Abstract
Cell-to-cell communication is crucial for the development of multicellular organisms, especially during the generation of new tissues and organs. Secondary growth—the lateral expansion of plant growth axes—is a highly dynamic process that depends on the activity of the cambium. The cambium is a stem cell–like tissue whose activity is responsible for wood production and, thus, for the establishment of extended shoot and root systems. Attempts to study cambium regulation at the molecular level have been hampered by the limitations of performing genetic analyses in trees and by the difficulty of accessing this tissue in model systems such as Arabidopsis thaliana. Here, we describe the roles of two receptor-like kinases, REDUCED IN LATERAL GROWTH1 (RUL1) and MORE LATERAL GROWTH1 (MOL1), as opposing regulators of cambium activity. Their identification was facilitated by a novel in vitro system in which cambium formation is induced in isolated Arabidopsis stem fragments. By combining this system with laser capture microdissection, we characterized transcriptome remodeling in a tissue- and stage-specific manner and identified series of genes induced during different phases of cambium formation. In summary, we provide a means for investigating cambium regulation in unprecedented depth and present two signaling components that control a process responsible for the accumulation of a large proportion of terrestrial biomass.
Author Summary
In contrast to animals, plants have the capacity to grow and form new organs throughout their entire life cycle, thereby building up some of the largest organisms on earth. This remarkable capacity is based on the activity of stem cell–like tissues—the meristems—located at shoot and root apices and, in a large repertoire of species, in lateral positions at the flanks of growth axes. In comparison to apical meristems, our knowledge of the molecular mechanisms controlling the activity of lateral meristems like the cambium is very limited. This is despite the fact that lateral growth is responsible for wood formation, and thus for the accumulation of large amounts of terrestrial biomass, and for fixation of atmospheric CO2. Here, we introduce an in vitro system by which cambium initiation can be stimulated under controlled conditions in stems of the reference plant Arabidopsis thaliana. By revealing genome-wide and tissue-specific alterations in transcript accumulation during cambium initiation, we identify two novel receptor-like kinases, namely MOL1 and RUL1, as opposing cambium regulators. These findings demonstrate that our in vitro system represents a valuable tool for studying cambium regulation and open up possibilities to dissect lateral growth in plants from novel perspectives.
Introduction
The development of multicellular organisms requires extensive cell-to-cell communication to integrate the activity of single cells into the context of the whole organism. Plant development is especially demanding in this respect due to the high degree of plasticity caused by their interdependence with external cues. The capacity to establish pluripotent and proliferating tissues - the meristems - from differentiated cells represents a remarkable example of this developmental plasticity, which has been the focus of extensive research in the past [1]–[3]. The formation of the interfascicular cambium (Figure 1A, 1B) is one of the few instances in which post-embryonic de novo-initiation of meristematic activity occurs during normal plant development [4], and thus serves as an attractive model for studying the complexity of cell fate regulation in general, and cambium regulation in particular.
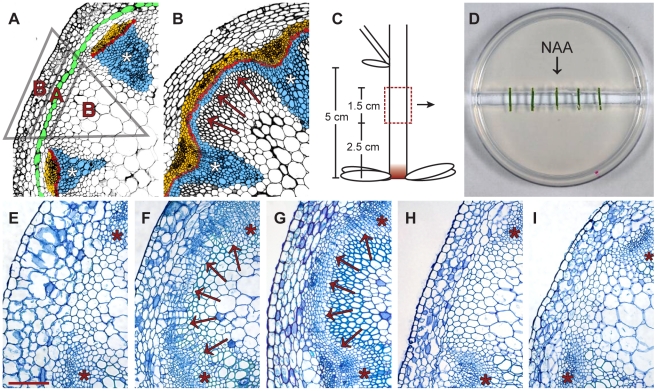
Figure 1. In vitro-induction of secondary growth (CIS–incubation).
(A and B) Comparison of cross-sections from a primary (A) and secondary (B) stem (IC: arrows in B). Blue: xylem/xylem fibers; red: fascicular and interfascicular cambium; yellow: phloem/phloem parenchyma; green: starch sheath. Triangle: see Figure 2H. (C) Origin of stem fragments for CIS-incubation. At the stage of collection, IC initiation was restricted to the region labeled in red [4]. (D) Experimental setup of CIS. (E–G) Stem fragments incubated without (E) and with (F) apically applied NAA in comparison to a stem immediately above the uppermost rosette leaf of a 15 cm tall plant (G). Arrows indicate dividing tissues in interfascicular regions. (H and I). Fragments incubated with basally applied NAA (H) and with apically applied NAA together with ubiquitously applied NPA (I, 1 µg/ml). Size bar in (E): 100 µm, same magnification in (E–I). The positions of primary vascular bundles are labeled by asterisks.
The cambium is a meristematic tissue found in a tube-like domain enclosing the center of the growth axes of dicotyledonous plants. It is responsible for the lateral expansion of plants by producing xylem and phloem - tissues involved in long-distance transport [5]. This process, also referred to as secondary growth, is the basis of wood formation, and is thus indispensable for extended plant growth and the accumulation of a large proportion of terrestrial biomass. In primary shoots of many species, including Arabidopsis, discrete vascular bundles are found, consisting of the fascicular cambium, the primary phloem, and the primary xylem (Figure 1A). These tissues originate from procambium strands established immediately below the shoot apical meristem. During secondary growth initiation, the meristematic character of the fascicular cambium located in the center of primary vascular bundles is extended into interfascicular regions. This process results in interfascicular cambium (IC) formation, and thereby in the establishment of a closed domain of meristematic activity important for coordinated lateral shoot growth (Figure 1B).
The unique features of this process, including de novo initiation of cambium identity from differentiated cells, and easy-to-follow changes in cellular patterns, allow IC formation to be used as a model to decipher cambium formation and activity [4]. During IC formation in Arabidopsis shoots, cell divisions are initiated primarily in the starch sheath - the innermost cell layer of the cortex that is important for directional shoot growth by sensing the gravity vector using sedimented amyloplasts (Figure 1A) [4], [6], [7]. There is strong evidence that IC formation is independent of cell identity in interfascicular regions. For example, procambium formation upon injury is initiated de novo at variable positions [8] and, depending on the developmental stage of the plant, the IC is established in different cell types [4]. Consequently, IC initiation seems to encompass a dramatic change in cell identity, and to depend on long- and/or short-distance derived signals of a mostly unidentified nature.
