rr47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutting GR. Modifier genetics: cystic fibrosis. Annu. Rev. Genomics Hum. Genet. 2005;6:237–260. doi: 10.1146/annurev.genom.6.080604.162254. [DOI] [PubMed] [Google Scholar]
- 6.Welsh MJ, Ramsey BW, Accurso FJ, Cutting GR. Cystic fibrosis. In: Scriver CR, Beaudet AL, Valle D, Sly WS, editors. The Metabolic and Molecular Bases of Inherited Disease. III. McGraw-Hill, Inc.; New York: 2001. pp. 5121–5188. [Google Scholar]
- 7.Anderson MP, Gregory RJ, Thompson S, et al. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science. 1991;253:202–205. doi: 10.1126/science.1712984. [DOI] [PubMed] [Google Scholar]
- 8.Cutting GR. Cystic fibrosis. In: Rimoin DL, Connor JM, Pyeritz RE, Korf BR, editors. Emery and Rimoin’s Principles and Practice of Medical Genetics. Vol. 2. Churchill Livingstone Elsevier; Philadelphia: 2007. pp. 1354–1394. [Google Scholar]
- 9.Cystic Fibrosis Foundation . Cystic Fibrosis Foundation Patient Registry Annual Data Report 2005. Cystic Fibrosis Foundation; Bethesda, MD: 2005. [Google Scholar]
- 10.Bobadilla JL, Macek M, Fine JP, Farrell PM. Cystic fibrosis: a worldwide analysis of CFTR mutations—correlation with incidence data and application to screening. Hum. Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 11.The Cystic Fibrosis Genotype-Phenotype Consortium Correlation between genotype and phenotype in patients with cystic fibrosis. N. Engl. J. Med. 1993;329:1308–1313. doi: 10.1056/NEJM199310283291804. [DOI] [PubMed] [Google Scholar]
- 12.Wilschanski M, Dupuis A, Ellis L, et al. Mutations in cystic fibrosis transmembrane regulator gene and in vivo transepithelial potentials. Am. J. Respir. Crit Care Med. 2006;78:787–794. doi: 10.1164/rccm.200509-1377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mickle JE, Cutting GR. Genotype-phenotype relationships in cystic fibrosis. Med. Clin. North Am. 2000;84:597–607. doi: 10.1016/s0025-7125(05)70243-1. [DOI] [PubMed] [Google Scholar]
- 14.Zielenski J. Genotype and phenotype in cystic fibrosis. Respiration. 2000;67:117–133. doi: 10.1159/000029497. [DOI] [PubMed] [Google Scholar]
- 15.Stecenko AA, Moran A. Update on cystic fibrosis-related diabetes. Curr. Opin. Pulm. Med. 2010;16:611–615. doi: 10.1097/MCP.0b013e32833e8700. [DOI] [PubMed] [Google Scholar]
- 16.Kerem E, Reisman J, Corey M, et al. Prediction of mortality in patients with cystic fibrosis. N. Engl. J. Med. 1992;326:1187–1191. doi: 10.1056/NEJM199204303261804. [DOI] [PubMed] [Google Scholar]
- 17.Kerem E, Corey M, Kerem B-S, et al. The relation between genotype and phenotype in cystic fibrosis—analysis of the most common mutation (deltaF508) N. Engl. J. Med. 1990;323:1517–1522. doi: 10.1056/NEJM199011293232203. [DOI] [PubMed] [Google Scholar]
- 18.Gan K-H, Veeze HJ, Van Den Ouweland AMW, et al. A cystic fibrosis mutation associated with mild lung disease. N. Engl. J. Med. 1995;333:95–99. doi: 10.1056/NEJM199507133330204. [DOI] [PubMed] [Google Scholar]
- 19.De Braekeleer M, Allard C, Leblanc J-P, et al. Genotype-phenotype correlation in cystic fibrosis patients compound heterozygous for the A455E mutation. Hum. Genet. 1997;101:208–211. doi: 10.1007/s004390050616. [DOI] [PubMed] [Google Scholar]
- 20.Falconer DS. Inheritance of liability to certain diseases estimated from incidence among relatives. Ann. Hum. Genet. 1965;29:51–71. [Google Scholar]
- 21.Spector TD. The history of twin and sibling-pair studies. In: Spector TD, Snieder H, MacGregor AJ, editors. Advances in Twin and Sib-Pair Analysis. Greenwich Medical Media Ltd.; London: 2000. pp. 1–10. [Google Scholar]
- 22.Mekus F, Ballmann M, Bronsveld I, et al. Categories of deltaF508 homozygous cystic fibrosis twin and sibling pairs with distinct phenotypic characteristics. Twin. Res. 2000;3:277–293. doi: 10.1375/136905200320565256. [DOI] [PubMed] [Google Scholar]
