Following the discovery of fetal DNA in maternal plasma different strategies have been reported for noninvasive prenatal diagnosis of genetic diseases (1). Despite the advances in improving the analytical sensitivity of methods it is still very challenging to distinguish between fetal and maternal sequences and noninvasive prenatal diagnosis of genetic diseases has not yet attained a clinical diagnostic routine application. An innovative strategy was reported based on co-amplification at lower denaturation temperature-PCR (COLD-PCR) which exploits differences in the melting temperature (Tm) of variant or mismatched sequences compared to wild-type ones and uses a critical denaturation temperature (Tc) lower than the melting temperature (Tm) to selectively amplify minority mutated alleles (2).
We developed assays for the identification of fetal paternally inherited mutations in maternal plasma using full COLD-PCR for the detection of IVSI.110 (G>A) and Cd39 (C>T) beta-globin (HBB) mutations causing beta-thalassemia.
Full COLD-PCR is based on the generation of heteroduplexes between mutant and wild-type sequences. These melt at lower temperatures than wild-type homoduplexes, and are selectively denatured at the Tc and subsequently amplified. Since it was demonstrated that both fetal and maternal DNA content in maternal plasma vary from pregnancy to pregnancy (3), a protocol specific for the identification of each mutation was performed on several replicates of the same sample by using a range of Tc values. To simplify the determination of the Tc, we amplified plasma DNA of wild-type controls using a conventional PCR protocol on several replicates of the same sample and by varying the denaturation temperature. The lowest denaturation temperature (81.9°C for IVSI.110; 83.8°C for Cd39) at which wild-type samples were still amplified was chosen as upper limit of a Tc range which spanned over 1°C. The denaturation in each of 8 replicates of the maternal plasma sample was scaled down by 0.1–0.2°C with respect to the previous one starting from the upper Tc value identified. By using a range of potential Tc values on several replicas, as opposed to a single Tc, we ensure that there is at least one replica where differential denaturation between mutant and wild type alleles occurs, still allowing robust PCR amplification.
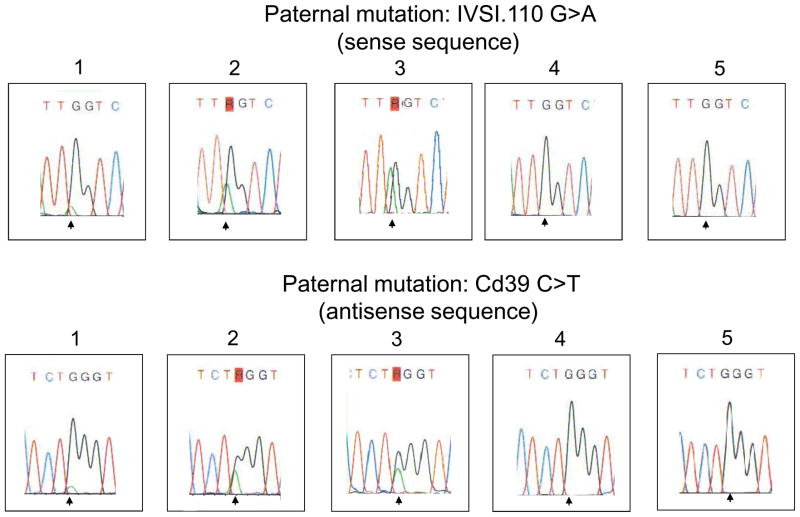
Six mL of maternal blood were collected prior to chorionic villus sampling in couples where parents carried different mutations after written informed consent approved by local Ethical Review Boards. DNA was extracted from 500 μL of plasma (4) and eluted in 60 μL. COLD-PCR was performed in 25 μL containing 5 μL of eluted DNA, 200 μM dNTPs, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 1.5 U of Fast Start (Roche, Mannheim, Germany) and 10 pmoles of each primer. Cycling conditions: 95°C, 4 min;25 cycles of (95°C, 15 s; 54°C, 30 s; 72°C, 1 min); 72°C, 10 min; 95°C, 4 min; 35 cycles of (95°C, 15 s; 70°C, 8 min for generating heteroduplexes; specific Tc-range: 3 s; 72°C, 1 min); 72°C, 7 min. For IVSI.110, Tc range: 81.9 – 80.9°C; for Cd39, Tc range: 83.8 – 82.8°C. Primers for IVSI.110: 5′-TAAGGAGACCAATAGAAACT-3′ (forward); 5′-GTAGACCACCAGCAGC-3′ (reverse); amplicon size: 119bp; for Cd39: 5′-GTCTATTTTCCCACCCTT-3′ (forward); 5′-AGCACTTTCTTGCCATGA-3′ (reverse); amplicon size: 134 