Our knowledge of the regulation of cambium initiation and activity, including its integration into general growth processes, at the cellular level is scarce. However, it is known that this process is modulated by a plethora of hormones [9]. In particular, IC initiation, and cambium activity in general, depend strongly on the basipetal transport of auxin along the shoot mediated by PIN auxin exporters [10]–[12]. The interplay between auxin transport and the establishment of vascular tissue has been studied most extensively during Arabidopsis leaf development. In this case, cell files become specialized for PIN1-mediated auxin transport stimulated by a positive feedback loop also involving the transcription factors MP and ATHB8 [13], [14]. In young leaf primordia, incipient broad expression domains of PIN1 and MP become restricted to narrow cell files marked and supported by ATHB8 expression. Subsequently, selected cell files transform into vascular bundles by employing unknown downstream signaling cascades [14], [15], recapitulating principles of the auxin canalization theory originally postulated by Tsvi Sachs [16].
Whether the same mechanisms identified for procambium formation during leaf development apply to IC formation is unknown. In this regard, it can be expected that the elucidation of transcriptional profiles in cells during IC formation will facilitate a comparison of both processes, and moreover will help to decipher the regulatory networks involved in cambium establishment and regulation. Previous transcriptional profiling did not have the spatial or temporal resolution required to identify genes associated with cambium initiation [4], [17]–[21]. In this context, an elegant approach elucidating the cambium-specific transcriptome from Populus tremula, was made possible by the availability of large amounts of starting material [22]. This approach showed that selected genes, or gene families, involved in apical meristem regulation are also expressed in the cambium, suggesting a high degree of similarity between the regulation of apical and lateral meristems [5], [22], [23].
PHLOEM INTERCALATED WITH XYLEM (PXY, also known as TDR), a cambium-specific receptor-like kinase, and its corresponding peptide ligands, CLE41/44, are important for cambium activity and the patterning of cambium-derived cells in Arabidopsis. This signaling module is the first cambium-related module characterized in detail at the molecular level [24]–[26]. The mechanistic similarities to the CLV1-CLV2/3 signaling module, important for shoot meristem regulation (reviewed in [27]), further suggest that similar molecular principles are involved in the regulation of both meristem types. Recently, the role of the homeobox transcription factor WUSCHEL-RELATED HOMEOBOX4 (WOX4) as an essential cambium regulator that is positively regulated by PXY has been revealed [28], [29]. This finding is reminiscent of the role of the WOX gene family members WUS and WOX5 in shoot and root apical meristems, respectively [30], [31], and provides another essential piece of evidence in support of the concept of regulatory similarities in indeterminate plant meristems.
Laser capture microdissection (LCM) is a valuable tool for harvesting material from specific tissues or cells, and for elucidating the transcript profiles of various plant tissues [32]–[35]. In comparison to fluorescent-activated cell sorting (FACS) [36], [37], LCM has the advantage of being able to harvest heterogeneous populations of cells, or cells that change their identity over time and cannot be labeled by a single fluorescent marker. Therefore, LCM-based transcript profiling represents an ideal approach to study IC formation because, in this case, a new tissue is established from clearly unrelated cell types. However, the advantages of LCM for characterizing early changes in transcript accumulation during this process are compromised by the restricted nature of IC formation in the Arabidopsis stem [4]. Furthermore, an exact time-course analysis is complicated by slight variations in the degree of IC formation between individuals, making it difficult to accurately predict when, and in which starch sheath cells, the IC will be initiated in each individual plant. Because only a small selection of plants can be analyzed by LCM, incorrect predictions for a single individual might cause substantial misinterpretation of the samples.
Here, we present an auxin-dependent cambium-inducing in vitro system (CIS) suitable for analyzing the molecular regulation of secondary growth in isolated Arabidopsis stem fragments that overcomes the obstacles of studying the process in intact plants. The comparison of molecular markers in intact plants and in the in vitro system confirms that the in planta situation is largely reflected by CIS, thus demonstrating that it represents a valuable tool for studying this process under highly controlled conditions. Using an LCM approach, we harvested cells from interfascicular regions at different time points during interfascicular cambium initiation resulting in the identification of both stage- and tissue-specific marker genes for different steps of the process. RNA in situ hybridization confirmed the cambium-specific expression of early markers in intact plants, demonstrating the benefits of the in vitro system, and suggesting that we have indeed revealed genome-wide changes in transcript accumulation in cells gaining cambium identity. Moreover, reverse genetics of the two uncharacterized receptor-like kinases RUL1 and MOL1, reveals the role of two novel signaling components in cambium regulation, further highlighting the benefits of CIS incubation.
Results
In Vitro–Incubated Stem Segments Behave As In Planta in Terms of Secondary Growth Initiation
To establish a cambium inducing system (CIS) in which controlled pharmacological manipulation of secondary growth in Arabidopsis stems could be performed, we adapted an experimental system designed for the in vitro incubation of excised stem fragments [38]. As starting material, we collected internodal fragments of 15–20 cm tall inflorescence stems which, under standard growth conditions, do not develop an IC in intact plants [4] (Figure 1C). These were incubated on split-plates with their apical and basal ends resting on separate halves of a plate of medium with no connection between them (Figure 1D), thus allowing the independent application of substances to the apical and basal sides of the incubated fragments. To verify that secondary growth can be induced in this system, we compared fragments incubated with and without apically applied synthetic auxin (NAA). Histological analyses after 5 days of incubation showed that only NAA-treated fragments initiated periclinal cell divisions in the fascicular cambium and in interfascicular regions (Figure 1E, 1F). The position and orientation of these divisions were comparable to those found in segments of intact stems displaying IC initiation (Figure 1G). When fragments were analyzed along their entire length, similar effects were observed at all positions (data not shown). We also found that, as in intact stems [4], cell divisions in interfascicular regions were initiated in the starch sheath (Figure S1C). Applying different NAA concentrations revealed a saturation of the response at 1 µg/ml NAA (Figure S1); therefore, subsequent experiments were conducted using this concentration.
To see whether incubated fragments lose their original polarity during CIS incubation, NAA was applied exclusively in the basal half of the split-plate. Histological analysis showed that no cell divisions were initiated in this case (Figure 1H), indicating that incubated fragments maintain their apical-basal polarity. Importantly, the addition of NPA to plates with apically applied auxin repressed the initiation of cell division (Figure 1I), demonstrating that the effect depends on active basipetal auxin transport along incubated fragments.