- 23.Vanscoy LL, Blackman SM, Collaco JM, et al. Heritability of lung disease severity in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2007;175:1036–1043. doi: 10.1164/rccm.200608-1164OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connor GT, Quinton HB, Kneeland T, et al. Median household income and mortality rate in cystic fibrosis. Pediatrics. 2003;111:e333–e339. doi: 10.1542/peds.111.4.e333. [DOI] [PubMed] [Google Scholar]
- 25.Schechter MS. Non-genetic influences on CF lung disease: the role of sociodemographic characteristics, environmental exposures and healthcare interventions. Pediatr. Pulmonol. Suppl. 2004;26:82–85. doi: 10.1002/ppul.70061. [DOI] [PubMed] [Google Scholar]
- 26.Rubin BK. Exposure of children with cystic fibrosis to environmental tobacco smoke. N. Engl. J. Med. 1990;323:782–788. doi: 10.1056/NEJM199009203231203. [DOI] [PubMed] [Google Scholar]
- 27.Goss CH, Newsom SA, Schildcrout JS, et al. Effect of ambient air pollution on pulmonary exacerbations and lung function in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2004;169:816–821. doi: 10.1164/rccm.200306-779OC. [DOI] [PubMed] [Google Scholar]
- 28.Collaco JM, Blackman SM, McGready J, et al. Quantification of the relative contribution of environmental and genetic factors to variation in cystic fibrosis. Lung Funct. J. Pediatr. 2010;157:802–807. doi: 10.1016/j.jpeds.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanke F, Becker T, Kumar V, et al. Genes that determine immunology and inflammation modify the basic defect of impaired ion conductance in bystic fibrosis epithelia. J. Med. Genet. 2010 doi: 10.1136/jmg.2010.080937. doi:10.1136/jmg.2010.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristidis P, Bozon D, Corey M, et al. Genetic determination of exocrine pancreatic function in cystic fibrosis. Am. J. Hum. Genet. 1992;50:1178–1184. [PMC free article] [PubMed] [Google Scholar]
- 31.Durie P, Forstner GG, Gaskin KJ, et al. Age-related alteration of immunoreactive pancreatic cationic trypsinogen in sera from cystic fibrosis patients with and without pancreatic insufficiency. Pediatr. Res. 1986;20:209–213. doi: 10.1203/00006450-198603000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Sontag MK, Corey M, Hokanson JE, et al. Genetic and physiologic correlates of longitudinal immunoreactive trypsinogen decline in infants with cystic fibrosis identified through newborn screening. J. Pediatr. 2006;149:650–657. doi: 10.1016/j.jpeds.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 33.Moran A, Dunitz J, Nathan B, et al. Cystic fibrosis-related diabetes: current trends in prevalence, incidence, and mortality. Diabetes Care. 2009;32:1626–1631. doi: 10.2337/dc09-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milla CE, Warwick WJ, Moran A. Trends in pulmonary function in patients with cystic fibrosis correlate with the degree of glucose intolerance at baseline. Am. J. Respir. Crit. Care Med. 2000;162:891–895. doi: 10.1164/ajrccm.162.3.9904075. [DOI] [PubMed] [Google Scholar]
- 35.Moran A, Hardin D, Rodman D, et al. Diagnosis, screening and management of cystic fibrosis related diabetes mellitus: a consensus conference report. Diabetes Res. Clin. Pract. 1999;45:61–73. doi: 10.1016/s0168-8227(99)00058-3. [DOI] [PubMed] [Google Scholar]
