Abstract
Late-onset Alzheimer's disease (LOAD) is the most common form of dementia in the elderly. The National Institute of Aging-Late Onset Alzheimer's Disease Family Study and the National Cell Repository for Alzheimer's Disease conducted a joint genome-wide association study (GWAS) of multiplex LOAD families (3,839 affected and unaffected individuals from 992 families plus additional unrelated neurologically evaluated normal subjects) using the 610 IlluminaQuad panel. This cohort represents the largest family-based GWAS of LOAD to date, with analyses limited here to the European-American subjects. SNPs near APOE gave highly significant results (e.g., rs2075650, p = 3.2×10−81), but no other genome-wide significant evidence for association was obtained in the full sample. Analyses that stratified on APOE genotypes identified SNPs on chromosome 10p14 in CUGBP2 with genome-wide significant evidence for association within APOE ε4 homozygotes (e.g., rs201119, p = 1.5×10−8). Association in this gene was replicated in an independent sample consisting of three cohorts. There was evidence of association for recently-reported LOAD risk loci, including BIN1 (rs7561528, p = 0.009 with, and p = 0.03 without, APOE adjustment) and CLU (rs11136000, p = 0.023 with, and p = 0.008 without, APOE adjustment), with weaker support for CR1. However, our results provide strong evidence that association with PICALM (rs3851179, p = 0.69 with, and p = 0.039 without, APOE adjustment) and EXOC3L2 is affected by correlation with APOE, and thus may represent spurious association. Our results indicate that genetic structure coupled with ascertainment bias resulting from the strong APOE association affect genome-wide results and interpretation of some recently reported associations. We show that a locus such as APOE, with large effects and strong association with disease, can lead to samples that require appropriate adjustment for this locus to avoid both false positive and false negative evidence of association. We suggest that similar adjustments may also be needed for many other large multi-site studies.
Author Summary
Genetic factors are well-established to play a role in risk of Alzheimer's disease (AD). However, it has been difficult to find genes that are involved in AD susceptibility, other than a small number of genes that play a role in early-onset, high-penetrant disease risk, and the APOE ε4 allele, which increases risk of late-onset disease. Here we use a European-American family-based sample to examine the role of common genetic variants on late-onset AD. We show that variants in CUGBP2 on chromosome 10p, along with nearby variants, are associated with AD in those highest-risk APOE ε4 homozygotes. We have replicated this interaction in an independent sample. CUGBP2 has one isoform that is expressed predominantly in neurons, and identification of such a new risk locus is important because of the severity of AD. We also provide support for recently proposed associated variants (BIN1, CLU, and partly CR1) and show that there are markers throughout the genome that are correlated with APOE. This emphasizes the need to adjust for APOE for such markers to avoid false associations and suggests that there may be confounding for other diseases with similar strong risk loci.
Introduction
Alzheimer's disease (AD, MIM 104300) is by far the most common form of dementia in the elderly. Late onset Alzheimer's disease (LOAD), defined by the onset of symptoms after age 60 years, has annual incidence rates increasing from 1% at 65–70 years to 6–8% at 85 years and older [1]. By age 85 years and up, prevalence is 10–30% or more [2]. While the underlying causes of LOAD are still unknown, there is ample evidence for genetic factors affecting risk, including high estimated heritability of LOAD (58–79%) [3], and evidence from both twin [4], [5] and family studies [6]–[9].
A small number of genes have been identified in which variation contributes to Alzheimer's disease risk. Multiplex early-onset Alzheimer's disease (EOAD) pedigrees [10]–[12] facilitated the identification of mutations in three genes: the amyloid precursor protein (APP) [13], presenilin 1 (PSEN1) [14] and presenilin 2 (PSEN2) [15]. In contrast to the success in familial EOAD, only one gene, APOE, is an unequivocally established “susceptibility” gene for LOAD [16], with the ε4 allele associated with increased risk in a dose-dependent manner and the ε2 allele with decreased risk [17]. There is incomplete lifetime penetrance even in the highest-risk APOE genotypes [18], and the fraction of genetic variance for LOAD risk attributed to APOE is estimated as only 10–20% [19], [20]. This, coupled with results of oligogenic segregation analyses supporting the presence of at least 4–6 additional major genes [21], [22], suggests that additional risk loci remain to be discovered.
Multiple approaches have been used to identify additional loci contributing to LOAD. Several regions have been implicated as a result of multiple linkage-based genome scans [23]–[32]. With the exception of the APOE gene region, there is only modest overlap among the chromosomal regions identified by different analyses [33], and it has been difficult to identify causal variants. Multiple genome-wide association studies (GWAS) of unrelated subjects have now also been carried out [34]–[44]. With the exception of single nucleotide polymorphisms (SNPs) near APOE, all associated SNPs in these studies have had small estimated effect sizes, with odds ratios reported in the range of 1.1 to 1.5, and also with little overlap among studies. Such estimated odds ratios are likely to be highly inflated, and the true effects much smaller [45], complicating replication and identification of causal variants. However, among recent GWAS studies, a small number of loci have shown some evidence for replication across samples including clusterin (CLU), phosphatidylinositol binding clathrin assembly protein (PICALM) and complement component (3b/4b) receptor 1 (CR1) [42]–[44].
The use of densely affected families with LOAD, which are expected to carry higher frequencies of risk alleles, is an excellent alternative method of identifying additional genes contributing to LOAD. For example, the APOE-ε4 frequency is higher in LOAD cases with a positive family history than in sporadic LOAD cases [46], [47]. Compared to the more typical use of unrelated subjects, often without a family history of LOAD, family-based designs may enrich for variants with higher penetrance and consequent increase in odds ratios, and thus increase the power for their detection [48]. Such families can be used in both linkage and association-based designs, with appropriate correction for inclusion of related individuals [38].
Here we present results from a GWAS in multiplex LOAD families. Unlike many other studies, unaffected relatives were also evaluated and are included to increase the amount of genetic information and to provide additional phenotypes that can be used in subsequent analyses. A supplementary control group consisting of unrelated individuals was also recruited and underwent the same phenotypic evaluation. Thus, this cohort represents the largest family-based GWAS of LOAD to date, allowing us to explore issues related to stratification as well as providing a powerful approach for detailed modeling of the effects of APOE in the search for other novel risk loci in LOAD. The genotypic and phenotypic data generated in this study are part of the NIA-LOAD/NCRAD family study (http://ncrad.iu.edu) and are available to the research community through dbGaP (http://www.ncbi.nlm.nih.gov/gap). Biological samples from these well-characterized individuals and families are also available through NCRAD. Our results implicate a new region on chromosome 10p in individuals with the APOE ε4/ε4 genotype, and provide support for some of the recently implicated loci. They also suggest that sample structure and ascertainment bias related to the strong APOE association with AD risk are important confounders. This affects the interpretation of some of the recently implicated loci as well as other GWAS studies of LOAD.
Methods
Subjects and Sample
Subjects
The patient sample contained individuals from families as well as unrelated individuals; however, all patients with LOAD had a family history of Alzheimer's disease. All patients were recruited after providing informed consent and with approval by the relevant institutional review boards, and the study was conducted according to the principles expressed in the Declaration of Helsinki. Regardless of the source (NIA-LOAD Family Study or NCRAD), patients and families were required to meet the same study criteria. In the families, probands were required to have a diagnosis of definite or probable LOAD [49] with onset >60 years of age and a sibling with definite, probable or possible LOAD with a similar age at onset. A third biologically-related family member was required, who could have been a first-, second-, or third-degree relative of the affected sibling pairs, and who was 60 years of age or older if unaffected, or 50 years of age if diagnosed with LOAD or mild cognitive impairment [50]. In these families, additional relatives over age 50 years were recruited regardless of cognitive status. Persons deemed unaffected (controls) were required to have had documented cognitive testing and clinical examination to verify this clinical designation. The largest component of the dataset consisted of 607 families (1,516 affected, 1,306 unaffected) from the NIA-LOAD Family Study and 138 families from the National Cell Repository for Alzheimer Disease (NCRAD; 337 affected, 166 unknown intermediate phenotypes; see Figure S1). We also included pairs of affected siblings whose third family member was either too young, not sampled at the time of the investigation, or had died before an evaluation and blood sample could be obtained. Another 471 unrelated patients from the NIA-LOAD Family Study and NCRAD were included in order to enhance sample size and because they had a well-documented family history of dementia, although no other participating family members had as yet been examined at the time of the investigation.
Unrelated controls were ascertained through three sources: the NIA-LOAD Family Study (n = 794), and NCRAD (n = 144), with the NCRAD controls including 141 subjects from the University of Kentucky. The controls recruited by NIA-LOAD and NCRAD did not have a family history of LOAD in a first degree relative, while those recruited by the University of Kentucky were not excluded if they had a family history of LOAD. All controls demonstrated or had a documented history of normal cognitive function for age, and were evaluated in person or had neuropathology that did not provide any evidence of LOAD.
Phenotypes
A minimal dataset was available for each person consisting of demographics, diagnosis, age at onset for cases, method of diagnosis, Clinical Dementia Rating Scale [51], and the presence of other relevant health problems. Each recruitment site used standard research criteria for the diagnosis of LOAD [49]. Participants with advanced disease or those living in a remote location that could not complete a detailed in-person evaluation contributed a blood sample, and the site investigator conducted a detailed review of medical records to document the presence or absence of LOAD based on the same criteria. The age at onset for patients with LOAD was the age at which the family first observed cognitive complaints. For controls, we used their age at the time of their examination confirming the absence of dementia. For deceased family members who had undergone a postmortem brain evaluation, results of neuropathology were used to document the diagnosis. In general the data from NCRAD was more limited because families were geographically scattered, requiring medical record review, telephone cognitive assessment, and neuropathology data from brain tissue. In total, neuropathological documentation was available for 306 cases from 199 families, and for 25 controls.
For the purpose of analyses, a clinical case was defined as any individual meeting NINCDS-ADRDA criteria for probable or possible AD [49]. We used the NINCDS-ADRDA criteria for definite AD when clinical and pathological criteria were met or CERAD pathological criteria [52] for AD when based on postmortem information alone. Individuals with unspecified dementia, mild cognitive impairment, or uncorroborated family reports of dementia were not used in the analyses. Controls were defined as any individual with no evidence of LOAD, as described above.
Genotypes
Genome-scan genotyping for all samples was provided by CIDR (http://www.cidr.jhmi.edu) as a single project using the Illumina Infinium II assay protocol with hybridization to Illumina Human610Quadv1_B BeadChips (Illumina, San Diego, CA, USA). This array contained 592,532 SNPs with a mean spacing of 5.8 kb, and the minimum genotype completion rate for any sample released by CIDR was 98.3%. Blind duplicate reproducibility was 99.99% based on 118 paired samples. We used only these directly-genotyped data for analysis, without adding further genotypes via imputation. Genotyping of APOE polymorphisms (based on SNPs rs7412 and rs429358) for all samples was performed at PreventionGenetics (http://www.preventiongenetics.com). Genotyping was carried out in array tape [53] using allele-specific PCR with universal molecular beacons [54], [55]. DNA sequencing of positive control DNA samples was completed to assure correct assignment of alleles.
Data Analysis
Overarching approach
Our initial aim was to carry out a GWAS in the complete European-American (EA) component of the sample, with and without adjustment for APOE genotype, while accommodating the presence of related subjects in the sample. To this end, we had three goals: (1) to try to confirm recent reports of associated SNPs in the complete EA component of the sample [42]–[44]; (2) to determine if reports of residual association with SNPs near APOE that implicated other genes in that region [56]–[58] were robust to full adjustment for APOE genotype; and (3) to identify new associations in the genome scan, either in the presence or absence of APOE adjustment. Towards these goals, we carried out two primary genome scans, based on absence or presence of adjustment for APOE genotype, with this adjustment taking into account the full APOE genotype, and not just presence or absence of the ε4 allele, as is commonly done. The main analysis was based on a comparison of allele frequencies between cases and controls with adjustment for the effects of related individuals.
As a consequence of evaluation of the diagnostic indicators of the validity of assumptions associated with the statistical analyses, we also investigated possible sources and effects of confounding. The observations that lead to these additional analyses included deviation from the expected genome-wide null distribution of the test statistics, absence of overlap in SNPs identified with evidence of association across APOE-genotype strata, identification of ethnic subgroups within the larger EA sample, and recognition that the APOE allele frequencies differed among these subgroups. We note that deviation from the expected genome-wide null distribution was less extreme than has been reported in some other recent investigations [42], [43], but was still sufficiently large to warrant investigation. Identification of the sources of confounding involved a number of additional genome scan analyses of the full sample. No correction for additional testing was imposed on these analyses, since their primary purpose was investigation of possible confounding effects, and not of identification of associated SNPs.
Finally, it is important to note that there were complications introduced by use of a sample containing related individuals, coupled with need to make comparisons with results from only the unrelated individuals as part of our broader investigations into confounding and covariate adjustment. The major advantage of the full sample is increased power: with the methods used and assuming a type-I error of 10−7, an allele frequency difference with ∼80% power of detection in the complete family-based sample had a power of only ∼20% in the smaller sample of unrelated subjects. However, use of the complete sample in some analyses as well as the unrelated subjects in other analyses introduced constraints, since the identical analysis approaches were either not always possible or were statistically inappropriate. To the extent possible, when analyses were carried out on different datasets or components of a dataset, the analytical methods chosen were selected to be as comparable as possible. The goal was to be able address the same question, even if the details behind the method of analysis differed.