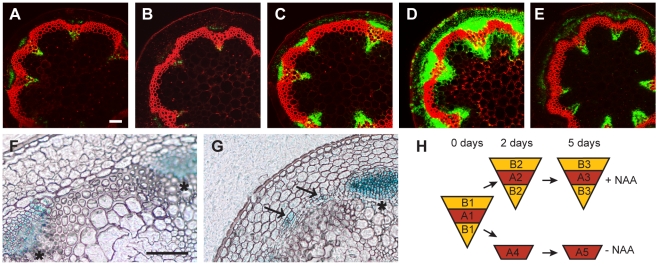
To characterize the distribution of auxin signaling in CIS-incubated fragments, we compared CIS-incubated fragments with fragments from intact plants derived from a DR5rev:GFP reporter line [39]. This analysis revealed a pattern of the GFP-derived signal in CIS-incubated fragments similar to that in intact stems for all time points, with enhanced signal intensity after 5 days of incubation (Figure 2A–2E). This result indicates that, although more auxin seems to be present in the in vitro-incubated stems after prolonged incubation, its distribution is comparable to that of intact stems. To confirm that interfascicular cell divisions represented secondary growth, we analyzed CIS-incubated stem fragments from a line carrying an APL:GUS reporter visualizing phloem identity [4]. APL, which encodes for a MYB transcription factor, is essential for the establishment of phloem cell characteristics, and is specifically expressed in the phloem throughout all growth stages and organs [40], [41]. APL:GUS-derived signals were found in interfascicular regions following NAA addition to the medium, and in groups of cells resembling cell clusters formed during phloem initiation in intact plants [4] (Figure 2F, 2G). This shows that interfascicular cell divisions lead to the formation of secondary vascular tissue. In summary, we conclude that CIS-incubated stem fragments behave in the same way as intact stems with respect to auxin-dependent secondary growth initiation [11], [42], thus making the system an ideal tool for studying the process of cambium regulation.
Figure 2. Marker analysis during CIS–incubation and sampling strategy.
(A–E) The activity of the DR5rev:GFP reporter in CIS incubated stems (A–D) without NAA after 2 (A) or 5 days (B), and with apically applied NAA after 2 (C) or 5 days (D) in comparison to a fragment taken from the base of the inflorescence stem of an 18 cm tall plant (E). (F and G) APL:GUS detection in mock-treated (F) and NAA-treated (G) samples after 5 days of CIS-incubation. Signals in interfascicular regions are indicated by arrows. (H) Sampling strategy by LCM as also indicated in Figure 1A. Size bars in (A,F): 100 µm, same magnification in (A–E) and (F–G). The positions of primary vascular bundles are labeled by asterisks.
Identification of Tissue-Specific Genes Induced during IC Initiation
Taking advantage of the inducibility of secondary growth by CIS-incubation, we identified genes that were up-regulated in a tissue-specific manner during IC initiation. Stem fragments were collected before and after 2 and 5 days of incubation, with and without the addition of NAA. Histological analyses showed that cell divisions were not yet initiated after an incubation of 2 days (data not shown). Subsequently, interfascicular regions were harvested from the resulting five groups of stem fragments by LCM (Figure 1A, Figure 2H and Figure S2). To identify genes with increased mRNA abundance specifically in IC initiating cells, we dissected two different types of tissue from the same sections. Type A represented the starch sheath and/or the dividing area in interfascicular regions. Type B represented the surrounding tissues, including the cortex and interfascicular fibers (Figure 1A, Figure 2H and Figure S2), and served as a control for tissue specificity. From samples incubated without NAA, only type A was dissected (Figure 2H). Thus, eight different samples, each with three biological replicates, were used for tissue collection. From these eight samples, RNA was prepared for micro-array analysis.
To identify genes up-regulated during cambium initiation, we first compared data sets A2 and A3 with data set A1 (Figure 2H, Comparison I, Dataset S1) resulting in 2217 genes whose transcript abundance in IC-initiating cells changed after 2 or 5 days (Table S1). We reasoned that a large fraction of these genes simply responded to our in vitro conditions and thus were not specific to cambium formation. These non-specific genes were identified by comparing data sets A4 and A5 with data set A1 (Figure 2H, Comparison II, Dataset S2), which was obtained from samples incubated without the presence of NAA. Furthermore, we identified genes not exclusively activated in cambium-initiating cells by comparing data sets B2 and B3 with data set B1 (Figure 2H, Comparison III, Dataset S3). We subtracted genes identified as being up-regulated in Comparisons II and III from our initial group of genes (Comparison I), leaving 1004 genes that were classified as being associated with a tissue-specific change in transcript abundance during IC initiation (Table S2). Interestingly, among this group, only 132 genes were found to be down-regulated, with 872 genes being up-regulated. We refer to the up-regulated genes as Group 1 (Table S2). Group 1 was further subdivided into ‘early’, ‘late’, or ‘transient’ genes according to their relative expression levels during our time course analysis (Table S2 and Figure S3). To select candidate genes for more detailed analysis, we further reduced Group 1 by applying more stringent selection criteria. To this end, we decreased the statistical thresholds for Comparisons II and III (fold change of 1.41), thereby subtracting more genes from Comparison I. This procedure resulted in a reduced list of 117 up-regulated genes, which we designated as Group 2 (Table S3). The relative expression levels of a subset of these genes were confirmed by qRT-PCR in all isolated samples (Figure S4).
Cambium-Inducing Cells Establish a Unique Transcriptional Profile
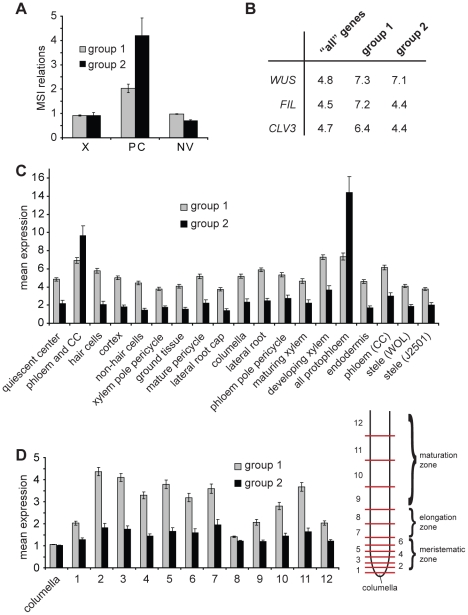
Next, we were interested to see whether there were indications that the identified genes were indeed cambium-specific. Therefore, we analyzed their expression values in different tissues employing data from tissue-specific transcriptional profilings described previously. Characterization of the transcript profile of a mixture of cambium and phloem tissues from hypocotyls represents the closest approximation to a cambium-specific profile described for Arabidopsis to date [21]. Calculating the relationship between mean signal intensities (MSI) in phloem/cambium, xylem, and non-vascular tissues [21] for genes from Group 1, we found a bias toward a phloem/cambium-specific expression (Figure 3A). The same analysis for genes from Group 2 showed an even stronger bias, suggesting that, by applying more stringent selection criteria, we increased tissue specificity within the group of selected genes (Figure 3A). Consistently, analysis of the expression of the selected genes according to high-resolution expression maps of the shoot apical meristem and the root tip [36], [37], revealed a bias towards expression in phloem tissues with a stronger bias for Group 2 (Figure 3C). Interestingly, we did not observe a bias toward a stem cell-specific expression in root or shoot apical meristems (Figure 3B–3D), arguing against a large overlap of gene expression profiles when comparing lateral and apical meristems. Importantly, the search for known cambium markers identified PXY and ATHB8 as members of Group 1 and 2. Both genes belong to the small group of genes described to be specifically expressed in (pro)cambium cells [24], [43], and their presence serves as an indicator for the tissue specificity of the identified group of genes in intact plants. Because WOX4, a cambium regulator identified recently [29], is not present on the ATH1 array, we performed qRT-PCR experiments in order to reveal WOX4 transcript abundance in our LCM-derived samples. This analysis showed that the WOX4 transcript accumulates preferentially in interfascicular regions of 5 day NAA-treated fragments (Figure S4). Taken together, we conclude that cambium identity is established in interfascicular regions of CIS-incubated fragments, and that our approach provides a series of genes expressed in a tissue specific manner at different stages during cambium formation in intact plants.