- 36.Blackman SM, Hsu S, Vanscoy LL, et al. Genetic modifiers play a substantial role in diabetes complicating cystic fibrosis. J. Clin. Endocrinol. Metab. 2009;94:1302–1309. doi: 10.1210/jc.2008-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blackman SM, Hsu S, Ritter SE, et al. A susceptibility gene for type 2 diabetes confers substantial risk for diabetes complicating cystic fibrosis. Diabetologia. 2009;52:1858–1865. doi: 10.1007/s00125-009-1436-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Escobar MA, Grosfeld JL, Burdick JJ, et al. Surgical considerations in cystic fibrosis: a 32-year evaluation of outcomes. Surgery. 2005;138:560–571. doi: 10.1016/j.surg.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 39.Hamosh A, King TM, Rosenstein BJ, et al. Cystic fibrosis patients bearing the common missense mutation Gly->Asp at codon 551 and the deltaF508 are indistinguishable from deltaF508 homozygotes except for decreased risk of meconium ileus. Am. J. Hum. Genet. 1992;51:245–250. [PMC free article] [PubMed] [Google Scholar]
- 40.Blackman SM, Deering-Brose R, McWilliams R, et al. Relative contribution of genetic and nongenetic modifiers to intestinal obstruction in cystic fibrosis. Gastroenterology. 2006;131:1030–1039. doi: 10.1053/j.gastro.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozmahel R, Wilschanski M, Matin A, et al. Modulation of disease severity in cystic fibrosis transmembrane conductance regulator deficient mice by a secondary genetic factor. Nat. Genet. 1996;12:280–287. doi: 10.1038/ng0396-280. [DOI] [PubMed] [Google Scholar]
- 42.Stoltz DA, Meyerholz DK, Pezzulo AA, et al. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci. Transl. Med. 2010;2:29ra31. doi: 10.1126/scitranslmed.3000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun X, Sui H, Fisher JT, et al. Disease phenotype of a ferret CFTR-knockout model of cystic fibrosis. J. Clin. Invest. 2010;120:3149–3160. doi: 10.1172/JCI43052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blackman SM, Vanscoy LL, Collaco JM, et al. Variability in body mass index in cystic fibrosis is determined partly by a genetic locus on chromosome 5. Pediatr. Pulmonol. Supp. 2008;31:271. 10–23-0008. [Google Scholar]
- 45.Bronsveld I, Mekus F, Bijman J, et al. The European CF Twin and Sibling Study Consortium Residual chloride secretion in intestinal tissue of deltaF508 homozygous twins and siblings with cystic fibrosis. Gastroenterology. 2000;119:32–40. doi: 10.1053/gast.2000.8524. [DOI] [PubMed] [Google Scholar]
- 46.Bronsveld I, Mekus F, Bijman J, et al. Chloride conductance and genetic background modulate the cystic fibrosis phenotype of Delta F508 homozygous twins and siblings. J. Clin. Invest. 2001;108:1705–1715. doi: 10.1172/JCI12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knowles MR, Paradiso AM, Boucher RC. In vivo nasal potential difference: techniques and protocols for assessing efficacy of gene transfer in cystic fibrosis. Hum. Gene Ther. 1995;6:445–455. doi: 10.1089/hum.1995.6.4-445. [DOI] [PubMed] [Google Scholar]
- 48.Wilschanski M, Zielenski J, Markiewicz D, et al. Correlation of sweat chloride concentration with classes of the cystic fibrosis transmembrane conductance regulator gene mutations. J. Pediatr. 1995;127:705–710. doi: 10.1016/s0022-3476(95)70157-5. [DOI] [PubMed] [Google Scholar]
- 49.Bartlett JR, Friedman KJ, Ling SC, et al. Genetic modifiers of liver disease in cystic fibrosis. JAMA. 2009;302:1076–1083. doi: 10.1001/jama.2009.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castaldo G, Fuccio A, Salvatore D, et al. Liver expression in cystic fibrosis could be modulated by genetic factors different from the cystic fibrosis transmembrane regulator genotype. Am. J. Med. Genet. 2001;98:294–297. doi: 10.1002/1096-8628(20010201)98:4<294::aid-ajmg1097>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 51.Davies JC, Griesenbach U, Alton E. Modifier genes in cystic fibrosis. Pediatr. Pulmonol. 2005;39:383–391. doi: 10.1002/ppul.20198. [DOI] [PubMed] [Google Scholar]
- 52.Knowles MR. Gene modifiers of lung disease. Curr. Opin. Pulm. Med. 2006;12:416–421. doi: 10.1097/01.mcp.0000245707.59138.40. [DOI] [PubMed] [Google Scholar]