Quality assessment of data and samples
Family structure both within and across pedigrees was checked and confirmed using Relative ver. 1.1 [59] (ftp://linkage.rockefeller.edu/software/relative and Prest ver. 3.0.2 [60] (http://fisher.utstat.toronto.edu/sun/Software/Prest). A genotyping error rate of 0.03 was used, with a likelihood ratio >1000 used to flag and review pairs with potential discrepancies from their stated relationships. For the analyses presented here, family structure was subsequently used only to identify a sample of unrelated individuals for use in a series of sub-analyses.
After stringent filtering on marker quality control indicators and eliminating monomorphic markers, 565,336 polymorphic autosomal markers from the Illumina panel were used for analysis. Incomplete genotyping (>2%) lead to elimination of 1.12% of markers. Additional metrics such as deviation from compatibility with Hardy Weinberg equilibrium (HWE) in unrelated individuals (0.2% of SNPs) were only used to further evaluate SNPs with evidence for association (p<0.00001), because presence of such disequilibrium is expected in regions with true association and can be informative in the search for gene-disease associations [61]–[63]. Of course such disequilibrium can also identify problem SNPs, but in the absence of evidence for association, such SNPs have little effect on the overall conclusions. No markers of greatest interest were eliminated because of such deviation from HWE, although a small number of such SNPs were eliminated from the tabulation of those SNPs yielding p<5×10−4.
Population structure
Two data sets of unrelated individuals were constructed, based on self-reported ethnic information combined with principal component analysis, described below. The first was drawn from the complete sample, and was of mixed origin, with all unrelated individuals used together. The sample was selected by choosing a random genotyped individual from each family. The second consisted of a sub-sample of individuals defined as “European-American-clustering”, based on the first analysis (henceforth referred to as European-Americans). From the latter sample of European-Americans, a dataset containing only unrelated cases and controls (CCun) was generated such that a single case was selected from each family. For this purpose, clinically defined cases were prioritized based on how AD was defined. Definite AD as defined by NINCDS-ADRDA [49] or CERAD [52] was selected over probable or possible AD. In instances where more than one individual met the most stringent criteria, the case with the lower age of onset was selected.
The complete sample of unrelated individuals was used for initial investigation of population structure in the sample using genotypes from the Illumina panel and smartpca from the Eigensoft package [64]. Initial cluster analysis was based on a principal component analysis (PCA) of the complete unrelated sample, with ethnic-specific clusters delineated based on self-declared ethnicity. This initial analysis was used to refine the cluster location of the European-American sample within the larger sample, and to classify subjects with undeclared ethnicity to define the final sample used for further analysis. For evaluation of cluster-membership of all subjects, the SNP weights for each eigenvector were then applied to all remaining family members. Self-described non-Hispanic European-Americans that clustered as part of the main European-American cluster were subsequently flagged and reanalyzed separately, first using unrelated European-Americans and then applying the SNP weights to all European-Americans. This separate PCA of the European-American sample was stimulated by results in the full sample, in which concerns arose about possible effects of additional stratification. This final PCA lead us to identify and further investigate and delineate three subgroups within the European-American sample.
We hypothesized that these subgroups might represent individuals of northwestern European, southeastern European, and Ashkenazi Jewish ancestry, based on other studies [65] coupled with the population makeup near the collection sites. We used 88 of 159 European-specific ancestry informative markers (AIMs) tailored to NW European (NW), SE European (SE), and Ashkenazi Jewish (AJ) ancestry [65] (http://genepath.med.harvard.edu/~reich/) that were available in our marker panel for evaluating this hypothesis, restricting analysis to the markers for which the minor allele frequency (MAF) was <0.4 in all three sub-populations. These were the markers that provided unambiguous matching of our alleles with those reported previously [65], and avoided potential allele mismatch due to unclear specification of allele calling procedures. For these markers, we obtained the population that best explained each subject by maximizing the likelihood over all markers under the assumption of independence of the markers and using the published allele frequencies [65]. The distributions of the difference in reported vs. observed marker allele frequencies in each cluster vs. known population were also investigated. Finally, after assigning subjects to their subgroup defined by their PCA clustering, allele frequencies were computed genome-wide for pairs of subpopulations, to determine whether known strong gene-frequency clines that characterize north-south gradients in Europe were apparent, such as the regions surrounding lactase and HLA [66], [67].
Kinship estimation
Kinship coefficients estimated from the data were used for several purposes. This included detecting cryptic relatedness, quantifying and comparing relatedness, and correcting for relatedness in statistical tests carried out on samples that included related individuals. We estimated kinship coefficients for pairs of individuals in the European-American sample with Kinship ver. 2.0 (http://faculty.washington.edu/wijsman/software.shtml) based on methods described in detail elsewhere [68]. In brief, for each pair of individuals, we maximized the likelihood for each of the nine condensed identity coefficients [69], and collapsed these into an estimate of the kinship coefficient. Use of a maximum likelihood estimate (MLE), rather than a moment estimate, provides the most accurate such kinship estimates [70], with superior power and control of type I error in hypothesis testing, although at modest increased computational cost. We estimated the kinship coefficients for two groups: the entire group of European-American subjects, including related individuals (referred to as the CCall sample), as well as for the three subgroups: the NW, SE and AJ, as determined by the analyses described above. Each of these four groups included both related and unrelated cases and controls. The estimates for the individual subgroups were used only in analyses that incorporated subgroup information; in all other situations, the estimates from the CCall sample were used.
For the purpose of estimating kinship coefficients, we used 11,471 SNPs chosen to be maximally informative and relatively-uniformly spaced on the reference sequence. Use of higher numbers of markers adds little additional precision, while adding unnecessary computational burden [68]. The panel of SNPs was chosen by restricting use to those with >99% data completion and a MAF>0.05 in the whole data set, with a minimum interval between markers of 100 KB, and an attempt to maximize the MAF. Ultimately, 87% of SNPs used for this purpose had MAF>0.45 and 95% had MAF>0.4.
Case-control analysis, full sample
All tests of allele frequency differences in the CCall sample were carried out with the program cor_chi (http://faculty.washington.edu/wijsman/software.shtml), using procedures that correct for relationships in the sample [68]. We used pair-wise kinship coefficients estimated from the marker data to correct the variance in a chi-square test of association, in a modification to the approach suggested by Bourgain et al [71]. Cases were defined as all affected individuals, and controls as all unaffected individuals, as described above under Phenotyping. This approach allows for variation in the realized IBD-sharing within families, as well as incorporating the effects of additional, unspecified relationships. This approach thus has somewhat better properties in the presence of related subjects than a test based only on pedigree-based expected kinship coefficients [68] or other relationship information from the pedigree structure alone [38]. The correction for kinship was carried out either under the assumption that the full European-American sample represents a single population, or under the assumption that each of three identified sub-populations represents a separate population.
For our primary genome scans, we carried out an analysis of the full sample without adjusting for APOE since this is a common approach [34], [42], as well as an analysis that controlled for APOE genotype effects. The method of analysis that corrected for the existence of relatives did not permit use of a covariate-adjusted model, which is the most common approach for controlling for specific genotype effects. Instead, we carried out a stratified analysis (e.g., structured association), which is also a well-established approach [72]. It is less often used than the former approach because of the need for large sample sizes, but the large size of the full sample available to us made this possible. The consequent number of strata that could be used also allowed a more subtle accommodation for known differences in LOAD risk among APOE genotypes [73] than simply adjusting for presence or absence of the ε4 allele. Attention to appropriate use of covariates is more critical for detection of small than large contributions to risk and to avoid spurious conclusions about association, so that this fuller adjustment for APOE genotype was expected to provide the better model in this context. The strata were based on four APOE genotype groups consisting of ε4/ε4, ε4/ε3, ε3/ε3, and the combined sample of ε3/ε2 and ε2/ε2 (full adjustment). The ε3/ε2 and ε2/ε2 genotypes were combined because they are each relatively infrequent low-risk genotypes for AD [17]. The ε4/ε2 genotype was omitted because of small sample size in analysis alone, and because it was unclear how to combine it with other genotype(s) to compensate for the small sample size. Analyses were performed within each stratum, with results also combined across strata in a Cochran-Mantel-Haenszel 1 df test. Results were combined across strata by weighting allele frequency differences and their variances proportional to sample sizes, as well as by assuming equal weights across strata. For APOE stratified analyses, for which there were relatives in different strata, this did not fully adjust for the effect of relationships across strata, resulting in modest genome-wide inflation, λ, of the observed test statistic relative to that expected, as quantified by the observed vs. expected median [74]. We therefore corrected the cross-strata tests by dividing by λ as a genome control correction factor [74].
Case-control analysis, unrelated sample
The CCun dataset was also used in the primary analyses to provide a comparison with the analysis of the full sample including the pedigree data, including different effects of adjusting for covariates. Analyses were carried out both without and with adjustment for APOE genotype. The primary analyses were based on logistic regression, using case status as the dependent variable with an additive model for the test SNP (0, 1 or 2 copies of the minor allele). Logistic regression, as opposed to an allele-based test, was chosen because it is insensitive to small deviations from HWE, and can include APOE genotypes as covariates, rather than requiring a less-ideal stratified analysis. The smaller CCun dataset was too small to carry out a stratified analysis, since the small within-stratum analysis made this statistically inappropriate. Therefore, to parallel the analysis of CCall as closely as possible given the different data structure, we included the number of APOE ε4 and ε2 alleles as covariates in an additive model (full-adjustment). This approach efficiently captures most available haplotype information with the cost of only two degrees of freedom [75], [76], and sufficiently captured the effects of APOE, as measured by residual association in the APOE region. For a small number of diagnostic comparisons of the tests used, we also carried out analyses with an allele-based test analogous to that used for analysis of CCall, but without the kinship correction, since there was little evidence of cryptic relatedness. Finally, further analyses were undertaken to probe interaction of SNP effects within the APOE ε4/ε4 genotype, in order to parallel results obtained in the CCall sample within ε4 homozygotes. For these analyses, we carried out focused analyses of specific SNPs, using logistic regression with APOE and test SNP main effects plus an interaction effect, since the analyses based on CCall suggested a statistical interaction. The main interest in these analyses was in the coefficient for interaction.
Adequacy and interpretation of analysis models
Adequacy of case-control tests was evaluated by computing λ [74], and by verifying the entire distribution of resulting p-values against expected quantiles. We used quantile difference plots, which better facilitate visual evaluation of the adequacy of the full distribution of results than do standard QQ plots, by plotting the difference in the negative logarithm of the observed and expected p-values against the negative logarithm of the expected p-values. Deviation from the null distribution over any part of the distribution is an indicator of possible violation of assumptions behind the test, and therefore interpretation of the results. A conservative Bonferroni correction was used initially to adjust for multiple testing, resulting in a threshold of p = 8.8×10−8 (−log(p) = 7.05) as a significance threshold that retains a genome-side significance level of 0.05, given the number of markers used for analysis. Plotting and evaluation of analysis results was carried out with R (http://www.r-project.org) and GnuPlot (http://www.gnuplot.info). In addition, for the analyses based on the CCun sample, we carried out permutation tests to define empirical confidence bounds under the null distribution. For these analyses we permuted the disease status, but kept the genome scan genotyping intact in order to maintain the LD structure in the genotype data. An equivalent estimate of the confidence bounds of the test results could not be obtained for the CCall sample because of the difficulty of carrying out the permutations under the constraints of relationships in the sample. However, since the CCall sample is larger than the CCun sample, with greater power, the confidence bounds for the CCun sample can be taken as a conservative estimate for the CCall sample.
Analysis of confounding in the NIA-LOAD/NCRAD sample
To evaluate possible sources and effects of confounding and different approaches to correcting for confounding, we carried out additional analyses in both the CCall and CCun samples. Evaluation of the efficacy of each approach to correcting for confounding was based on examination of the genome-wide p-value distribution, as a metric of adequacy of the correction. For comparison to the full-adjustment for APOE, we carried out a stratified analysis (CCall) and a covariate analysis (CCun) based only on presence/absence of APOE ε4 (ε4-adjustment), since this is a common approach for APOE adjustment [42], [43], [57]. Results were combined across strata as described above. In addition to genome-wide distributions of p-values, particular attention was paid to the effects of this intermediate adjustment on evidence for association with SNPs in the APOE region because of interest in additional potential risk loci in this region [56]–[58]. In one set of analyses of CCall, we carried out a stratified analysis based on the three identified European-American sub-populations, using kinship coefficients estimated separately within each group. In one analysis of CCun we included covariate adjustment for the loadings on the first four European-American-specific principal components [64] as an evaluation of correction for stratification in the sample.
Direct evaluation of the APOE ε4 allele as a source of confounding was obtained through two analyses, after eliminating from the analysis all SNPs within 300 KB of APOE. We examined ε4 allele frequency as a function of PCA loading. We also carried out a genome-wide case-only analysis to compare SNP allele frequencies in ε4 carriers vs. non-carriers among cases from the CCall sample. This eliminated association attributable to case status so that residual association, manifested as either genome-wide deviation of the test results from that expected under the null distribution or for specific SNPs of interest, could therefore be attributed to APOE-associated confounding or similar sources of population structure.
Bioinformatics Analysis
We performed bioinformatics analysis to identify genes that are located near the top SNP signals from our own GWAS, or genes that are biologically related to the genes at the SNP locations. For this purpose, we used SNAP and GRAIL (http://www.broadinstitute.org/mpg/grail/grail.php) to identify candidate genes. SNAP (http://www.broadinstitute.org/mpg/snap/ldsearch.php) identifies genes, extending in both directions until r2<0.5, while GRAIL searches for genes that are in the SNP region and that are biologically related to each other based on the published literature.