Figure 3. Expression of identified genes in various tissues.
(A) Average of mean array signal intensity (MSI) relations as described [21] for genes present in Groups 1 and 2 comparing xylem (X), phloem/cambium (PC) and non-vascular (NV) tissues from hypocotyls. (B) Percentage of genes classified as being specifically expressed in WUS, FIL or CLV3 expression domains (Supplementary Table 5 in [37]) comparing ‘all’ genes present in the genome, genes found in Group 1, and in Group 2, respectively. (C and D) Radial (C) and longitudinal (D) distribution of expression levels of genes from Group 1 and 2 in root meristems based on the values for the top 50% of varying probe sets described in Supplementary Table 12 in [36]. Average expression levels of genes listed in Table S2 (Group 1) and listed in Table S3 (Group 2) are shown. In (C), domains are defined by the expression of GFP marker lines. For (D), roots were dissected at different longitudinal positions resulting in samples representing subsequent developmental stages (see [36] for details). Sample 1 contains the quiescent center and tissue-specific stem cells. CC = companion cells.
Identified Genes Are Expressed in the Cambium of Intact Plants
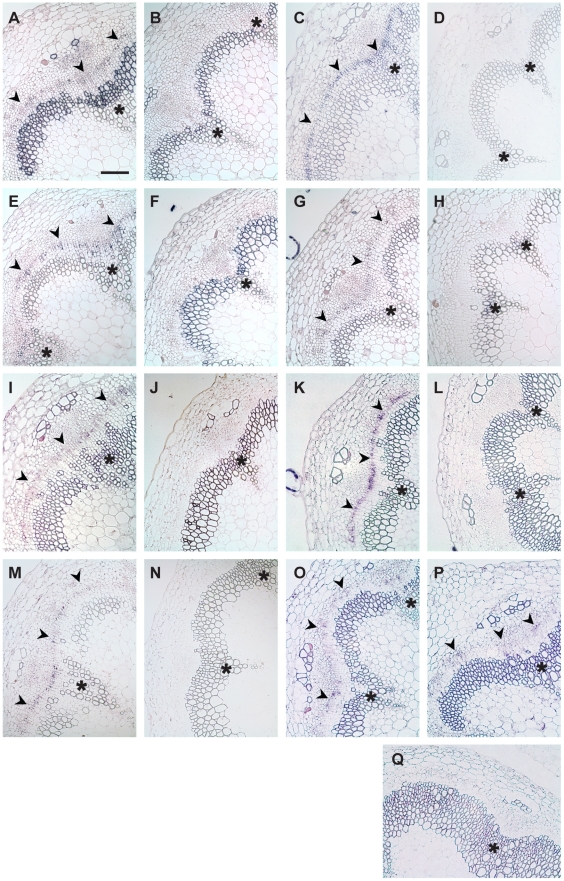
To validate the above conclusion, we concentrated on ‘early’ genes from Group 2, categorized as being involved in developmental processes, signal transduction, and transcription (see GO annotations, http://www.arabidopsis.org/tools/bulk/go/index.jsp, for a more detailed analysis). As we were primarily interested in genes specifically regulating secondary growth, we eliminated those genes for which severe embryo defects in corresponding mutants had been described [44], leaving us with 13 genes (Table 1). Expression of these genes and WOX4 was analyzed by RNA in situ hybridization (RISH) with the exception of AHP3 and ATSEN1, for which no specific RNA probes could be designed due to extended sequence similarity to close homologues. For the remaining genes, sense and antisense probes were hybridized to cross-sections taken from immediately above the uppermost rosette leaf of 30 cm-tall inflorescence stems. For those genes for which GUS reporter lines were available [45]–[48], corresponding reporter lines were analyzed by using a GUS-specific RNA probe. As a result, for all but three cases in which no mRNA accumulation could be detected (Table 1), mRNA was found to accumulate in a continuous domain consisting of one to three cells in a radial orientation located between xylem and phloem tissue (Figure 4). The domain of RNA accumulation detected for all positive genes was similar to that of the cambium-specific genes PXY and WOX4, suggesting that all genes are similarly cambium-specific or, at least, transcribed in close proximity. This verification of the cambium-specific mRNA accumulation of a large proportion of the genes identified suggests that we had indeed elucidated the mRNA profile established during cambium formation in planta, and that CIS incubation can be used for the identification and analysis of cambium regulators.
Table 1. Genes selected for RISH analysis.
| AGI code | Name/description | FC, 2 days | FC, 5 days | Cambium expression (RISH) | Reference |
| AT5G05160 | RUL1 | 2.25 | 12.91 | ✓ | this work |
| AT1G46480 | WOX4 | 0.92 | 4.23 | ✓ | [28], [29] |
| AT1G52340 | ABA2 | 1.46 | 3.83 | - | [47] |
| AT5G61480 | PXY | 1.56 | 3.77 | ✓ | [24]-[26] |
| AT4G32880 | ATHB8 | 1.06 | 3.68 | ✓ | [43] |
| AT5G51350 | MOL1 | 1.45 | 3.31 | ✓ | this work |
| AT5G39340 | AHP3 | 1.84 | 3.17 | no probe | [71] |
| AT5G57110 | ACA8 | 1.56 | 2.94 | ✓ | [46] |
| AT2G26070 | RTE1 | 1.83 | 2.89 | - | [48] |
| AT3G45590 | ATSEN1 | 1.72 | 2.85 | no probe | [72] |
| AT2G27040 | AGO4 | 1.46 | 2.61 | ✓ | [73] |
| AT5G57130 | unknown protein | 1.33 | 2.56 | ✓ | |
| AT1G11130 | SCM | 1.72 | 2.30 | ✓ | [45] |
| AT2G41070 | ATBZIP12 | 1.47 | 2.12 | - |
Figure 4. Transcript detection by RISH using antisense and sense probes.