- 53.Collaco JM, Cutting GR. Update on gene modifiers in cystic fibrosis. Curr. Opin. Pulm. Med. 2008;14:559–566. doi: 10.1097/MCP.0b013e3283121cdc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zielenski J, Corey M, Rozmahel R, et al. Detection of a cystic fibrosis modifier locus for meconium ileus on human chromosome 19q13. Nat. Genet. 1999;22:128–129. doi: 10.1038/9635. [DOI] [PubMed] [Google Scholar]
- 55.Garred P, Pressler T, Madsen HO, et al. Association of mannose-binding lectin gene heterogeneity with severity of lung disease and survival in cystic fibrosis. J. Clin. Invest. 1999;104:431–437. doi: 10.1172/JCI6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlsson M, Sjoholm AG, Eriksson L, et al. Deficiency of the mannan-binding lectin pathway of complement and poor outcome in cystic fibrosis: bacterial colonization may be decisive for a relationship. Clin. Exp. Immunol. 2005;139:306–313. doi: 10.1111/j.1365-2249.2004.02690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drumm ML, Konstan MW, Schluchter MD, et al. Gene modifiers of lung disease in cystic fibrosis. N. Engl. J. Med. 2005;353:1443–1453. doi: 10.1056/NEJMoa051469. [DOI] [PubMed] [Google Scholar]
- 58.Choi EH, Ehrmantraut M, Foster CB, et al. Association of common haplotypes of surfactant protein A1 and A2 (SFTPA1 and SFTPA2) genes with severity of lung disease in cystic fibrosis. Pediatr. Pulmonol. 2006;41:255–262. doi: 10.1002/ppul.20361. [DOI] [PubMed] [Google Scholar]
- 59.Faria EJ, Faria IC, Ribeiro AF, et al. Association of MBL2, TGF-beta1 and CD14 gene polymorphisms with lung disease severity in cystic fibrosis. J. Bras. Pneumol. 2009;35:334–342. doi: 10.1590/s1806-37132009000400007. [DOI] [PubMed] [Google Scholar]
- 60.McDougal KE, Green DM, Vanscoy LL, et al. Use of a modeling framework to evaluate the effect of a modifier gene (MBL2) on variation in cystic fibrosis. Eur. J. Hum. Genet. 2010;18:680–684. doi: 10.1038/ejhg.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olesen HV, Jensenius JC, Steffensen R, et al. The mannan-binding lectin pathway and lung disease in cystic fibrosis–disfunction of mannan-binding lectin-associated serine protease 2 (MASP-2) may be a major modifier. Clin. Immunol. 2006;121:324–331. doi: 10.1016/j.clim.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 62.Gabolde M, Guilloud-Bataille M, Feingold J, Besmond C. Association of variant alleles of mannose binding lectin with severity of pulmonary disease in cystic fibrosis: cohort study. Br. Med. J. 1999;319:1166–1167. doi: 10.1136/bmj.319.7218.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davies JC, Turner MW, Klein N. Impaired pulmonary status in cystic fibrosis adults with two mutated MBL-2 alleles. Eur. Respir. J. 2004;24:798–804. doi: 10.1183/09031936.04.00055404. [DOI] [PubMed] [Google Scholar]
- 64.Yarden J, Radojkovic D, De Boeck K, et al. Polymorphisms in the mannose binding lectin gene affect the cystic fibrosis pulmonary phenotype. J. Med. Genet. 2004;41:629–633. doi: 10.1136/jmg.2003.017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pulleyn LJ, Newton R, Adcock IM, Barnes PJ. TGFbeta1 allele association with asthma severity. Hum. Genet. 2001;109:623–627. doi: 10.1007/s00439-001-0617-y. [DOI] [PubMed] [Google Scholar]
- 66.Wu L, Chau J, Young RP, et al. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax. 2004;59:126–129. doi: 10.1136/thorax.2003.005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silverman ES, Palmer LJ, Subramaniam V, et al. Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. Am. J. Respir. Crit. Care Med. 2004;169:214–219. doi: 10.1164/rccm.200307-973OC. [DOI] [PubMed] [Google Scholar]
- 68.Akhurst RJ. TGF beta signaling in health and disease. Nat. Genet. 2004;36:790–792. doi: 10.1038/ng0804-790. [DOI] [PubMed] [Google Scholar]