Replication Sample and Analysis
As an independent replication of the CUGBP2 association identified in the main analysis in the presence of APOE ε4 homozygotes, we examined a combined sample consisting of a Caribbean Hispanic cohort, and subjects from the combined Washington University case-control dataset and the Alzheimer's Disease Neuroimaging Initiative (WU-ADNI) [77]. These datasets were chosen because all had been genotyped on one of the Illumina platforms and shared multiple SNPs. The Caribbean Hispanic sample comprised 549 cases and 544 controls from two studies, including the Washington Heights-Inwood Columbia Aging Project (WHICAP) study [78] and the Caribbean Hispanic family study of familial AD [79]. The WU-ADNI data set comprised 788 EA cases and 643 EA controls. For the replication analysis, we used a conservative sample of 231 cases and 187 controls from the Caribbean Hispanic sample and 386 cases and 386 controls from the WU-ADNI sample, restricting subjects to homozygotes for each of the APOE ε4 and ε3 alleles, respectively, to avoid the heterogeneity caused by pooling different APOE genotypes that was identified in our primary analysis. While the Caribbean Hispanic sample is ethnically different that the European-American NIA-LOAD sample, this is advantageous since it reduces the probability of inflated evidence for association due simply to the shared ancestry of repeated samples from the same population [80]. SNP genotypes were from the Illumina HumanHap 650Y panel (Caribbean Hispanics) and several different Illumina platforms (WU-ADNI). We evaluated a total of all 24 SNPs in CUGBP2 that were genotyped in all of these samples as well as in the NIA LOAD sample. APOE genotyping was based on the same method as that for the NIA-LOAD cohort, or as described elsewhere [57].
We tested possible association of SNPs in CUGBP2 on LOAD risk in joint analyses across cohorts. For our primary analysis, we analyzed only rs201119 in the independent replication cohorts because this SNP gave the strongest genome-wide-significant evidence for association in the NIA-LOAD/NCRAD sample in the APOE ε4/ε4 stratum. For this analysis we did not apply a multiple testing correction because it was the single primary SNP tested for replication. In a second analysis we carried out analysis of all 24 SNPs that were available in both the replication and original cohorts, using a Bonferroni correction for multiple testing. In a final analysis, we combined the CCun component of the original NIA LOAD/NCRAD cohort with the replication sample, and carried out the same joint analysis using all of the cohorts. The analysis model used was stimulated by the APOE genotype-specific association identified in the main sample, which suggested an interaction between APOE ε4/ε4 and rs201119: we used logistic regression with an additive model for cohort, number of SNP alleles, APOE genotype (ε3/ε3 vs. ε4/ε4), and an interaction between the SNP and APOE, testing for both a SNP main effect and an interaction with APOE genotype. The component of the analysis of interest here was the interaction coefficient, given the original results that suggested such an interaction.
Results
Sample Used for Analysis
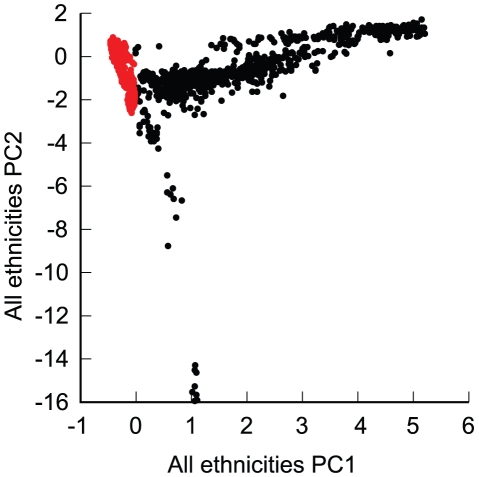
The final genotyped NIA-LOAD/NCRAD cohort consisted of 5,220 subjects. The complete sample was ethnically diverse, with 4,232 who were self-declared European-Americans, and the remainder 180 self-declared African American subjects, 309 Hispanic subjects, 300 subjects with other backgrounds, and 199 subjects with no self-reported race and ethnic information. Some individuals clustered with a group other than their self-reported group, leaving 3839 individuals (Table 1) that clustered as European-Americans based on a principal components analysis of all unrelated subjects (Figure 1) and were used in the CCall sample. Of the 3,839 European-American subjects, 993 cases and 884 controls were used in the CCun sample. As expected in a geographically distributed sample from North America, the fraction of subjects from any one self-reported ethnic group varied across collection site.
Table 1. European-American–clustering individuals used for analysis.
| Ascertainment | Study | Affected individualsa | Unaffected individuals | Total | ||||
| Subjects | Families | Mean age at onset (SD) | Subjects | Families | Mean age at exam (SD) | |||
| Multiplex fams | LOADb | 1138 | 475 | 73.6 (7.2) | 986 | 216 | 63.8 (10.8) | 2124 |
| NCRAD | 325 | 133 | 72.4 (6.2) | 0 | 0 | 325 | ||
| Unrelated | LOADb | 184 | 184 | 73.6 (7.7) | 1002 | 1002 | 75.6 (8.6) | 1186 |
| NCRAD | 201 | 201 | 72.4 (6.1) | 3 | 3 | 75.7 (2.5) | 204 | |
| Totals | 1848 | 993 | 1991 | 1221 | 3839 | |||
Definite, probable, and possible AD diagnoses were 28%, 64%, and 8% of the total sample, respectively, and 34.6%, 63.9%, and 1.5% among affected individuals used in the CCun sample.
Includes 135 control subjects and 6 cases from University of Kentucky.
Figure 1. Principal components analysis of the complete sample, based on all ethnicities.
Red: European-American subjects. PC1 and PC2: first and second principal component.
Population Structure
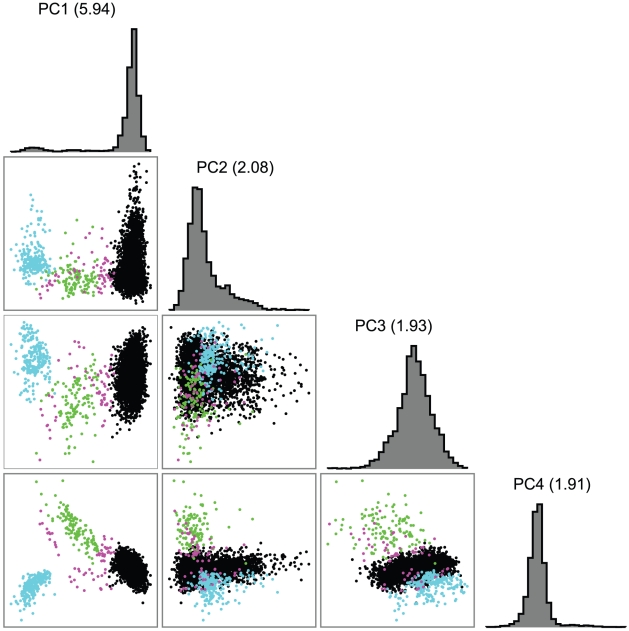
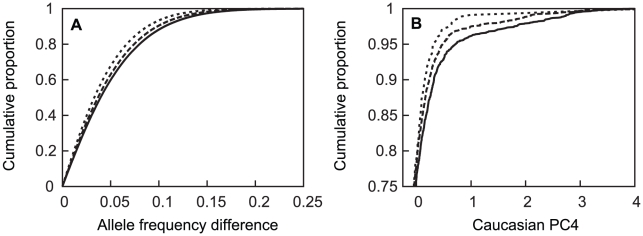
The European-American-specific principal components (PCs) revealed substructure within the sample. Although apparent with the first two principal components (PC1 and PC2), three subgroups were most clearly defined by the first and fourth principal components (Figure 2). Estimated fractions of each subpopulation varied across sites (Table 2), with the NW group the largest (90.2%) sample (Table 2). A few subjects fell between the main clusters, and were excluded in subsequent subgroup analyses (Figure 2). Subgroup assignments were strongly supported by likelihood computations based on European subgroup-specific AIMs, and by comparison of allele frequencies in the three groups with those of the AIMs. Large between-group allele frequency differences between the NW and other groups near lactase on chromosome 2 and HLA on chromosome 6 [81] further supported these subgroup assignments: e.g., allele frequency differences >0.55 for SNPs near lactase, as do overall comparison of allele frequency differences between pairs of populations. Although the median allele frequency difference was relatively low (<0.04) for all three pairs of populations (Figure 3A), 7%, 9% and 12% of the markers had a substantial allele frequency difference of >0.1 in the NW-SE, AJ-SE, and NW-AJ comparison, respectively. These larger allele frequency differences coupled with varying fractions of cases from the different contributing sites (Table 2) predispose to confounding.
Figure 2. First four principal components (PCs) in the European-American sample alone.
Colors represent inferred ancestry. Black: northwest (NW) Europe; green: southeast (SE) Europe; cyan: Ashkenazi Jewish (AJ); magenta: indeterminate (omitted from subpopulation analyses).
Table 2. Estimated subpopulation membership for the 17 largest individual contributing sites.
| Subpopulation % | |||||
| Sitea | Nb | NW | SE | AJ | case % |
| A | 662 | 93.8 | 0.8 | 5.4 | 99.4 |
| B | 504 | 96.8 | 1.8 | 1.4 | 42.3 |
| C | 478 | 96.7 | 1.7 | 1.7 | 10.9 |
| D | 410 | 76.6 | 2.4 | 21.0 | 40.2 |
| E | 398 | 93.2 | 1.5 | 5.3 | 33.8 |
| F | 361 | 97.5 | 1.1 | 1.4 | 38.2 |
| G | 351 | 97.2 | 1.4 | 1.4 | 49.8 |
| H | 222 | 94.6 | 1.8 | 3.6 | 70.0 |
| I | 147 | 98.0 | 0.7 | 1.4 | 35.5 |
| J | 144 | 99.3 | 0.7 | 0.0 | 52.7 |
| K | 139 | 95.0 | 3.6 | 1.4 | 45.1 |
| L | 92 | 64.1 | 17.4 | 18.5 | 32.5 |
| M | 87 | 75.9 | 12.6 | 11.5 | 39.1 |
| N | 67 | 76.1 | 19.4 | 4.5 | 30.8 |
| O | 65 | 89.2 | 1.5 | 9.2 | 62.7 |
| P | 65 | 70.8 | 9.2 | 0.0 | 43.5 |
| Q | 65 | 83.1 | 16.9 | 0.0 | 49.2 |
Sites that each contributed >50 European-American subjects.
Number of subjects.
Figure 3. Cumulative distribution of absolute value of allele frequency differences between subpopulations and APOE genotypes.
Panel A: subjects from NW and SE (dotted line), AJ and SE (dashed line), and NW and AJ (solid line) groups. Panel B: cumulative distribution of European-American PC4 values as a function of APOE genotype for ε4 homozygotes (dotted line), ε4 heterozygotes (dashed line); genotypes with no ε4 (solid line). In panel A, the horizontal axis is truncated at 0.25 despite a few rare allele frequency differences that extend to 0.59; in panel B the vertical axis is only presented for the upper quartile of the distributions, where the curves are differentiated.
APOE allele frequencies also differed among the three sub-groups, along with a higher fraction of cases relative to the subgroup sample size drawn from the AJ and SE sub-groups than the NW sub-group (Table 3). The allele frequencies in the unrelated controls varied in a manner that is consistent with a known north-south ε4 allele frequency gradient, with higher ε4 allele frequencies in northern than southern European populations [82]–[84], and with lower ε4 frequencies reported in Jewish populations [85], [86]. In these unrelated controls, the ε4 allele frequency was higher in subjects of NW ancestry (0.139) than in subjects of SW (0.109) or AJ (0.092) ancestry, with the same allele frequency patterns also apparent in the unrelated (control) family members, and in the affected individuals (cases). The cumulative distribution of European-American PC4 values in the whole European-American sample differed among APOE genotypes in a manner that was also consistent with existence of sub-structure (Figure 3B), with similar results observed in the NW group alone (not shown).
Table 3. APOE allele frequencies in European-American subjects.
| Unrelated controls | Related controls | Cases | ||||||||
| Subjects | Counta | Allele | Count | Allele | Count | Allele | ||||
| Sample | N | 2N | ε2 | ε4 | 2N | ε2 | ε4 | 2N | ε2 | ε4 |
| CCallb | 3839 | 1402 | 0.097 | 0.138 | 2566 | 0.054 | 0.255 | 3678 | 0.023 | 0.465 |
| NW | 3446 | 1244 | 0.100 | 0.139 | 2336 | 0.054 | 0.264 | 3312 | 0.024 | 0.472 |
| SE | 110 | 46 | 0.043 | 0.109 | 64 | 0.047 | 0.141 | 110 | 0.018 | 0.291 |
| AJ | 214 | 76 | 0.079 | 0.092 | 124 | 0.048 | 0.185 | 228 | 0.017 | 0.448 |
Number of alleles.
A few subjects were missing APOE genotype data.