(A and B) AT5G05160/RUL1, (C and D) PXY, (E and F) ATHB8, (G and H) AT5G51350/MOL1, (I and J) AGO4, (K and L) AT5G57130, (M and N) WOX4, (O) SCM (GUS probe), (P) ACA8 (GUS probe), (Q) GUS sense probe on ACA8:GUS line. (A, C, E, G, I, K, M, O, P) show results using antisense probes, (B, D, F, H, J, L, N, Q) using sense probes. Arrowheads indicate cambium-specific mRNA accumulation and asterisks label the position of primary vascular bundles. Stem sections come from immediately above the uppermost rosette leaf of 15 cm tall plants. Size bar in (A): 100 µm, same magnification in (A–Q).
MOL1 and RUL1 Function As Opposing Regulators of Cambium Activity
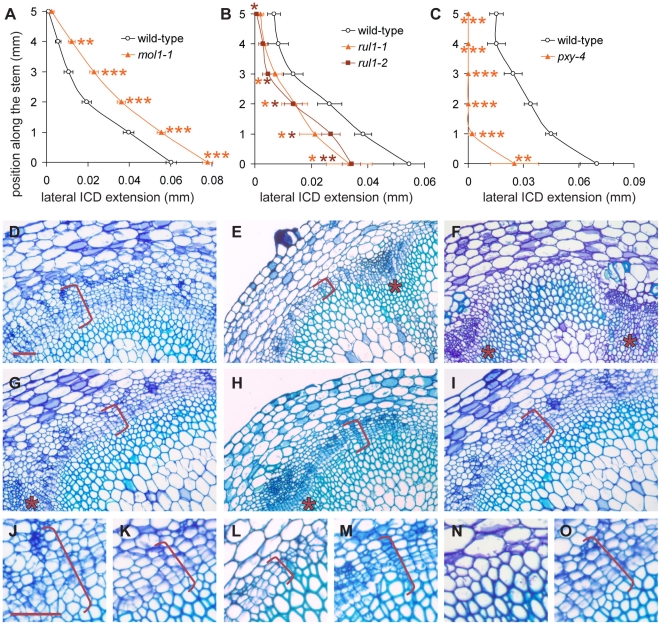
As IC initiation is a highly dynamic process likely to involve extensive cell-to-cell communication, we predicted that the identified signaling components play an important role in cambium regulation. To test this assumption, we analyzed lines impaired in the expression of the uncharacterized leucine-rich repeat receptor-like kinases (LRR-RLKs) present on our reduced list of genes (Table 1, Figure S5) and which we designated MORE LATERAL GROWTH1 (MOL1) and REDUCED IN LATERAL GROWTH1 (RUL1). As a reference for plants affected in cambium activity, we also included pxy-4 mutants [24] in our analysis. All homozygous T-DNA insertion lines (Figure S5) were indistinguishable from wild-type with respect to their overall growth behavior. Histological analyses showed that most pxy-4 mutant plants did not initiate IC formation (Figure 5), thus confirming the role of PXY in secondary growth and demonstrating that pxy-specific defects [26] prevent the establishment of a closed cambium cylinder in the stem. In contrast, IC was detected in the two other mutants with no sign of altered tissue patterning (Figure 5). However, mol1 mutants displayed an enhanced formation of secondary vascular tissue in fascicular and interfascicular regions that exceeded the wild-type by 30%, indicating a substantial increase in cambium activity. By contrast, IC-based tissue formation was decreased by 40% in rul1 mutants, suggesting a reduction in cambium activity in rul1 mutant backgrounds (Figure 5). These results indicate that both genes regulate the production of secondary vascular tissues with MOL1 functioning as a repressor, and RUL1 as an activator, of cambium activity.
Figure 5. Quantification of IC activity.
(A–C) Lateral extension of the IC together with the IC-derived tissue (ICD) in different genetic backgrounds at different positions along the shoot base. See Figure S5 for allele characterization. Significance levels are indicated by asterisks with the corresponding color. (D–I) Histological representations of mol1-1 (D), rul1-2 (E), and pxy-4 (F) mutants in comparison to corresponding wild-type plants (G, H, I). Brackets indicate the extension of the ICD. (J–O) Higher magnification of mol1-1 (J), rul1-2 (H), pxy-4 (N) mutants and the corresponding wild-type plants (K, M, O) as shown in (D–I). Size bar in (D) and (J): 50 µm, same magnification in (D–I) and in (J–O). Sections from immediately above the uppermost rosette leaf are shown (i.e. position 0 in A–C). Positions of primary vascular bundles are labeled by asterisks in (E–I).
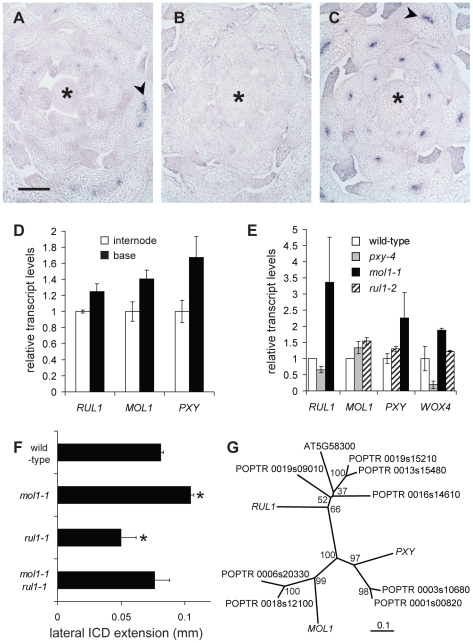
To analyze the interaction of both signaling components, we first determined whether they are generally co-expressed by performing RISH in vegetative shoot tips. This revealed that RUL1 mRNA localizes to the (pro)cambium mainly in more mature leaf primordia (Figure 6A). In contrast, no MOL1 mRNA accumulation was detected in the apex, suggesting that it is specific to the cambium in the stem (Figure 6B). Both expression domains are unlike the PXY expression domain, which is found in procambium strands starting in very young leaf primordia (Figure 6C). However, qRT-PCR analyses showed that the mRNA abundance of all three genes correlates positively with the progression of secondary growth along the stem (Figure 6D). Analysis of transcript accumulation in the respective mutant backgrounds revealed an increase of RUL1 and PXY mRNA abundance in mol1-1 (Figure 6E). As a possible downstream target of the identified receptors, we also analyzed WOX4 transcript abundance in rul1-2, mol1-1, and pxy-4 backgrounds. Consistent with a role of PXY as an activator of WOX4 [29], the analysis revealed decreased WOX4 transcript levels in pxy-4 mutants (Figure 6E). In contrast, as for RUL1 and PXY mRNAs, an increase of WOX4 mRNA levels in mol1-1 backgrounds was detected, whereas the level was almost unchanged in rul1-2 (Figure 6E). The up-regulation of this selection of cambium-expressed genes in mol1 confirms a role of MOL1 as a negative regulator of cambium activity, at least partly, upstream of RUL1 and PXY. To test whether MOL1 and RUL1 function in a linear mode of action or rather in parallel pathways, we analyzed the genetic interaction between MOL1 and RUL1. The analysis of the mol1-1 rul1-1 double mutant revealed a wild-type-like rate of vascular tissue production in interfascicular regions in comparison to the respective single mutants (Figure 6F). Taken together, we conclude that there is no direct and exclusive dependence between MOL1 and RUL1 activity, but that both genes function in parallel in the cambium to balance the production of secondary vascular tissues.