- 69.Bremer LA, Blackman SM, Vanscoy LL, et al. Interaction between a novel TGFB1 haplotype and CFTR genotype is associated with improved lung function in cystic fibrosis. Hum. Mol. Genet. 2008 doi: 10.1093/hmg/ddn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dorfman R, Sandford A, Taylor C, et al. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J. Clin. Invest. 2008;118:1040–1049. doi: 10.1172/JCI33754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Collaco JM, Vanscoy L, Bremer L, et al. Interactions between secondhand smoke and genes that affect cystic fibrosis lung disease. JAMA. 2008;299:417–424. doi: 10.1001/jama.299.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brazova J, Sismova K, Vavrova V, et al. Polymorphisms of TGF-beta1 in cystic fibrosis patients. Clin. Immunol. 2006;121:350–357. doi: 10.1016/j.clim.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 73.Arkwright PD, Laurie S, Super M, et al. TGF-beta(1) genotype and accelerated decline in lung function of patients with cystic fibrosis. Thorax. 2000;55:459–462. doi: 10.1136/thorax.55.6.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gu Y, Harley IT, Henderson LB, et al. Identification of IFRD1 as a modifier gene for cystic fibrosis lung disease. Nature. 2009;458:1039–1042. doi: 10.1038/nature07811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hillian AD, Londono D, Dunn JM, et al. Modulation of cystic fibrosis lung disease by variants in interleukin-8. Genes Immun. 2008;9:501–508. doi: 10.1038/gene.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bonfield TL, Konstan MW, Berger M. Altered respiratory epithelial cell cytokine production in cystic fibrosis. J. Allergy Clin. Immunol. 1999;104:72–78. doi: 10.1016/s0091-6749(99)70116-8. [DOI] [PubMed] [Google Scholar]
- 77.Chmiel JF, Berger M, Konstan MW. The role of inflammation in the pathophysiology of CF lung disease. Clin. Rev. Allergy Immunol. 2002;23:5–27. doi: 10.1385/CRIAI:23:1:005. [DOI] [PubMed] [Google Scholar]
- 78.Corvol H, Boelle PY, Brouard J, et al. Genetic variations in inflammatory mediators influence lung disease progression in cystic fibrosis. Ped. Pulm. 2008;43:1224–1232. doi: 10.1002/ppul.20935. [DOI] [PubMed] [Google Scholar]
- 79.Darrah R, McKone E, O’Connor C, et al. EDNRA variants associate with smooth muscle mRNA levels, cell proliferation rates, and cystic fibrosis pulmonary disease severity. Physiol. Genomics. 2010;41:71–77. doi: 10.1152/physiolgenomics.00185.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knowles MR. CF genome-wide association study: Update from NA Consortium. Pediatric Pulm. 2009;32(Supplement):165–166. [Google Scholar]
- 81.Cutting GR, CF Modifier Consortium A genome-wide association and linkage study (GWALS) identifies two loci modifying lung disease severity in cystic fibrosis: North American CF Modifier Consortium; American Society of Human Genetics Annual Meeting-60th Annual Meeting Program Guide; 2010; Available at www.ashg.org/2010meeting. [Google Scholar]
- 82.Knowles MR. Genetic Modifiers in cystic fibrosis; American Society of Human Genetics Annual Meeting-60th Annual Meeting Program Guide; 2010; Available at www.ashg.org/2010meeting. [Google Scholar]
- 83.Haston CK, Tsui LC. Loci of intestinal distress in cystic fibrosis knockout mice. Physiol. Genomics. 2003;12:79–84. doi: 10.1152/physiolgenomics.00114.2002. [DOI] [PubMed] [Google Scholar]