Case-Control Analyses
Genome scans, no APOE adjustment
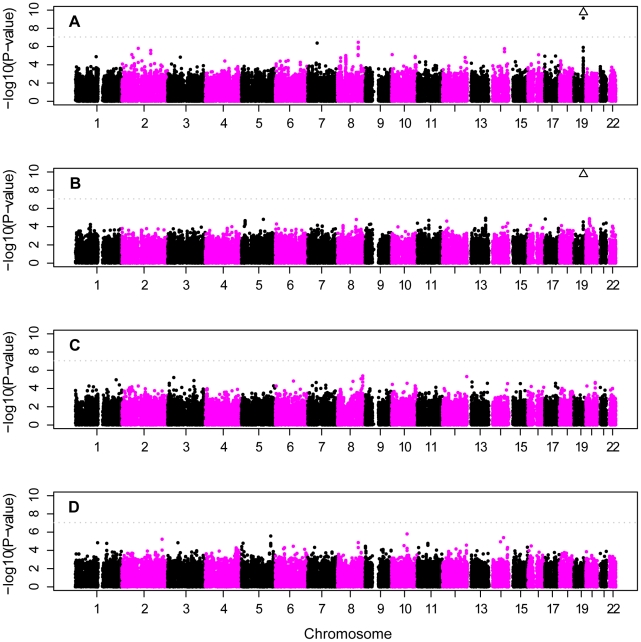
As expected, SNPs near APOE provided the strongest genome-wide evidence for association in the unadjusted analyses (Figure 4A, Table 4). In the primary analyses, SNP rs2075650 in TOMM40, which is in strong linkage disequilibrium with rs429358 in our sample (D′ = 0.70; r2 = 0.45; using the 884 unrelated controls), and which tags the APOE ε4 allele, gave highly significant results in analysis of both the CCall and CCun sample (p = 3.2×10−81 and p = 6.3×10−77, respectively). The secondary analyses gave similar results with rs2075650 in the analysis of the ethnic-stratified analysis for the unweighted combined results, the NW sample, and the AJ sample (p = 1.2×10−15, p = 3.2×10−73, and p = 3.7×10−8, respectively). In each of these analyses, six additional SNPs near APOE also provided very strong support for this association (e.g., in the CCall sample, p-values ranged from p = 4.9×10−10 to p = 2.9×10−24). Only in the small SE sample was the evidence for association with rs2075650 merely suggestive (p = 0.03), consistent with the reduced inflation, in this sample, of the ε4 frequency in cases relative to that observed in the other subgroups (Table 3). For this SE sample, rs7007878 on chromosome 8 at ∼29 MB provided the strongest evidence of association (p = 6.5×10−6).
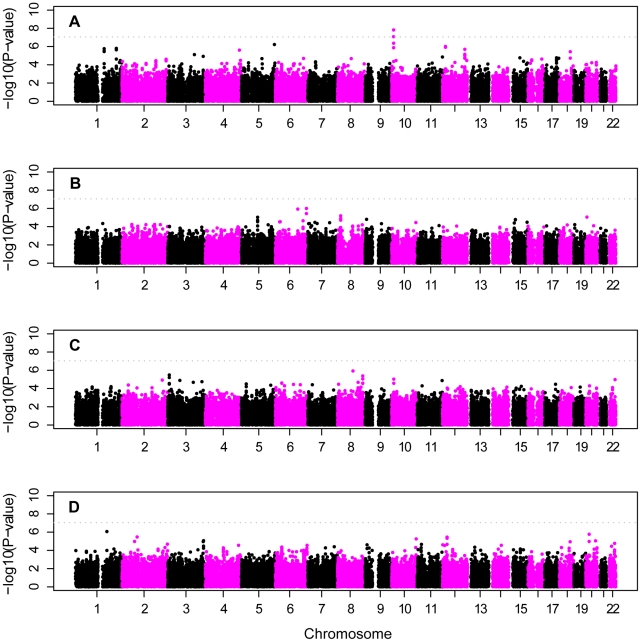
Figure 4. Genome scan of European-American subjects.
Panel A: CCall sample analyzed as a single population; panel B: stratified analysis of CCall sample that accounts for three subpopulations (NW, SE, AJ); panel C: stratified analysis of CCall sample across four APOE genotypes; panel D: CCun sample, with covariate adjustment for the number of ε2 and ε4 alleles. Plots have been truncated at −log10p = 10 on the vertical axis to more easily visualize results for most of the genome. Multiple SNPs near APOE on chromosome 19 yielded −log10p≫10 in the analyses that did not control for APOE (Panels A and B, see text for details), and are represented by a single triangle at the top of each such panel. Horizontal line shows genome-wide significance level.
Table 4. SNPs with strongest evidence for association under each analysis condition.
| SNP | Group | p-value | Chra | Position (bp) | Alleleb | Freq-control | Freq-case | ORc | Genes |
| rs2075650 | CCall | 2.95×10−81 | 19 | 50087459 | A | 0.820 | 0.615 | 0.35 | APOE, APOC1, APOC4 TOMM40, PVRL2 |
| rs2075650 | Ethnic stratified | 1.18×10−15 | 19 | 50087459 | A | APOE, APOC1, APOC4 TOMM40, PVRL2 | |||
| rs2075650 | AJ | 3.74×10−8 | 19 | 50087459 | A | 0.875 | 0.639 | 0.25 | APOE, APOC1, APOC4 TOMM40, PVRL2 |
| rs2075650 | NW | 3.21×10−73 | 19 | 50087459 | A | 0.814 | 0.608 | 0.35 | APOE, APOC1, APOC4 TOMM40, PVRL2 |
| rs7007878 | SE | 6.53×10−6 | 8 | 29098372 | A | 0.446 | 0.755 | 3.83 | KIF13B |
| rs10489216 | APOE ε3ε2+ε2ε2 | 8.44×10−7 | 1 | 166869486 | G | 0.780 | 0.5 | 0.28 | DPT, XCL1 |
| rs7814569 | APOE ε3ε3 | 1.18×10−6 | 8 | 81252421 | A | 0.930 | 0.871 | 0.51 | ZBTB10, TPD52 |
| rs4145454 | APOE ε4ε3 | 1.03×10−6 | 6 | 164492026 | G | 0.706 | 0.786 | 1.53 | |
| rs201119 | APOE ε4ε4 | 1.52×10−8 | 10 | 11089983 | A | 0.793 | 0.939 | 4.02 | CUGBP2, PITRM1 |
| rs2673604 | APOE stratified | 2.60×10−6 | 8 | 133480789 | A | KCNQ3 |
Chromosome.
Allele depicted is the allele on the Illumina TOP strand.
Odds ratio with respect to allele noted.
Other than SNPs in the APOE region, the region with the strongest evidence for association in the primary analysis spanned 109.2–109.8 MB on chromosome 8, in which several SNPs (e.g., rs1975804, rs1679666, rs1789964) came close to achieving genome-wide significance in either the CCun or CCall sample (Tables S1, S2). Both samples gave similar results: p-values 1.6×10−6 to 3.3×10−7 in the CCall sample, and 9×10−6 to 5.7×10−7 in the CCun sample. A few regions of the genome yielded marginally stronger evidence than the unadjusted analysis for association in the analysis that stratified on ethnic subgroup (Figure 4B), but no regions other than the APOE region reached genome-wide significance. A portion of this sample has been used previously to investigate 29 SNPs as part of focused followup analyses [38], [40]. However, only two of these previously-investigated SNPs overlap with our current study, and neither of these SNPs gave significant results in either the earlier [38] or current analyses.
Genome scans, APOE full adjustment
In contrast to unadjusted analysis, the analyses based on a full-adjustment for APOE genotype identified no SNPs with genome-wide significance in either the full CCun or CCall samples (Figure 4C, 4D). Complete adjustment for APOE genotype accounted for most association in the APOE region. Evidence for association with rs2075650, which had the strongest evidence for association in the unadjusted analysis, fell precipitously after adjustment: only modest evidence for association remained for the CCun sample (p = 6.6×10−4, Table S3) and evidence for association was eliminated in the CCall sample (p = 0.15).
Analysis of individual APOE genotype strata led to identification of one novel region with genome-wide-significant evidence of association on chromosome 10p14 (Figure 5A, Table 4). SNP rs201119 provided strong genome-wide-significant evidence of association within the APOE ε4/ε4 stratum (p = 1.5×10−8). Surrounding SNPs also gave strong results within this stratum (Figure 5A, Table S4), including rs201099, which also reached genome-wide significance (p = 8.3×10−8). Even stronger genome-wide-significant results were obtained when analysis was confined to the APOE ε4/ε4 individuals in the NW subgroup (p = 6.6×10−9 and 2.2×10−8 for rs201119 and rs201099, respectively), eliminating the possibility that this association was explained by the existence of the AJ and SE subjects in the sample. SNPs on chromosomes 8 and 6 gave suggestive evidence for association in the APOE ε3/ε4 (Figure 5B, p = 1.0×10−6, Table S5) and APOE ε3/ε3 (Figure 5C, p = 1.2×10−6, Table S6) strata. Finally, a region on chromosome 1 was identified with strong evidence for association in the APOE ε3/ε2+ε2/ε2 stratum (Figure 5D, p = 8.4×10−7).
Figure 5. Stratified analysis of APOE-defined subgroups of all European-American subjects.
Panels A: ε4/ε4 genotype, B: ε3/ε4 genotype; C: ε3/ε3 genotype; and D: ε2/ε2+ε2/ε3 combined genotype. Horizontal line shows genome-wide significance level.
Bioinformatics results
Table 4 lists genes located on or near the SNPs with the strongest p-value for each analysis. Several genes emerged (Table 4), in addition to APOE and related genes (e.g., APOC1). The most promising candidate is CUGBP2 (CUG triplet repeat, RNA binding protein 2; 11,087,265–11,509,495 bp) on chromosome 10p14, which was associated with the top SNP, rs201119, identified from the APOE ε4/ε4 restricted analysis. This SNP is in the middle of this gene, as is SNP rs201099, also with genome-wide significant evidence for association.
Replication analysis of CUGBP2
The primary analysis of CUGBP2 in the replication cohorts supported rs201119 as associated with LOAD in the presence of APOE ε4/ε4 (Table 5). Targeted analysis gave a significant interaction effect with APOE (p = 0.048, OR = 1.43, 95% CI 1.0–2.03 for the same allele as showed a higher frequency in cases than controls in the CCall ε4/ε4 sample). The NIA-LOAD/NCRAD CCun sample similarly gave significant evidence for an interaction with the same model (p = 0.00016), consistent with the results from the stratified analysis of the larger CCall sample. Among the 24 SNPs evaluated in the joint analyses of the replication cohorts, 6 SNPs provided nominal p-values below 0.05 for interaction with APOE, with p = 5.6×10−4 for rs62209 (OR = 1.75, 95% CI 1.27–2.41), which is significant at the 1% level after Bonferroni correction. SNP rs201099, which also provided genome-wide significant evidence for association in the NIA-LOAD scan, provided nominal evidence of an interaction in the replication sample (p = 9.1×10−3). In the joint analysis of the NIA LOAD/NCRAD, Caribbean Hispanic, Washington University, and ADNI samples, 12 SNPs were significant for interaction with APOE at a nominal 5% level, and 6 SNPs remained significant after Bonferonni correction (p = 2.08×10−3, Table 5). The cohorts used for the replication analysis had similar patterns of linkage disequilibrium between pairs of SNPs in CUGBP2 (Figure S2), further supporting the evidence for replication. These results in joint analysis of the replication samples provide further support for a statistical interaction of SNPs in CUGBP2 with APOE ε4/ε4 in conferring AD risk.
Table 5. Significance of SNPs in CUGBP2 in prediction of disease risk.
| CCall APOE ε4/ε4 stratum | Caribbean Hispanic+WU+ADNI | CCun+Hispanic+WU+ADNI | |||||||||
| SNP | Allelea | Nb | Pc | Nb | ORd | 95% CI | Pe | Nb | ORd | 95% CI | Pe |
| rs11256915 | A | 408 | 0.286 | 1435 | 1.25 | 0.95–1.68 | 0.125 | 2382 | 1.43 | 1.13–1.83 | 3.3×10−3 |
| rs2146551 | A | 407 | 0.055 | 1447 | 1.16 | 0.86–1.56 | 0.331 | 2393 | 1.35 | 1.05–1.73 | 0.018 |
| rs731229 | G | 408 | 0.140 | 1444 | 1.16 | 0.83–1.62 | 0.378 | 2390 | 1.22 | 0.93–1.60 | 0.142 |
| rs992193 | A | 408 | 0.313 | 1446 | 1.12 | 0.84–1.52 | 0.439 | 2391 | 1.20 | 0.95–1.53 | 0.118 |
| rs2146553 | G | 408 | 0.033 | 1445 | 0.83 | 0.60–1.15 | 0.259 | 2392 | 0.81 | 0.63–1.05 | 0.106 |
| rs10795839 | G | 408 | 0.488 | 1435 | 0.86 | 0.3–1.18 | 0.361 | 2382 | 0.86 | 0.66–1.14 | 0.305 |
| rs1928724 | A | 407 | 0.698 | 1447 | 0.93 | 0.69–1.26 | 0.646 | 2394 | 0.94 | 0.72–1.23 | 0.655 |
| rs1928722 | A | 408 | 0.789 | 1434 | 0.89 | 0.65–1.23 | 0.493 | 2381 | 0.88 | 0.66–1.16 | 0.371 |
| rs201070 | A | 408 | 0.201 | 1446 | 0.96 | 0.71–1.29 | 0.793 | 2392 | 1.11 | 0.87–1.41 | 0.406 |
| rs201071 | A | 407 | 0.018 | 1447 | 1.19 | 0.79–1.81 | 0.403 | 2394 | 1.49 | 1.06–2.11 | 0.022 |
| rs201074 | A | 408 | 4.5×10−3 | 1433 | 0.69 | 0.48–1.00 | 0.052 | 2380 | 0.68 | 0.52–0.91 | 7.9×10−3 |
| rs62209 | C | 402 | 3.8×10−3 | 1439 | 1.75 | 1.27–2.41 | 5.6×10−4 | 2371 | 2.04 | 1.56–2.67 | 1.6×10−7 |
| rs201082 | G | 408 | 0.096 | 1435 | 1.61 | 1.14–2.29 | 7.5×10−3 | 2382 | 1.79 | 1.35–2.38 | 5.1×10−5 |
| rs201097 | A | 408 | 0.509 | 1447 | 0.83 | 0.43–1.58 | 0.561 | 2394 | 0.75 | 0.46–1.22 | 0.248 |
| rs201099 | A | 408 | 8.3×10−8 | 1447 | 1.56 | 1.12–2.18 | 9.1×10−3 | 2393 | 1.92 | 1.47–2.56 | 3.4×10−6 |
| rs201100 | G | 408 | 0.019 | 1433 | 0.65 | 0.45–0.95 | 0.024 | 2380 | 0.68 | 0.52–0.89 | 4.9×10−3 |
| rs201119 | A | 408 | 1.5×10−8 | 1447 | 1.43 | 1.00–2.03 | 0.048 | 2394 | 2.08 | 1.53–2.79 | 2.1×10−6 |
| rs201124 | A | 408 | 1.3×10−6 | 1423 | 1.43 | 1.00–2.06 | 0.048 | 2370 | 1.82 | 1.34–2.46 | 1.2×10−4 |
| rs7099713 | A | 408 | 2.3×10−4 | 1435 | 1.16 | 0.65–2.06 | 0.618 | 2381 | 1.45 | 0.94–2.22 | 0.096 |
| rs1547221 | G | 408 | 4.4×10−5 | 1447 | 1.43 | 0.88–2.31 | 0.153 | 2394 | 1.82 | 1.24–2.64 | 1.9×10−3 |
| rs913918 | A | 408 | 0.565 | 1434 | 1.47 | 1.04–2.09 | 0.029 | 2380 | 1.47 | 1.11–1.98 | 7.8×10−3 |
| rs11256951 | G | 408 | 0.154 | 1435 | 0.95 | 0.70–1.28 | 0.740 | 2382 | 1.06 | 0.84–1.36 | 0.604 |
| rs1999207 | A | 408 | 0.914 | 1447 | 0.92 | 0.67–1.24 | 0.568 | 2394 | 0.88 | 0.69–1.15 | 0.361 |
| rs932918 | A | 407 | 0.908 | 1447 | 1.22 | 0.85–1.74 | 0.279 | 2392 | 1.12 | 0.84–1.50 | 0.420 |
Allele depicted is the allele with higher frequency in the APOE ε4/ε4 cases than ε4/ε4 controls, as denoted on the Illumina TOP strand.