Figure 6. Characterization of RUL1, MOL1, and PXY expression.
(A–C) Detection of RUL1 (A), MOL1 (B) and PXY (C) transcripts by RISH on cross sections of vegetative shoot tips. Asterisks label the apical meristem, arrowheads the cambium-specific signals in leaf bundles. Size bar in (A): 100 µm, same magnification in (A–C). (D) qRT-PCR-based analysis of transcript accumulation in the bottom-most centimeter of the stem and a fragment 3 cm further apically (compare Figure 1C). (E) qRT-PCR-based analysis of RUL1, MOL1, PXY, and WOX4 mRNA abundance in the first internode above the rosette of the corresponding mutants. (F) Genetic interaction between MOL1 and RUL1. Lateral extension of the ICD immediately above the uppermost rosette leaf is shown. (G) Unrooted tree for RUL1, MOL1, and PXY protein sequences, and their closest homologs from Populus trichocarpa based on full length protein sequences. There are two homologs each for MOL1 and PXY with a similar degree of sequence similarity. For RUL1, the situation is less straightforward as another Arabidopsis protein (AT5G58300) belongs to the same sub-clade partly displaying low bootstrap values. The scale bar represents 0.1 amino acid substitutions per position. Bootstrap values are given in percentage.
Discussion
We have characterized genome-wide transcriptome remodeling during cambium formation in a stage- and tissue-specific manner, and identified two novel signaling components involved in the regulation of cambium activity. Cambium analysis has been hampered in the past by the lack of a defined model system in a background in which extensive molecular and genetic tools are available. The formation of procambial strands during Arabidopsis leaf formation is an appropriate model for the establishment of vascular tissue identity and patterning [13], [14]. However, due to a lack of a continuous and indeterminate stem cell activity in leaves, the analysis of cambium regulation is inevitably restricted to shoots and roots. IC formation represents an attractive target for addressing various aspects of cambium regulation, mainly due to its traceability by prominent histological landmarks and the de novo initiation of the cambium-specific stem cell niche from differentiated and unrelated cell types [4]. These features allow detailed analysis of the sequence of molecular events resulting in the establishment of cambium identity, potentially allowing the identification of key regulatory steps. With respect to the availability of tools, Arabidopsis represents the ideal species for application of this approach [49] and these tools can be exploited even more efficiently when the process of interest can be tightly controlled by an appropriate experimental system. By introducing CIS-incubation of Arabidopsis stem fragments, we have established such a system for cambium formation. Marker gene analysis, transcript profiling, and reverse genetics support the conclusion that CIS accurately reflects the situation in intact plants with respect to secondary growth regulation and provides experimental controllability of a highly complex developmental process. By following this strategy, we present ways of using the most established plant model as an efficient tool for deciphering the mechanisms of cambium regulation. These findings are especially significant as the cambium is considered to be the least studied, and least understood, indeterminate plant meristem [9], [50].
Ex planta tissue-culture systems have been used very successfully for analyzing various plant developmental processes [38], [42], [51], [52], demonstrating the robustness of plant organ dynamics when exposed to selected key signals. In our case, the pivotal factor required to induce cambium activity is auxin applied apically to isolated stem fragments. The specific responsiveness of such fragments provides another example of the key role of basipetal auxin transport in cambium initiation and activity [11], [53], [54]. Surprisingly, we did not find that genes classified as being either auxin-inducible or auxin signaling components were overrepresented among the genes identified as being induced during IC initiation. This might suggest that the processes involved in IC initiation do not depend on basipetal auxin transport directly, but rather represent tissue-specific processes initiated upon auxin-dependent activation of the fascicular cambium. This is supported by previous analyses showing that the fascicular cambium is activated before IC initiation occurs [4]. Consistent with our findings, genes expressed in gradients along the radial sequence of tissues in Populus stems, and peaking in the cambium, have no preference for being auxin responsive suggesting that only a few of the cambium-specific genes directly depend on auxin signaling [42]. Whether the influence of other hormonal pathways or environmental conditions [9], [12], [19] can be reflected by CIS remains to be seen.
Importantly, this study provides the first tissue-specific transcriptome profiling performed at different stages of cambium formation, and will allow specific steps of the process, such as dedifferentiation, or the establishment of stem cell characteristics, to be addressed in the future. The elucidation of the cambium-specific transcript profile allowed us to compare the profiles of lateral and apical meristems. In contrast to previous concepts [18], [50], we did not find a large overlap between genes expressed in apical and lateral meristems, arguing that the cambium-specific expression profile is highly unique (Figure 3). However, this does not necessarily mean that general mechanisms involved in the regulation of meristematic activity differ in both meristem types. As shown for the case of WOX genes [29], [31], [55], different members of a gene family might be active only in a specific meristem, making an overall comparison of gene expression profiles less straightforward. However, the small group of regulators found to be expressed in different types of stem cells (Figure 3) might provide a good set of candidates for general meristem regulators.