bp. Following COLD-PCR, samples were direct sequenced (ABI 3730 DNA Analyzer, Applied Biosystems, Foster City, CA, USA). In total, 35 diagnoses were performed including 21 cases where the father carried the Cd39 mutation (in 10 of these the fetus inherited the paternal mutation) and 14 cases where the father carried the IVSI.110 mutation (in 12 of these the fetus inherited the paternal mutation). In all cases the fetal paternal mutated allele was not detectable with conventional PCR while it became evident after COLD-PCR. In Figure 1, an example of samples with absence of paternally inherited mutated allele and the lowest and the highest enrichment of fetal paternally inherited mutated alleles is shown for both mutations. Results obtained with COLD-PCR were in complete concordance with those obtained on fetal DNA extracted from chorionic villi (Figure 1).
Figure 1.
Sequence analysis in maternal plasma samples. The lowest (1) and the highest (2) enrichment of fetal mutated allele are shown following COLD-PCR-sequencing; results on fetal DNA extracted from chorionic villi are depicted in (3); the fetal mutated allele was not detectable using conventional PCR-sequencing (4); no mutated peak is visible when the fetus inherited the paternal wild-type allele (5).
Du et al reported in a conference the use of fast COLD-PCR to detect congenital paternal abnormalities using fetal circulating DNA in one subject (5). However, fast COLD-PCR cannot detect all mutations. To our knowledge, the present report is the first to indicate the feasibility of using full COLD-PCR for prenatal diagnosis, in a comprehensive study. We provide evidence that COLD-PCR enables straightforward and reliable identification of inherited mutated alleles without the need for sophisticated advanced and costly equipment. The method might be extended to noninvasive prenatal diagnosis of genetic diseases and has the potential to be easily transferable to clinical diagnostic laboratories.
Acknowledgments
Supported by the Ministero della Salute, Programma Strategico, Salute della donna/Area materno infantile, RFPS-2007-4-638637.
List of abbreviations
- COLD-PCR
co-amplification at lower denaturation temperature-PCR
- Tc
critical denaturation temperature
- Tm
melting temperature
- dNTPs
deoxyribonucleotide triphosphate
References
- 1.Lo YM. The quest for accurate measurement of fetal DNA in maternal plasma. Clin Chem. 2010 Sep 13; doi: 10.1373/clinchem.2010.149757. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 2.Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med. 2008;14:579–84. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- 3.Galbiati S, Smid M, Gambini D, Ferrari A, Restagno G, Viora E, et al. Fetal DNA detection in maternal plasma throughout gestation. Hum Genet. 2005;117:243–8. doi: 10.1007/s00439-005-1330-z. [DOI] [PubMed] [Google Scholar]
- 4.Galbiati S, Foglieni B, Travi M, Curcio C, Restagno G, Sbaiz L, et al. Peptide-nucleic acid-mediated enriched polymerase chain reaction as a key point for non-invasive prenatal diagnosis of beta-thalassemia. Haematologica. 2008;93:610–4. doi: 10.3324/haematol.11895. [DOI] [PubMed] [Google Scholar]
- 5.Du J, Zou X, Pan Y, Lu G-X. Noninvasive prenatal diagnosis of congenital adrenal hyperplasia in cell-free fetal DNA with COLD-PCR. Fertility and Sterility. 2009;92(3, Supplement):S199–S200. [Google Scholar]