Sample size.
P-value for relationship-corrected chi-square test of allele frequency differences.
Interaction odds ratio with respect to allele noted based on logistic regression model.
P-value for interaction coefficient of logistic model.
Replication of prior GWAS results
SNPs in four genes that were recently implicated [42]–[44] each provided initial nominal evidence of association in our dataset in analysis of the unadjusted data (Table 6), with the same allele associated with disease risk in our sample as was previously reported. An additional SNP, rs597668, near EXOC3L2 [44], was also considered, but because of its proximity to APOE, was examined in less detail. Two of the three SNPs highlighted initially with strongest evidence for association - rs3818361 in CR1 and rs11136000 in CLU - each reached nominally significant (p<0.05) evidence of association in the unstratified or unadjusted analyses of the CCun and CCall samples. SNP rs3851179 in PICALM achieved evidence for association at this nominal level only in analysis of CCall. The gene encoding bridging integrator 1 (BIN1) was also noted initially at a lower threshold of significance [42] with stronger results in a recent replication analysis [44], and gave nominally significant results in the unstratified and unadjusted analyses of our sample for rs7561528 in both CCun and CCall, and for rs744373 in CCun (Table 6). In addition, five of the six additional SNPs that were in these four genes in our genotyping panel achieved nominal significance in the unstratified analysis of the CCall sample, and two SNPs achieved nominally significant results in unadjusted analysis of the CCun sample (Table 6).
Table 6. P-values for candidate SNPs based on genes previously reported with genome-wide significant results.
| Covariate | Analysis | Sample size | Gene | |||||||||||
| Group | case | cont | CR1 SNPs | CLU SNPs | PICALM SNPs | BIN1 SNPs | ||||||||
| rs3818361a | rs6701710 | rs1408077 | rs11136000a | rs7012010 | rs541458 | rs543293 | rs7941541 | rs3851179a | rs744373a | rs7561528a | ||||
| None | CCall | 1848 | 1991 | 0.0273 | 0.0263 | 0.0478 | 0.0083 | 0.2786 | 0.01 | 0.0189 | 0.0479 | 0.039 | 0.0671 | 0.0299 |
| CCun | 993 | 884 | 0.0278 | 0.0278 | 0.0288 | 0.0019 | 0.0614 | 0.0628 | 0.1706 | 0.1556 | 0.1183 | 0.0007 | 0.0003 | |
| APOE ε4 | ε4− | 464 | 1208 | 0.5006 | 0.5006 | 0.4204 | 0.0956 | 0.9852 | 0.8416 | 0.5992 | 0.6839 | 0.7988 | 0.1872 | 0.0438 |
| ε4+ | 1384 | 783 | 0.0619 | 0.0581 | 0.1125 | 0.032 | 0.1174 | 0.0752 | 0.014 | 0.0338 | 0.1177 | 0.0894 | 0.1704 | |
| stratb | 1848 | 1991 | 0.1496 | 0.1461 | 0.1540 | 0.0159 | 0.4873 | 0.5398 | 0.5174 | 0.5497 | 0.6316 | 0.0565 | 0.0182 | |
| CCun | 993 | 884 | 0.0379 | 0.0379 | 0.0317 | 0.0108 | 0.1205 | 0.7050 | 0.7098 | 0.6120 | 0.6292 | 0.0007 | 0.0008 | |
| APOE geno | 3/2+2/2 | 43 | 207 | 0.4441 | 0.4441 | 0.4742 | 0.2433 | 0.0469 | 0.2723 | 0.967 | 0.7203 | 0.6769 | 0.5146 | 0.1459 |
| 3/3 | 421 | 1001 | 0.6813 | 0.6813 | 0.5579 | 0.1705 | 0.4181 | 0.5731 | 0.6405 | 0.7951 | 0.6534 | 0.2720 | 0.0959 | |
| 4/3 | 997 | 645 | 0.0509 | 0.0475 | 0.0739 | 0.088 | 0.3719 | 0.1903 | 0.045 | 0.0596 | 0.1987 | 0.0237 | 0.1179 | |
| 4/4 | 338 | 70 | 0.7887 | 0.7887 | 0.8674 | 0.9933 | 0.9897 | 0.8201 | 0.3667 | 0.8046 | 0.9759 | 0.9727 | 0.6974 | |
| stratb | 1799 | 1923 | 0.1262 | 0.1233 | 0.1199 | 0.0227 | 0.4962 | 0.5151 | 0.5430 | 0.6484 | 0.6949 | 0.0383 | 0.0090 | |
| CCun | 993 | 884 | 0.0237 | 0.0237 | 0.0184 | 0.0029 | 0.0785 | 0.7780 | 0.7007 | 0.5948 | 0.6552 | 0.0007 | 0.0010 | |
| Ethnicity | NW | 1664 | 1797 | 0.0832 | 0.0805 | 0.1293 | 0.0096 | 0.378 | 0.0053 | 0.0086 | 0.0243 | 0.0152 | 0.1282 | 0.0801 |
| stratb | 1834 | 1952 | 0.0071 | 0.0071 | 0.0168 | 0.4876 | 0.6502 | 0.4065 | 0.3494 | 0.4482 | 0.8509 | 0.2269 | 0.0377 | |
Bold: nominal significance p<0.05.
Cells show p-values for CCall sample, except where noted as CCun sample.
combined results across individual strata: presence vs. absence of APOE ε4; APOE genotype; or ethnic group (NW, SE, AJ).
Confounding
The distribution of p-values obtained from the unadjusted genome scans deviated from a uniform distribution, suggesting the presence of uncorrected confounding. This effect was mild near the median test value (λ = 0.97 for CCun, λ = 1.02 for CCall) but more apparent in the tails of the distribution, providing evidence for potential confounding in analysis of both samples (Figure 6A, 6B; magenta points; Figure S2). Some of the deviation from the null distribution is likely to be attributable to the greater sensitivity to HWD for the allele-based tests than for logistic regression in CCun (Figures S2, S3, CCun results). However, deviation from the null distribution in the direction of an increased type I error over the nominal level was especially marked in the upper 0.1% of the tail of the distribution for the unadjusted analysis of the CCun sample even under analysis with logistic regression (Figure S4), and in the upper 1% for the CCall sample (Figure 6A, 6B, magenta points). The excess fraction of small p-values was not explained by SNPs in the APOE region (Figure 6C), some of which had, as expected, much more extreme p-values. This deviation from the null distribution was not explained by inadequate correction for relationships in the CCall sample since the same excess pattern of extreme p-values occurred in the analysis of both the CCun and CCall samples, and over a wider range of p-values when the CCun sample was analyzed with a chi-square test instead of with logistic regression (Figure S2). Control for test statistic inflation was also not achieved by incorporation of the first four principal components as covariates [64] (Figure 6A, grey points; Figure S3), or by restricting analysis to the more uniform NW group (Figure 6B, grey points).
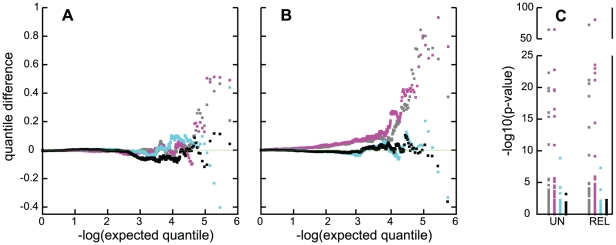
Figure 6. Quality control evaluation of association tests in the CCun and CCall samples.
Panels A, B: Quantile difference plots for association tests excluding SNPs in the APOE region; and panel C: −log10(p) for the same analyses for the 95 SNPs in the APOE region. For panels A and B, results are shown, for N tests, as the difference of the ith of N ordered observed (−log10(pi)) and expected (−log10(i/N)) quantiles plotted against the expected quantiles. A: results for the CCun sample, with grey: PCA adjusted; magenta: unadjusted analysis; cyan: ε4 adjustment; black: full adjustment. B: results for the sample containing related individuals; grey: unadjusted analysis of NW subgroup; magenta: unadjusted analysis of CCall; cyan: ε4-stratified analysis of CCall; black: full adjustment. C: UN depicts results for analysis of CCun; REL depicts results for analysis of the larger sample, in both cases for the same four conditions and colors as in panels A and B.
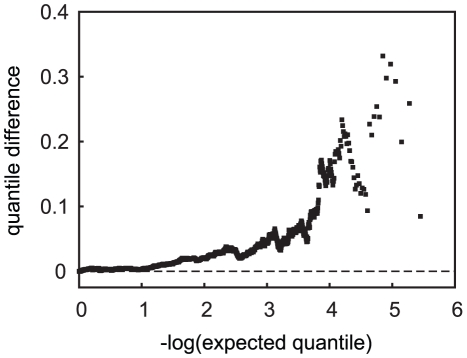
Two sources of evidence suggested that an important source of potential confounding was APOE genotype. The first was the effect of adjustment for APOE genotype, which had a notable effect on the distribution of resulting genome-wide p-values. Simple adjustment of APOE through binary ε4-status yielded a distribution of p-values that was closer to a uniform distribution than was obtained from unadjusted analysis. However, deviation from the expected null distribution was still evident (Figure 6A, 6B, cyan points), and there was still evidence for association with SNP rs2075650 near APOE (Figure 6C) in both the unrelated and related samples (p = 1.5×10−9 for CCun, and p = 1.2×10−7 for CCall). The full APOE adjustment achieved the best control of the null distribution of p-values (Figure 6A, 6B, black points), and produced close to the expected uniform distribution of p-values under the null distribution (Figure S3). Addition of the PCs as covariates alone did not produce the desired distribution of p-values (Figure S4, Table S7) and in addition to the full APOE adjustment in the CCun sample did not provide further improvement to the distribution of p-values over the APOE adjustment (Table S8 versus Table S3). This analysis also eliminated all statistically significant association with SNPs in the APOE region (Figure 6C), and evidence for adequate genomic control within each APOE stratum was reasonable (λ = 0.997, 1.02, 1.009, 1.003 for the ε4/ε4, ε4/ε3, ε3/ε3 and ε3/ε2+ε2/ε2 strata, respectively). A second source of evidence for confounding or population stratification was obtained from the results from the case-only analysis: the genome-wide distribution of p-values from the allele frequency comparison in ε4 carriers vs. non-carriers in the case-only sample also showed an overall deviation from the expected null distribution in the direction of an excess of small p-values (Figure 7). This indicates that there are many markers that are correlation with APOE in the highly-ascertained case sample.
Figure 7. Quantile difference plot of tests of allele frequency differences in APOE ε4-carrier versus non-carrier cases.
SNPs in the APOE region are not included.
Effect of confounding on replication SNPs
Association between LOAD and a subset of the replication SNPs showed evidence of APOE-induced confounding. Evidence for association with SNPs in PICALM was highly sensitive to adjustment for APOE genotype (Table 6), suggesting the possibility of confounding. Results from the case-only analysis supported this interpretation: differences in allele frequencies among cases who were ε4 carriers vs. non-carriers were nominally significant (p<0.05) or close to significant for all of the four SNPs evaluated in PICALM: p = 0.0047, 0.0184, 0.0524, and 0.0222 for rs541458, rs543293, rs7941541, and rs3851179, respectively. Furthermore, for all four SNPs, the pattern of allele frequencies in cases and controls, and among the individual APOE genotypes in the cases, was consistent with such confounding: the allele that was associated with higher risk of case-status in the original case-control analysis always had the highest allele frequency among ε4/ε4 cases and the lowest allele frequency among ε3/ε3 cases, with the allele frequency intermediate in the ε3/ε4 cases. For example, for rs3851179, the major allele, C, had a frequency of 0.655 in cases and 0.634 in controls, and had allele frequencies of 0.618, 0.665, and 0.67 in ε3/ε3, ε3/ε4, and ε4/ε4 cases, respectively. Similar results were obtained for rs597668 near EXOC3L2, with evidence for association in the absence of APOE adjustment (p = 0.0007 in CCall) weakening considerably with full adjustment for APOE (p = 0.67), and with very strong evidence for allele frequency differences between ε4 carriers and non-carriers in the case-only analysis (p = 6.79×10−8). In contrast, there were no significant allele frequency differences identified in case-only analyses for the SNPs in Table 6 for CR1, CLU and BIN1 (p = 0.31–0.74).