PXY and WOX4 are the only cell-autonomous cambium regulators described to date [24]–[26], [29]. Both genes belong to the group of genes identified in the current study. Interestingly, PXY expression was already detected at early stages of IC formation (Table 1) and we observed an almost complete absence of IC formation in pxy mutants (Figure 5). However, whether the function of PXY is fundamental for IC formation directly, or whether the pxy phenotype is simply due to a reduction of the activity of the fascicular cambium [26], thus preventing a spread of cambium activity into interfascicular regions, remains to be clarified. Even though PXY is expressed early during procambium formation in young leaf primordia (Figure 6) [26], no defect in pxy mutants during procambium formation has been reported [24]–[26]. Therefore, PXY seems to specifically activate and organize the stem cell population in the cambium. The early expression during IC formation might be essential for this reason. The temporal relationship between PXY and ATHB8 expression [14], [43] during procambium formation in leaves remains to be elucidated. The detection of ATHB8 transcription in cells gathering IC identity suggests some regulatory parallels to procambium formation. In contrast, PIN1 and MP, two genes working together with ATHB8 in a concerted manner during procambium formation in leaves, and expressed similarly in a very localized manner [13]–[15], were not detected. Although the dynamics of PIN1 protein accumulation [14], [15], and transcript accumulation could differ, this might argue for an alternative mechanism functioning during early phases of IC formation, similarly resulting in ATHB8 activation.
By performing reverse genetics on the two uncharacterized signaling components in our reduced list of genes (Table 1), we identified RUL1 and MOL1 - two receptor-like kinases - as regulators of cambium activity, each with opposite effects (Figure 5, Figure 6). RUL1 and MOL1 belong to the subgroups LRR-III and LRR-XIV, respectively, of the LRR-RLK family [56]. Based on knowledge about other LRR-RLKs, it is attractive to hypothesize that the products of these two genes might function to recognize and communicate long- and/or short-range signals to cambium cells. In addition to the cambium of the shoot, the positive regulator RUL1 is also expressed in procambium cells of leaf primordia (Figure 6). Because the overlap of expression domains is only partial, a direct and exclusive mutual regulation of the expression of both factors seems to be unlikely. A negative effect of MOL1 on RUL1, PXY (LRR-XI subgroup) and WOX4 expression in the shoot was detected, confirming the role of MOL1 as a repressor of cambium activity that seems to be, at least partly, upstream of the other three factors (Figure 6). However, the genetic interaction between MOL1 and RUL1 rather argues for a parallel mode of action and suggests that the increased RUL1 transcription in the MOL1-defective background is a secondary effect based on enhanced cambium activity. Interestingly, the negative effect of MOL1 on WOX4 expression shows parallels to the situation in shoot and root apical meristems in which CLV and ACR4-dependent signaling pathways inhibit WUS and WOX5 expression, respectively [57], [58].
As cambium activity is regulated by various endogenous as well as environmental stimuli [9], [12], [19], the presence of numerous parallel, interacting and counteracting pathways mediating these inputs is expected. For example, in addition to activating pathways, the inhibitory role of the two MADS box transcription factors SOC1 and FUL on secondary growth has been reported [59], demonstrating that active repression of cambium activity exists in Arabidopsis. To understand the role of both novel receptors in detail, the nature of the input that is recognized by them, especially their ligands and the origin of these ligands, has to be identified. In addition to MOL1, RUL1 and PXY, we also found SCM - an LRR-RLK originally described to control epidermal patterning in roots [45], [60] - as being transcribed in the cambium (Table 1). Furthermore, more LRR-RLKs are induced during IC formation in a slightly less specific manner (Table S2), including BAM1, BAM2, and BAM3, which, among other functions, promote stem cell activity in the shoot apical meristem and are involved in the regulation of leaf venation [61]. Considering the potential crosstalk between corresponding signaling pathways, including the putative formation of higher order complexes between LRR-RLKs [62]–[64], we envisage a highly intricate cambium regulation at the level of membrane-bound receptors.
Interestingly, orthologs of MOL1 and RUL1 seem to exist in Populus (Figure 6G), and therefore it would be of great interest to see whether gene functions are conserved between herbaceous and woody species. The identification of specific factors up- and downstream of RUL1 and MOL1 will be the next step towards integrating their function into the network of systems regulating wood production. As wood is an important source of sustainable energy and an important sink for atmospheric CO2 [65], a better understanding of the molecular mechanisms regulating its production is highly desirable.
Materials and Methods
Plant Materials, Growth Conditions, and Histology
Arabidopsis thaliana (L.) Heynh. plants (Col-0 accession) were used for all experiments unless stated otherwise. T-DNA insertion lines were obtained from the Nottingham Arabidopsis Stock Center (NASC). Plants were grown for 3 weeks under short-day conditions (8 h light (10,000 LUX), 16 h dark, 21°C, 60% humidity), and then shifted to long-day conditions (16 h light (21°C, 10,000 LUX), 8 h dark (16°C), 60% humidity) to induce flowering. For histological analyses of intact plants, individuals 15–20 cm in height in which the first internode was at least 3.5 cm long were selected, unless stated otherwise. Except for LCM analysis, stem fragments were embedded in paraffin, sectioned, stained, and analyzed, including GUS reporter analysis, as previously described [4]. For quantitative analyses, at least five plants were evaluated for each data point. The standard errors of means were used to visualize variation. Histological data were subjected to statistical analysis using a two-tailed independent Student’s t test with SPSS 18.0 software (http://www.spss.com). Significance levels of P<0.05, P<0.01 and P<0.001 are indicated by single, double, and triple asterisks, respectively.
CIS–Incubation
Standard Petri dishes (9 cm in diameter) were filled with 45 ml of ½ MS. A 6 mm-wide strip was removed from the center of the plate. Suitable volumes of stock solutions (1 mg/ml) of NAA or NPA were applied on the media halves at standardized positions. Plates were left at room temperature for 3 days to allow diffusion. For stem fragment preparation, the entire first internode, which was at least 5 cm in length, was collected from plants 15–20 cm tall. After both ends were sealed with liquid wax, samples were surface sterilized by 70% ethanol (45 s) and washed four times with sterile water (1 min each). Stem fragments of 1.5 cm in length located 2.5 cm apically from the base were excised (Figure 1C) with a scalpel and transferred to the ‘split-plates’. After 2 or 5 days of incubation, the central area (3 mm) of these samples, which had no direct contact with the medium, was collected for further analysis.