- 84.Norkina O, De Lisle RC. Potential genetic modifiers of the cystic fibrosis intestinal inflammatory phenotype on mouse chromosomes 1, 9, and 10 BMC. Genet. 2005;6:29. doi: 10.1186/1471-2156-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Young FD, Newbigging S, Choi C, et al. Amelioration of cystic fibrosis intestinal mucous disease in mice by restoration of mCLCA3. Gastroenterology. 2007;133:1928–1937. doi: 10.1053/j.gastro.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 86.Henderson LB, Doshi V, Blackman SM, et al. A haplotype in the MSRA gene confers decreased risk of meconium ileus in cystic fibrosis; American Society of Human Genetics Annual Meeting—59th Annual Meeting Program Guide; 2009; Available at www.ashg.org/2009meeting. [Google Scholar]
- 87.Dorfman R, Li W, Sun L, et al. Modifier gene study of meconium ileus in cystic fibrosis: statistical considerations and gene mapping results. Hum. Genet. 2009;126:763–778. doi: 10.1007/s00439-009-0724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 89.Cauchi S, El Achhab Y, Choquet H, et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J. Mol. Med. 2007;85:777–782. doi: 10.1007/s00109-007-0203-4. [DOI] [PubMed] [Google Scholar]
- 90.Marshall BC, Butler SM, Stoddard M, et al. Epidemiology of cystic fibrosis-related diabetes. J. Pediatr. 2005;146:681–687. doi: 10.1016/j.jpeds.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 91.Liu Z, Habener JF. Glucagon-like peptide-1 activation of TCF7L2-dependent Wnt signaling enhances pancreatic beta-cell proliferation. J. Biol. Chem. 2008;283:8723–8735. doi: 10.1074/jbc.M706105200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wegner L, Hussain MS, Pilgaard K, et al. Impact of TCF7L2 rs7903146 on insulin secretion and action in young and elderly Danish twins. J. Clin. Endocrinol. Metab. 2008;93:4013–4019. doi: 10.1210/jc.2008-0855. [DOI] [PubMed] [Google Scholar]
- 93.Stoltz DA, Meyerholz DK, Pezzulo AA, et al. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci. Trans. Med. 2010;2:29ra31. doi: 10.1126/scitranslmed.3000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trevisiol C, Boniotto M, Giglio L, et al. MBL2 polymorphisms screening in a regional Italian CF Center. J. Cyst. Fibros. 2005;4:189–191. doi: 10.1016/j.jcf.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 95.Doring G, Krogh-Johansen H, Weidinger S, Hoiby N. Allotypes of alpha 1-antitrypsin in patients with cystic fibrosis, homozygous and heterozygous for deltaF508. Pediatr. Pulmonol. 1994;18:3–7. doi: 10.1002/ppul.1950180104. [DOI] [PubMed] [Google Scholar]
- 96.Henry MT, Cave S, Rendall J, et al. An alpha(1)-antitrypsin enhancer polymorphism is a genetic modifier of pulmonary outcome in cystic fibrosis. Eur. J. Hum. Genet. 2001;9:273–278. doi: 10.1038/sj.ejhg.5200623. [DOI] [PubMed] [Google Scholar]
- 97.Frangolias DD, Ruan J, Wilcox PJ, et al. Alpha 1-antitrypsin deficiency alleles in cystic fibrosis lung disease. Am. J. Respir. Cell Mol. Biol. 2003;29:390–396. doi: 10.1165/rcmb.2002-0271OC. [DOI] [PubMed] [Google Scholar]
- 98.de Faria EJ, de Faria IC, Alvarez AE, et al. Association between alpha 1 antitrypsin deficiency and cystic fibrosis severity. J. Pediatr. (Rio J) 2005;81:485–490. doi: 10.2223/JPED.1423. [DOI] [PubMed] [Google Scholar]
- 99.Mahadeva R, Westerbeek RC, Perry DJ, et al. Alpha1-antitrypsin deficiency alleles and the Taq-I G–>A allele in cystic fibrosis lung disease. Eur. Respir. J. 1998;11:873–879. doi: 10.1183/09031936.98.11040873. [DOI] [PubMed] [Google Scholar]
- 100.Meyer P, Braun A, Roscher AA. Analysis of the two common alpha-1-antitrypsin deficiency alleles PiMS and PiMZ as modifiers of Pseudomonas aeruginosa susceptibility in cystic fibrosis. Clin. Genet. 2002;62:325–327. doi: 10.1034/j.1399-0004.2002.620413.x. [DOI] [PubMed] [Google Scholar]
- 101.Arkwright PD, Pravica V, Geraghty PJ, et al. End-organ dysfunction in cystic fibrosis: association with angiotensin I converting enzyme and cytokine gene polymorphisms. Am. J. Respir. Crit. Care Med. 2003;167:384–389. doi: 10.1164/rccm.200204-364OC. [DOI] [PubMed] [Google Scholar]