The evidence for association with SNPs in some of these four genes was also dependent on ethnic stratification, suggesting a further or alternative source of confounding. For all tested SNPs in CR1, evidence for association in the CCall sample, while modest in each sub-population, was consistent across subpopulations and therefore strengthened in analysis that stratified on ethnic subgroup (Table 6). Evidence for association with rs7561528 in BIN1 also remained present in the ethnically-stratified analysis. In contrast, evidence for association with CLU and PICALM was only present in the unstratified analysis of the full sample, and in the NW group, with no support from the AJ and SE samples or from the ethnically-stratified analysis. Adjustment for ethnic subgroup did not fully correct for residual sources of correlation, as judged by the quantile difference plot for the NW sample (Figure 6B), suggesting that residual sources of correlation exist, even after correcting for relationship information. Similarly, evaluation of the quantile difference plot for the analysis of the CCun sample, using principal component loadings to correct for possible ethnic variability, also failed to produce the desired genome-wide quantile difference plot (Figure 6A).
Discussion
Analysis of the NIA-LOAD/NCRAD sample indicates that unraveling susceptibility to LOAD is complex even when individuals from genetically-loaded multiplex families are included. As with other studies, support for the association between LOAD and SNPs near APOE was strong. By taking advantage of this association, we were able to identify a potential novel locus, CUGBP2, on chromosome 10p14 with genome-wide significant evidence of association within the highest-risk APOE ε4/ε4 stratum, with replication in an independent sample. We also found support for association with recently-reported SNPs in CLU and BIN1, and to a lesser extent with CR1. However, we found that the strong APOE association also introduced a source of structure into the sample that had effects that were detectable through standard evaluation of analysis results. Our results provide strong evidence that this correlation with APOE explains the association in this sample with some, but not all, previously-noted SNPs, including PICALM and the recently-proposed association near EXOC3L2, both of which have significantly different allele frequencies in AD cases who are carriers vs. non-carriers of the APOE ε4 allele.
Detection of true risk loci in a GWAS of LOAD requires careful attention to potential sampling biases [87]. Large samples such as ours are necessary for detecting modest associations, but such samples usually involve multiple collection sites, introducing the potential for confounding or other complications. Consistent with this, across our participating sites we found variability in the numbers of cases and controls, the fraction of underlying identifiable ethnic subgroups, differences among subgroups in terms of APOE genotype frequencies, and differences in APOE genotype distributions as a function of an indicator of genetic differentiation. None of this is surprising, given the history of US colonization and immigration coupled with differentiation among European populations [81], [88]. Other large samples in Europe and other locations are likely to have similar issues, as suggested by genome-wide inflation factors reported by recent studies [42], [43] that were higher than those in our study. Appropriate accommodation for confounding or structure when it is present can provide both protection against false positive associations, as well as increased power to detect associations that are confined to a subset of the sample, as we have demonstrated as part of our investigations surrounding the influence of APOE on our results. We also found that common methods failed to provide the necessary correction for APOE-induced associations, including use of principal components adjustment [64] and genomic control [74]. Together these observations have important implications for interpretation of results from other large combined samples.
Accommodation for APOE genotype was key for obtaining appropriate genomic control in our sample. Incorporation of individual APOE genotypes, as opposed to the more typical use of presence or absence of ε4, resulted in the closest approximation to a uniform distribution of p-values over a wide range of the test results. This likely resulted in a reduction in false positive association results since such control must be achieved before accepting evidence of association. Not only were our genome-wide results impacted by adjustment for APOE genotypes, but the support for some SNP associations from previous studies was similarly affected. For the SNPs that were most sensitive to APOE-adjustment, the allele frequencies differed among cases as a function of APOE genotype, suggesting a relatively simple diagnostic for which SNPs require adjustment for APOE as part of the analysis: for such SNPs, a full adjustment for APOE genotype may be critical for genomic control in part because of allele frequency differences among populations [82], [89]. These differences could lead to structure in the ascertained sample through variability in disease risk or survival in underlying subpopulations, as seen across the subpopulations identified in this sample. It thus may represent a corollary to confounding through ascertainment of cases, possibly related to the effects discussed by Voight and Pritchard [90]. Alternatively, it may represent statistical interaction resulting from population stratification, which can create mild linkage-disequilibrium between many markers that are on different chromosomes, with the strongest such LD occurring between loci with the largest frequency differences across populations. Such genome-wide effects of population stratification have recently been demonstrated both in simulated data, and in breast cancer, where there is association, detectable in cases, between SNPs in LCT and genome-wide SNPs, with a similar genomewide shift in the distribution of p-values [91]. Such adjustments for loci with strong effects may also be important in other diseases with such strong risk loci.
Stratification on APOE genotype did facilitate the identification of a novel region with genome-wide significant evidence for association on chromosome 10p14, which replicated in a second sample consisting of three additional cohorts. This region was identified only in the APOE ε4/ε4 stratum or in a logistic analysis that contrasted ε4 and ε3 homozygotes in a model with an interaction term with APOE. The relative infrequency of ε4 homozygotes means that these results will need to be further investigated in other large data sets to determine its importance. Data sets that consist of high-risk families, such as our sample and the NIMH AD sample [92], may be preferable in such analyses, since such sample ascertainment may have contributed to the detection of this locus through the resulting presence of a relatively high fraction of APOE ε4 homozygotes. It is also worth noting that an earlier linkage analysis of a subset of the families used here, based on the Illumina 6K mapping panel, obtained lod scores for rs1537626 of 2.35 in the whole sample and 1.6 in an analysis that retained only APOE ε4-positive cases. This SNP is within 10 cM of rs201119 [93]. This SNP was not on the marker panel used here, nor was rs201119 on the earlier 6K marker panel, preventing further comparison of results. It is also possible that analysis within the high-risk APOE ε4/ε4 genotype improved detection of this region in the current study by increasing the within-genotype penetrance, possibly by affecting age-at onset. If so, this would be similar to the strategy of identifying risk- or age-at-onset modifier loci on a background of a single, early-onset AD mutation [94]–[96]. The implicated region on chromosome 10p14 contains the genes CUGBP2 and PITRM1. CUGBP2 has one isoform that is expressed predominantly in neurons, with experimental evidence suggesting involvement in apoptosis in the hippocampus [97], with both these observations consistent with a role in pathogenesis of Alzheimer's disease. PITRM1 can degrade amyloid β4 APP protein when it is accumulated in mitochondria [98].
Our results both support and refute recently proposed association with SNPs in several genes [42]–[44]. Evidence for association with SNPs previously reported in each of BIN1, CLU, and CR1 was relatively robust to APOE adjustment within this European-American sample, with evidence for BIN1 and CR1 also obtained across an analysis that conditioned on ethnic background. Recent reports by others that include portions of the sample we used here also report evidence for association with PICALM [99], [100], but did not report the results of quality control analyses that allow evaluation of adequacy of correction for confounding. In our analyses, with correction for sources of confounding, evidence for association with SNPs in PICALM and EXOC3L2 was much less convincing than for these other three loci because of the exquisite sensitivity to APOE adjustment. One interpretation of sensitivity of these associations to APOE adjustment is that this statistical interaction is indicative of biological interaction in an analysis that includes a subset of the current sample [99]. However, the differences in SNP allele frequencies across APOE strata within cases that we showed here coupled with information demonstrating the existence of population stratification raise concerns that the original associations for these latter SNPs may represent confounding or other aspects of sample or population structure. This could include linkage disequilibrium with APOE, even for unlinked markers. Further investigation in genetically more diverse populations will still be necessary to clarify even the role of SNPs with positive evidence for association, because shared history can lead to spurious replication in samples drawn from the same population [80].
The results presented here and in other GWAS reports of LOAD underscore the view that such studies do not necessarily identify the specific genetic alterations contributing to disease risk. Rather, they are useful in identifying genes or gene pathways involved in disease pathogenesis or risk. In that sense, GWAS represents a method of screening the genome for genes that may also contain rare variants. While the large number of subjects in current GWAS provides a benefit in terms of perceived statistical power, it comes at a price. For example, despite the very low p-values representing genome-wide statistical significance, the effect sizes in most recent GWAS involving LOAD are small. It has also been suggested that different significance thresholds as a function of sample size are needed in order to balance power against the false-discovery rate [101], with very large studies requiring more stringent thresholds. This means that subtle differences in the genetic architecture of either the cases or the controls become more important with increasing sample sizes. In this situation some of the “significant” differences in allele frequency may also represents differences in ancestral origins rather than disease phenotype-genotype associations, and would likely not lead to further biological insights. As we have shown here, genetic variability within European-American groups exists and can affect analyses of association. Moving forward, GWAS in LOAD should consider more detailed care to control for population stratification or APOE genotypes prior to drawing firm conclusions about associations. In this sense bigger studies of LOAD or of other diseases with similar influential risk loci may not always be better, if the increases in sample size result in added data structure or confounding.
Supporting Information
Flow chart of source and number of subjects used to generate the NIA-LOAD/NCRAD resource sample.
(0.09 MB PDF)
Linkage disequilibrium among all pairs of 24 SNPs in CUGBP2 on chromosome 10 in the NIA LOAD/NCRAD sample, the Caribbean Hispanic sample, and the Washington University/ADNI sample. For SNP pairs with a log10(Lr)>2 supporting D′>0, where Lr is the likelihood ratio under the estimated D′ versus absence of disequilibrium, bright red indicates D′ = 1, while shades of red/pink indicate D′<1, with the value as indicated. For SNP pairs with a log10(Lr)<2 supporting D′>0, blue represents D′ = 1, and white represents other values of D′.
(0.24 MB PDF)
QQ (top) and quantile difference (bottom) plots for analysis of unadjusted European-American samples for all but 95 SNPs near APOE. For quantile difference, vertical axis is the negative logarithm of the difference in the observed and expected p-values. CCall: grey diamonds and cyan squares, showing results without and with adjustment for relationships, respectively, illustrating the large deviation of the p-value distribution that occurs in the absence of adjustment for relationships. CCun: black diamonds and magenta circles, representing chi-square tests of allele frequency differences and logistic regression, respectively, illustrating deviation from the null distribution in the chi-square analysis compared to the trend test from logistic regression. Green line: expectation under the null distribution.
(1.22 MB PDF)
Quantile difference plot of analysis model results of the CCun sample. Points represent the negative logarithm of the difference between observed and expected p-values, and lines represent the upper 95% confidence bounds. Analyses are represented by black triangles and dashed line: test of allele frequency difference; magenta circles and line: logistic regression with no adjustment for covariates; grey diamonds: logistic regression with adjustment for the first four principal components; cyan squares: logistic regression with adjustment for the number of APOE ε2 and ε4 alleles; and the blue line: confidence bound for logistic regression with adjustment for covariates (confidence bounds for all logistic regression conditions were essentially identical).
(0.25 MB PDF)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the full European-American sample (CCall), after eliminating SNPs with low genotype completion rates or that failed the test for Hardy-Weinberg equilibrium (7 SNPs) as described in the quality control analyses under Methods. Results were further restricted to SNPs with a total count of the minor allele across both cases and controls that exceeds 20, since caution in the interpretation of results is required in any case where the total number of minor alleles is small. The Odds Ratio (OR) shown is the allele-based OR for the allele shown (the Illumina TOP strand allele), and the pair of allele counts for each diagnostic class of individuals gives the count for the allele on the TOP strand first.
(0.03 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the unrelated European-American sample, CCun, for analysis based on logistic regression without additional covariates. Results were further restricted to SNPs with a total count of the minor allele across both cases and controls that exceeds 20, as in Table S1. The Odds Ratio (OR) shown is the OR attributable to 1 copy of the A1 allele. The two possible alleles (A1 and A2) are shown with both the forward strand and Illumina TOP strand coding. Genotypes for the three numbers for each of the cases and controls are the number of such individuals who are homozygous for the A1 allele, heterozygous, and homozygous for the A2 allele, respectively. P-values for tests for deviation for HWE in controls are also shown.
(0.04 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the unrelated European-American sample, CCun, for analysis based on logistic regression with adjustment for the number of APOE ε2 and ε4 alleles. See legend to Table S2 for further explanation of columns.
(0.04 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the APOE ε4/ε4 homozygotes in the European-American sample, CCall. No SNPs were removed because of deviation from HWE. Conditions for presentation of SNPs are described in legend to Table S1.
(0.03 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the APOE ε4/ε3 heterozygotes in the European-American sample, CCall. 5 SNPs were removed because of deviation from HWE. Conditions for presentation of SNPs are described in legend to Table S1.
(0.02 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the APOE ε3/ε3 homozygotes in the European-American sample, CCall. 5 SNPs were removed because of deviation from HWE. Conditions for presentation of SNPs are described in legend to Table S1.
(0.02 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the the unrelated European-American sample, CCun, for analysis based on logistic regression with adjustment for the first four principal components. Additional conditions are as described in Table S2.
(0.05 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the the unrelated European-American sample, CCun, for analysis based on logistic regression with adjustment for the first four principal components and for the number of APOE e4 and e2 alleles. Additional conditions are as described in Table S2.
(0.04 MB TXT)
Acknowledgments
We thank contributors who collected samples used in this study, and we particularly thank the patients and their families, whose help and participation made this work possible. Susan LaRusse Eckert and Stephanie Doan (Columbia University), and Michele Goodman (Indiana University), helped coordinate the project across the United States. Creighton H. Phelps, PhD, Marcelle Morrison-Bogorod, PhD, and Marilyn Miller, PhD, at the NIA provided guidance. Samples used in this study were obtained from the National Cell Repository for Alzheimer's Disease (NCRAD).
The following investigators and Alzheimer's Disease Centers participated in the Study: Boston University Robert Green, Neil Kowall, Lindsay Farrer; Columbia University Jennifer Williamson, Vincent Santana; Duke University Donald Schmechel, Perry Gaskell, Kathleen Welsh-Bohmer, Margaret Pericak-Vance; Indiana University, Bernardino Ghetti, Martin R. Farlow, Kelly Horner; Massachusetts General Hospital John H. Growdon, Deborah Blacker, Rudolph E. Tanzi, Bradley T. Hyman; Mayo Clinic-Rochester Bradley Boeve, Karen Kuntz, Lindsay Norgaard, Nathan Larson; Mayo Clinic-Jacksonville Dana Kistler, Fracine Parfitt, Jenny Haddwow; Mount Sinai School of Medicine Jeremy Silverman, Michal Schnaider Beeri, Mary Sano, Joy Wang, Rachel Lally; Northwestern University Nancy Johnson, Marcel Mesulum, Sandra Weintraub, Eileen Bigio; Oregon Health and Science University Jeffery Kaye, Patricia Kramer, Jessica Payne-Murphy; Rush University David Bennett, Holli Jacobs, Jeen-Soo Chang, Danielle Arends; University of Alabama at Birmingham Lindy Harrell; University of California, Los Angeles George Bartzokis, Jeffery Cummings, Po H Lu, Usha Toland; University of Kentucky William Markesbery, Charles Smith, Alise Brickhouse; University of Pennsylvania John Trojanowski, Vivianna Van Deerlin, Elisabeth McCarty Wood; University of Pittsburgh Steven DeKosky, Robert Sweet, Elise Weamer; University of Southern California I Helena Chui, Arousiak Varpetian; University of Texas Southwestern Ramon Diaz-Arrastia, Roger Rosenberg, Barbara Davis; University of Washington Thomas Bird, Malia Rumbaugh, Gerard D. Schellenberg, Murray Raskind; Washington University at St Louis Alison Goate, John Morris, Joanne Norton, Denise Levitch, Betsy Grant, Mary Coats.
Footnotes
The authors have declared that no competing interests exist.
This study was supported by the following federal grants from the National Institute of Aging: U24AG026395 (NIA-LOAD Family Study); U24AG021886 (National Cell Repository for Alzheimer's Disease); R01AG027224 and P50AG005133 University of Pittsburgh; P30AG10161 Rush University Medical Center; P30AG013846 Boston University; P50AG08702, P01AG07232, and R37AG015473 Columbia University; P30AG028377 and P50AG05128 Duke University; P30AG010133 Indiana University; P50AG05134 Massachusetts General Hospital; P50AG165574 Mayo Clinic, Rochester, and Mayo Clinic, Jacksonville; P01AG05138, P01AG02219, and P50AG05138 Mount Sinai School of Medicine; P30AG13854 Northwestern University Medical School; P30AG008017 Oregon Health and Science University; P50AG016582 University of Alabama at Birmingham; P50AG016579 David Geffen School of Medicine, University of California Los Angeles; P30AG028383 University of Kentucky, Lexington; P30AG10124 University of Pennsylvania; P50AG05142 University of Southern California; P30AG12300 The University of Texas Southwestern Medical Center; P50AG05136 University of Washington; and P50AG05681 and P01AG03991 Washington University School of Medicine. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript, but did review the manuscript prior to submission.
References
- 1.Mayeux R. Epidemiology of neurodegeneration. Annu Rev Neurosci. 2003;26:81–104. doi: 10.1146/annurev.neuro.26.043002.094919. [DOI] [PubMed] [Google Scholar]
- 2.Fratiglioni L, De Ronchi D, Aguero-Torres H. Worldwide prevalence and incidence of dementia. Drugs & Aging. 1999;15:365–375. doi: 10.2165/00002512-199915050-00004. [DOI] [PubMed] [Google Scholar]
- 3.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 4.Bergem ALM, Lannfelt L. Apolipoprotein E type epsilon 4 allele, heritability and age at onset in twins with Alzheimer's disease. Clin Genet. 1997;52:408–413. doi: 10.1111/j.1399-0004.1997.tb04362.x. [DOI] [PubMed] [Google Scholar]
- 5.Meyer JM, Breitner JCS. Multiple threshold model for the onset of Alzheimer's disease in the NAS-NRC twin panel. Am J Med Genet. 1998;81:92–97. doi: 10.1002/(sici)1096-8628(19980207)81:1<92::aid-ajmg16>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 6.Akesson HO. A Population Study of Senile and Arteriosclerotic Psychoses. Hum Hered. 1969;19:546. doi: 10.1159/000152266. [DOI] [PubMed] [Google Scholar]
- 7.Breitner JCS, Folstein MF, Murphy EA. Familial Aggregation in Alzheimer Dementia .1. a Model for the Age-Dependent Expression of an Autosomal Dominant Gene. J Psychiatr Res. 1986;20:31–43. doi: 10.1016/0022-3956(86)90021-x. [DOI] [PubMed] [Google Scholar]
- 8.Mohs RC, Breitner JCS, Silverman JM, Davis KL. Alzheimers-Disease - Morbid Risk among 1st-Degree Relatives Approximates 50-Percent by 90 Years of Age. Arch Gen Psychiatry. 1987;44:405–408. doi: 10.1001/archpsyc.1987.01800170019003. [DOI] [PubMed] [Google Scholar]
- 9.Lautenschlager NT, Cupples LA, Rao VS, Auerbach SA, Becker R, et al. Risk of dementia among relatives of Alzheimer's disease patients in the MIRAGE study: What is in store for the oldest old? Neurology. 1996;46:641–650. doi: 10.1212/wnl.46.3.641. [DOI] [PubMed] [Google Scholar]
- 10.Van Broeckhoven C, Genthe AM, Van den Berghe A, Horsthemke B, Backhovens H, et al. Failure of Familial Alzheimers-Disease to Segregate with the A4-Amyloid Gene in Several European Families. Nature. 1987;329:153–155. doi: 10.1038/329153a0. [DOI] [PubMed] [Google Scholar]
- 11.Bird TD, Lampe TH, Nemens EJ, Miner GW, Sumi SM, et al. Familial Alzheimer's disease in american descendants of the Volga Germans: probable genetic founder effect. Ann Neurol. 1988;23:25–31. doi: 10.1002/ana.410230106. [DOI] [PubMed] [Google Scholar]
- 12.St. George-Hyslop PH, Myers RH, Haines JL, Farrer LA, Tanzi RE, et al. Familial Alzheimers-Disease - Progress and Problems. Neurobiol Aging. 1989;10:417–425. doi: 10.1016/0197-4580(89)90082-1. [DOI] [PubMed] [Google Scholar]
- 13.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 14.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 15.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 16.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 17.Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, et al. Protective Effect of Apolipoprotein-E Type-2 Allele for Late-Onset Alzheimer-Disease. Nat Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 18.Myers RH, Schaefer EJ, Wilson PWF, Dagostino R, Ordovas JM, et al. Apolipoprotein E epsilon 4 association with dementia in a population-based study: The Framingham study. Neurology. 1996;46:673–677. doi: 10.1212/wnl.46.3.673. [DOI] [PubMed] [Google Scholar]
- 19.Bennett CL, Crawford F, Osborne A, Diaz P, Hoyne J, et al. Evidence that the APOE locus influences rate of disease progression in late-onset familial Alzheimer's-disease but is not causative. Am J Med Genet B Neuropsychiatr Genet. 1995;50:1–6. doi: 10.1002/ajmg.1320600102. [DOI] [PubMed] [Google Scholar]
- 20.Slooter AJC, Cruts M, Kalmijn S, Hofman A, Breteler MMB, et al. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: The Rotterdam study. Arch Neurol. 1998;55:964–968. doi: 10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- 21.Daw EW, Heath SC, Wijsman EM. Multipoint oligogenic analysis of age-at-onset data with applications to Alzheimer's disease pedigrees. Am J Hum Genet. 1999;64:839–851. doi: 10.1086/302276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, et al. The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet. 2000;66:196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pericak-Vance MA, Grubber JM, Bailey LR, Hedges D, West S, et al. Identification of Novel Genes in Late-Onset Alzheimer's Disease. Exp Gerontol. 2000;35:1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 24.Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, et al. Results of a high-resolution genome screen of 437 Alzheimer's Disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 25.Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, et al. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12:415–422. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- 26.Scott WK, Hauser ER, Schmechel DE, Welsh-Bohmer KA, Small GW, et al. Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. Am J Hum Genet. 2003;73:1041–1051. doi: 10.1086/379083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wijsman EM, Daw EW, Yu CE, Payami H, Steinbart EJ, et al. Evidence for a novel late-onset Alzheimer's disease locus on chromosome 19p13.2. Am J Hum Genet. 2004;75:398–409. doi: 10.1086/423393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, et al. Family-based association between Alzheimer's disease and variants in UBQLN1. N Engl J Med. 2005;352:884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 29.Rademakers R, Cruts M, Sleegers K, Dermaut B, Theuns J, et al. Linkage and association studies identify a novel locus for Alzheimer disease at 7q36 in a Dutch population-based sample. Am J Hum Genet. 2005;77:643–652. doi: 10.1086/491749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahs DW, McCauley JL, Crunk AE, McFarland LL, Gaskell PC, et al. A genome-wide linkage analysis of dementia in the Amish. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:160–166. doi: 10.1002/ajmg.b.30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JH, Cheng R, Santana V, Williamson J, Lantigua R, et al. Expanded genomewide scan implicates a novel locus at 3q28 among Caribbean Hispanics with familial Alzheimer disease. Arch Neurol. 2006;63:1591–1598. doi: 10.1001/archneur.63.11.1591. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Grupe A, Rowland C, Nowotny P, Kauwe JS, et al. DAPK1 variants are associated with Alzheimer's disease and allele-specific expression. Hum Mol Genet. 2006;15:2560–2568. doi: 10.1093/hmg/ddl178. [DOI] [PubMed] [Google Scholar]
- 33.Butler AW, Ng MYM, Hamshere ML, Forabosco P, Wroe R, et al. Meta-analysis of linkage studies for Alzheimer's disease-A web resource. Neurobiol Aging. 2009;30:1037–1047. doi: 10.1016/j.neurobiolaging.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 35.Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, et al. Evidence for novel susceptibility genes for late-onset Alzheimer's disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 36.Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, et al. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, et al. A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Medical Genomics. 2008;1:44. doi: 10.1186/1755-8794-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, et al. Genome-wide Association Analysis Reveals Putative Alzheimer's Disease Susceptibility Loci in Addition to APOE. Am J Hum Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, et al. Genome-wide Association Study Implicates a Chromosome 12 Risk Locus for Late-Onset Alzheimer Disease. Am J Hum Genet. 2009;84:35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrasquillo MM, Zou FG, Pankratz VS, Wilcox SL, Ma L, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poduslo SE, Huang R, Huang J, Smith S. Genome screen of late-onset Alzheimer's extended pedigrees identifies TRPC4AP by haplotype analysis. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:50–55. doi: 10.1002/ajmg.b.30767. [DOI] [PubMed] [Google Scholar]
- 42.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;62:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambert JC, Heath S, Even G, Campion D, Sleegers K, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 44.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, et al. Genome-wide Analysis of Genetic Loci Associated With Alzheimer Disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong H, Prentice RL. Correcting “‘Winner's Curse” in Odds Ratios from Genomewide Association Findings for Major Complex Human Diseases. Genet Epidemiol. 2010;34:78–91. doi: 10.1002/gepi.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jarvik GP, Larson EB, Goddard KAB, Schellenberg GD, Wijsman EM. Influence of apolipoprotein E genotype on the transmission of Alzheimer disease in a community-based sample. Am J Hum Genet. 1996;58:191–200. [PMC free article] [PubMed] [Google Scholar]
- 47.Slooter AJC, vanDuijn CM. Genetic epidemiology of Alzheimer disease. Epidemiol Rev. 1997;19:107–119. doi: 10.1093/oxfordjournals.epirev.a017933. [DOI] [PubMed] [Google Scholar]
- 48.McCarthy M, Kruglyak L, Lander E. Sib-pair collection strategies for complex diseases. Genet Epidemiol. 1998;15:317–340. doi: 10.1002/(SICI)1098-2272(1998)15:4<317::AID-GEPI1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 49.McKhann G, Drachman D, Folstein M, Katzman R, Price D, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 50.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, et al. Mild cognitive impairment - Clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 51.Hughes CP, Berg L, Danziger WL, Coben LA, Margin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- 52.Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 53.Chudyk A, Masiuk M, Myslak M, Domanski L, Sienko J, et al. Soluble HLA class I molecules exert differentiated influence on renal graft condition. Transplant Proc. 2006;38:90–93. doi: 10.1016/j.transproceed.2005.11.090. [DOI] [PubMed] [Google Scholar]
- 54.Myakishev MV, Khripin Y, Hu S, Hamer DH. High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res. 2001;11:163–169. doi: 10.1101/gr.157901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hawkins JR, Khripin Y, Valdes AM, Weaver TA. Miniaturized sealed-tube allele-specific PCR. Hum Mutat. 2002;19:543–553. doi: 10.1002/humu.10060. [DOI] [PubMed] [Google Scholar]
- 56.Bekris LM, Millard SP, Galloway NM, Vuletic S, Albers JJ, et al. Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J Alzheimers Dis. 2008;13:255–266. doi: 10.3233/jad-2008-13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, et al. Hippocampal Atrophy as a Quantitative Trait in a Genome-Wide Association Study Identifying Novel Susceptibility Genes for Alzheimer's Disease. PLoS ONE. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roses AD. An Inherited Variable Poly-T Repeat Genotype in TOMM40 in Alzheimer Disease. Arch Neurol. 2010;67:536–541. doi: 10.1001/archneurol.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Göring HHH, Ott J. Relationship estimation in affected rib pair analysis of late-onset diseases. Eur J Hum Genet. 1997;5:69–77. [PubMed] [Google Scholar]
- 60.Sun L, Wilder K, McPeek MS. Enhanced pedigree error detection. Hum Hered. 2002;54:99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- 61.Nielsen DM, Ehm MG, Weir BS. Detecting marker-disease association by testing for Hardy-Weinberg disequilibrium at a marker locus. Am J Hum Genet. 1998;63:1531–1540. doi: 10.1086/302114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:967–986. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zou GY, Donner A. The merits of testing Hardy-Weinberg equilibrium in the analysis of unmatched case-control data: A cautionary note. Ann Hum Genet. 2006;70:923–933. doi: 10.1111/j.1469-1809.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 64.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 65.Price AL, Butler J, Patterson N, Capelli C, Pascali VL, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harvey CB, Hollox EJ, Poulter M, Wang Y, Rossi M, et al. Lactase haplotype frequencies in Caucasians: association with the lactase persistence/non-persistence polymorphism. Ann Hum Genet. 1998;62:215–223. doi: 10.1046/j.1469-1809.1998.6230215.x. [DOI] [PubMed] [Google Scholar]
- 67.Hollox EJ, Poulter M, Zvarik M, Ferak V, Krause A, et al. Lactase haplotype diversity in the Old World. Am J Hum Genet. 2001;68:160–172. doi: 10.1086/316924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi Y, Wijsman EM, Weir BS. Case-control Association Testing in the Presence of Unknown Relationships. Genet Epidemiol. 2009;35:668–678. doi: 10.1002/gepi.20418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jacquard A. Genetic Information Given by a Relative. Biometrics. 1972;28:1101–1114. [PubMed] [Google Scholar]
- 70.Milligan BG. Maximum-likelihood estimation of relatedness. Genetics. 2003;163:1153–1167. doi: 10.1093/genetics/163.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bourgain C, Hoffjan S, Nicolae R, Newman D, Steiner L, et al. Novel case-control test in a founder population identifies P-selectin as an atopy-susceptibility locus. Am J Hum Genet. 2003;73:612–626. doi: 10.1086/378208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pritchard JK, Donnelly P. Case-control studies of association in structured or admixed populations. Theor Popul Biol. 2001;60:227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- 73.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 74.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 75.Chapman JM, Cooper JD, Todd JA, Clayton D. Detecting disease associations due to linkage disequilibrium using haplotype tags: A class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- 76.Yu CE, Seltman H, Peskind ER, Galloway N, Zhou PX, et al. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer's disease: Patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89:655–665. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cruchaga C, Kauwe JSK, Mayo K, Spiegel N, Bertelsen S, et al. SNPs Associated with Cerebrospinal Fluid Phospho-Tau Levels Influence Rate of Decline in Alzheimer's Disease. PLoS Genet. 2010;6:e1001101. doi: 10.1371/journal.pgen.1001101. doi: 10.1371/journal.pgen.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang MX, Stern Y, Marder K, Bell K, Gurland B, et al. The APOE-epsilon 4 allele and the risk of Alzheimer disease among African Americans, Whites, and Hispanics. JAMA. 1998;279:751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 79.Romas SN, Santana V, Williamson J, Ciappa A, Lee JH, et al. Familial Alzheimer disease among Caribbean Hispanics - A reexamination of its association with APOE. Arch Neurol. 2002;59:87–91. doi: 10.1001/archneur.59.1.87. [DOI] [PubMed] [Google Scholar]
- 80.Rosenberg NA, VanLiere JM. Replication of Genetic Associations as Pseudoreplication due to Shared Genealogy. Genet Epidemiol. 2009;33:479–487. doi: 10.1002/gepi.20400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heath SC, Gut IG, Brennan P, McKay JD, Bencko V, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- 82.Lucotte G, Loirat F, Hazout S. Pattern of gradient of apolipoprotein E allele *4 frequencies in Western Europe. Hum Biol. 1997;69:253–262. [PubMed] [Google Scholar]
- 83.Corbo RM, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’ allele? Ann Hum Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 84.Singh PP, Singh M, Mastana SS. APOE distribution in world populations with new data from India and the UK. Ann Hum Biol. 2006;33:279–308. doi: 10.1080/03014460600594513. [DOI] [PubMed] [Google Scholar]
- 85.Rosenmann H, Meiner Z, Kahana E, Aladjem Z, Friedman G, et al. An association study of the codon 72 polymorphism in the pro-apoptotic gene p53 and Alzheimer's disease. Neurosci Lett. 2003;340:29–32. doi: 10.1016/s0304-3940(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 86.Dresner-Pollak R, Kinnar T, Friedlander Y, Sharon N, Rosenmann H, et al. Estrogen Receptor Beta Gene Variant Is Associated with Vascular Dementia in Elderly Women. Genet Test Mol Biomarkers. 2009;13:339–342. doi: 10.1089/gtmb.2008.0129. [DOI] [PubMed] [Google Scholar]
- 87.Little J, Higgins JPT, Ioannidis JPA, Moher D, Gagnon F, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE Statement. Hum Genet. 2009;125:131–151. doi: 10.1007/s00439-008-0592-7. [DOI] [PubMed] [Google Scholar]
- 88.Nelis M, Esko T, Magi R, Zimprich F, Zimprich A, et al. Genetic Structure of Europeans: A View from the North-East. PLoS ONE. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zekraoui L, Lagarde JP, Raisonnier A, Gerard N, Aouizerate A, et al. High frequency of the apolipoprotein E *4 allele in African pygmies and most of the African populations in sub-Saharan Africa. Hum Biol. 1997;69:575–581. [PubMed] [Google Scholar]
- 90.Voight BF, Pritchard JK. Confounding from cryptic relatedness in case-control association studies. PLoS Genet. 2005;1:e32. doi: 10.1371/journal.pgen.0010032. doi: 10.1371/journal.pgen.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bhattacharjee S, Wang Z, Ciampa J, Kraft P, Chanock S, et al. Using Principal Components of Genetic Variation for Robust and Powerful Detection of Gene-Gene Interactions in Case-Control and Case-Only Studies. Am J Hum Genet. 2010;86:331–342. doi: 10.1016/j.ajhg.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, et al. Results of a high resolution genome screen in 443 Alzheimer's disease families: the NIMH Genetics Initiative. Am J Hum Genet. 2001;69:498–498. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 93.Lee JH, Cheng R, Graff-Radford N, Foroud T, Mayeux R. Analyses of the National Institute on Aging Late-Onset Alzheimer's Disease Family Study Implication of Additional Loci. Arch Neurol. 2008;65:1518–1526. doi: 10.1001/archneur.65.11.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pastor P, Roe C, Villegas A, Bedoya G, Chakraverty S, et al. Apolipoprotein E ε4 modifies Alzheimer's disease onset in an E280A PS1 Kindred. Ann Neurol. 2003;54:163–169. doi: 10.1002/ana.10636. [DOI] [PubMed] [Google Scholar]
- 95.Wijsman EM, Daw EW, Yu X, Steinbart EJ, Nochlin D, et al. APOE and other loci affect age-at-onset in Alzheimer's disease families with PS2 mutation. Am J Med Genet B Neuropsychiatr Genet. 2005;132B:14–20. doi: 10.1002/ajmg.b.30087. [DOI] [PubMed] [Google Scholar]
- 96.Marchani EE, Bird TD, Steinbart EJ, Rosenthal E, Yu CE, et al. Evidence for three loci modifying age-at-onset of Alzheimer's disease in early-onset PSEN2 families. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1031–1041. doi: 10.1002/ajmg.b.31072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pacini A, Toscano A, Cesati V, Cozzi A, Meli E, et al. NAPOR-3 RNA binding protein is required for apoptosis in hippocampus. Brain Res Mol Brain Res. 2005;140:34–44. doi: 10.1016/j.molbrainres.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 98.Falkevall A, Alikhani N, Bhushan S, Pavlov PF, Busch K, et al. Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J Biol Chem. 2006;281:29096–29104. doi: 10.1074/jbc.M602532200. [DOI] [PubMed] [Google Scholar]
- 99.Jun G, Naj AC, Beecham GW, Wang LS, Buros J, et al. Meta-analysis confirms CR1, CLU, and PICALM as Alzheimer disease risk loci and reveals interactions with APOE genotypes. 2010. Arch Neurol Sept. 3 Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 100.Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, et al. Replication of CLU, CR1, and PICALM Associations With Alzheimer Disease. Arch Neurol. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wakefield J. Bayes Factors for Genome-Wide Association Studies: Comparison with P-values. Genet Epidemiol. 2009;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow chart of source and number of subjects used to generate the NIA-LOAD/NCRAD resource sample.
(0.09 MB PDF)
Linkage disequilibrium among all pairs of 24 SNPs in CUGBP2 on chromosome 10 in the NIA LOAD/NCRAD sample, the Caribbean Hispanic sample, and the Washington University/ADNI sample. For SNP pairs with a log10(Lr)>2 supporting D′>0, where Lr is the likelihood ratio under the estimated D′ versus absence of disequilibrium, bright red indicates D′ = 1, while shades of red/pink indicate D′<1, with the value as indicated. For SNP pairs with a log10(Lr)<2 supporting D′>0, blue represents D′ = 1, and white represents other values of D′.
(0.24 MB PDF)
QQ (top) and quantile difference (bottom) plots for analysis of unadjusted European-American samples for all but 95 SNPs near APOE. For quantile difference, vertical axis is the negative logarithm of the difference in the observed and expected p-values. CCall: grey diamonds and cyan squares, showing results without and with adjustment for relationships, respectively, illustrating the large deviation of the p-value distribution that occurs in the absence of adjustment for relationships. CCun: black diamonds and magenta circles, representing chi-square tests of allele frequency differences and logistic regression, respectively, illustrating deviation from the null distribution in the chi-square analysis compared to the trend test from logistic regression. Green line: expectation under the null distribution.
(1.22 MB PDF)
Quantile difference plot of analysis model results of the CCun sample. Points represent the negative logarithm of the difference between observed and expected p-values, and lines represent the upper 95% confidence bounds. Analyses are represented by black triangles and dashed line: test of allele frequency difference; magenta circles and line: logistic regression with no adjustment for covariates; grey diamonds: logistic regression with adjustment for the first four principal components; cyan squares: logistic regression with adjustment for the number of APOE ε2 and ε4 alleles; and the blue line: confidence bound for logistic regression with adjustment for covariates (confidence bounds for all logistic regression conditions were essentially identical).
(0.25 MB PDF)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the full European-American sample (CCall), after eliminating SNPs with low genotype completion rates or that failed the test for Hardy-Weinberg equilibrium (7 SNPs) as described in the quality control analyses under Methods. Results were further restricted to SNPs with a total count of the minor allele across both cases and controls that exceeds 20, since caution in the interpretation of results is required in any case where the total number of minor alleles is small. The Odds Ratio (OR) shown is the allele-based OR for the allele shown (the Illumina TOP strand allele), and the pair of allele counts for each diagnostic class of individuals gives the count for the allele on the TOP strand first.
(0.03 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the unrelated European-American sample, CCun, for analysis based on logistic regression without additional covariates. Results were further restricted to SNPs with a total count of the minor allele across both cases and controls that exceeds 20, as in Table S1. The Odds Ratio (OR) shown is the OR attributable to 1 copy of the A1 allele. The two possible alleles (A1 and A2) are shown with both the forward strand and Illumina TOP strand coding. Genotypes for the three numbers for each of the cases and controls are the number of such individuals who are homozygous for the A1 allele, heterozygous, and homozygous for the A2 allele, respectively. P-values for tests for deviation for HWE in controls are also shown.
(0.04 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the unrelated European-American sample, CCun, for analysis based on logistic regression with adjustment for the number of APOE ε2 and ε4 alleles. See legend to Table S2 for further explanation of columns.
(0.04 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the APOE ε4/ε4 homozygotes in the European-American sample, CCall. No SNPs were removed because of deviation from HWE. Conditions for presentation of SNPs are described in legend to Table S1.
(0.03 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the APOE ε4/ε3 heterozygotes in the European-American sample, CCall. 5 SNPs were removed because of deviation from HWE. Conditions for presentation of SNPs are described in legend to Table S1.
(0.02 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the APOE ε3/ε3 homozygotes in the European-American sample, CCall. 5 SNPs were removed because of deviation from HWE. Conditions for presentation of SNPs are described in legend to Table S1.
(0.02 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the the unrelated European-American sample, CCun, for analysis based on logistic regression with adjustment for the first four principal components. Additional conditions are as described in Table S2.
(0.05 MB TXT)
SNPs with p<5×10−4 for allele frequency comparisons in cases versus controls from the the unrelated European-American sample, CCun, for analysis based on logistic regression with adjustment for the first four principal components and for the number of APOE e4 and e2 alleles. Additional conditions are as described in Table S2.
(0.04 MB TXT)