LCM, RNA Extraction, and RNA Amplification
For LCM analysis, samples were snap-frozen in OCT embedding medium (Labonord Cryoblock, France) in Peel-A-Way disposable histology moulds (Polysciences Inc., Warrington, PA, USA) and stored at −80°C. Transverse sections (5 µm) were cut with an HM550 cryostat (Microm, Walldorf, Germany) at −20°C. Cryosections were mounted on PET-membrane-coated stainless steel slides (Leica Microsystems, Wetzlar, Germany) and processed as described previously [33]. A Leica AS Laser Microdissection system (Leica Microsystems, Wetzlar, Germany) was used to harvest cells. In each case, three biological replicas consisting of five plants each were analyzed. In total, ∼30,000 cells per biological repetition were dissected. The settings for dissection were: 10× magnification; aperture: 15; intensity: 45; speed: 6; bridge: medium; offset: 22. Microdissected tissue was collected in the lid of a 0.5 ml microtube containing RLT buffer from the RNeasy Plus Micro Kit (Qiagen, Hilden, Germany). Total RNA was extracted using the same kit following the manufacturer’s instructions. Two RNA amplification rounds were performed using the TargetAm 2-Round Aminoallyl-aRNA Amplification Kit (EPICENTRE Biotechnologies, Madison, USA) following the manufacturer’s instructions. The quality of the amplified RNA was evaluated by OD260/OD280 measurements and agarose gel electrophoresis.
Microarray Hybridizations and Analyses
For Microarray hybridization, we used the hybridization services of NASC (http://affymetrix.arabidopsis.info/), which employ the ATH1 array (Affymetrix, Santa Clara, US). The Robust Multi-Array (RMA) method from the Bioconductor software package [66] was used for normalization and analysis. A P value of 0.05 and a fold-change of 2 were chosen as initial thresholds for selecting differentially expressed genes.
Accessing Microarray Data
Raw data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [67] and are accessible through GEO Series accession number GSE22947 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22947).
Analyses of Transcript Accumulation
For RNA extraction from whole stem samples, a standard Trizol-based (Invitrogen, Carlsbad, USA) protocol was used. Subsequently, RNA was purified using RNA-MiniElute columns (Qiagen, Hilden, Germany). Genomic DNA was eliminated using RNase-free DNase (Qiagen, Hilden, Germany) by column purification according to the manufacturer’s instructions and the quality of purified RNA was tested using the OD260/OD280 ratio and gel electrophoresis. In all cases cDNA templates were produced using the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany). Real-time quantitative PCR analysis was performed using the SensiMix SYBR & ROX kit (Peqlab, Erlangen, Germany), following the manufacturer’s instructions. 15 µl-volume reactions were performed utilizing an iQTM5 optical system (Bio-Rad, Hercules, USA). In order to transform fluorescence intensity data into cDNA levels a standard curve with a 10-fold dilution series of a single cDNA sample was constructed. For each sample, results obtained for a given gene were normalized using the value of an internal standard gene (EIF4A1; AT3G13920) based on the comparative threshold (CT) method as described by Perkin-Elmer Applied Biosystems (http://www.perkinelmer.com). After that, the value for the gene of interest in the control sample was set to 1.0 and relative values for the rest of the samples were normalized accordingly. The specificity of the amplification reactions was assessed using postamplification dissociation curves. In all cases, at least two biological and two technical replicates were carried out, resulting in four qRT-PCR reactions per gene. RISH was carried out as explained previously [3]. For probe synthesis, PCR products generated using cDNA as a template were cloned into the pGEM-T vector (Promega, Madison, USA) and used as a template for transcription from the T7 or SP6 promoter. Primers employed for generating PCR products for probe synthesis and qRT-PCR are depicted in Table S4.
Sequence Comparisons
Populus trichocarpa sequences were identified using the BLAST tool available on the Phytozome portal (http://www.phytozome.net/poplar). Full length protein sequences were aligned by ClustalW (http://clustalw.ddbj.nig.ac.jp/top-e.html) [68] using the default parameters (protein weight matrix: blosum; gap opening penalty: 10; gap extension penalty: 0.2; gap separation distance: 8) and visualized by TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Following this approach, the phylogenetic tree is constructed by using the neighbor-joining (NJ) method [69] and the Kimura method for estimating the number of amino acid substitutions between sequences [70]. Bootstrap values were obtained by 1,000 bootstrap replicates.
Supporting Information
Expression data for all genes spotted on the ATH1 array in comparison I.
(9.56 MB TXT)
Expression data for all genes spotted on the ATH1 array in comparison II.
(9.56 MB TXT)
Expression data for all genes spotted on the ATH1 array in comparison III.
(9.56 MB TXT)
Dose-response analysis of CIS-incubation. (A–F) Stem fragments were incubated for 5 days on split-plates containing 0 (A), 0.03 (B), 0.1 (C), 0.3 (D), 1 (E) and 3 (F) µg/ml NAA in the apical half of the plate. Expansion of the IC-derived tissue is indicated by brackets. An enhancement of the effect up to 1 µg/ml was observed. The positions of primary bundles are labeled by asterisks. Size bar in (A): 100 µm, same magnification in (A–F).
(6.38 MB TIF)
LCM sampling of tissues. (A and B) The starch sheath/dividing zone was labeled (A) and dissected by the laser beam (B). (C and D) Subsequently, the surrounding tissue was labeled (C) and dissected (D). Positions of primary vascular bundles are labeled by asterisks.
(5.51 MB TIF)
Classification of genes found in group 1 into (A) "early" (309), (B) "late" (276), and (C) "transient" (287) genes. See Table S2 for gene identifications.
(7.40 MB TIF)
Validation of microarray-based detection of mRNA abundance and analysis of WOX4 mRNA abundance in LCM-derived samples by qRT-PCR. (A) ACA8 (AT5G57110), (B) AT2G41230, (C) AT2G47180, (D) AT1G02850, (E) MOL1 (AT5G51350), (F) PXY (AT5G61480), (G) RTE (AT2G26070), (H) RUL1 (AT5G05160), (I) SCM (AT1G11130), (J) AT5G54690, (K) WOX4 (AT1G46480). Values obtained by qRT-PCR are in grey, values obtained by microarray analysis are in black. For sample labeling, see Figure 1A and Figure 2H.
(5.38 MB TIF)
Schematic representation of MOL1 (A) and RUL1 (C) gene and protein structure and gene expression in corresponding mutants. The upper panels show the genomic organization, with the transcribed region in gray and white, and exons in white; the lower panels show protein domains as elucidated using CLC Main Workbench 5.5 software. T-DNA insertions are indicated by triangles. (B and D) Transcript levels of MOL1 and RUL1 in the corresponding mutants as revealed by qRT-PCR. Primer positions used for the RT-PCR are indicated by small arrows above the schemes in (A and C). In addition to mol1-1, we analyzed the T-DNA insertion line SAIL_79_A04 as a potential second mutant allele for MOL1. However, we were unable to confirm that the line carries the T-DNA insertion.
(1.51 MB TIF)
Expression data for genes identified as being differentially expressed in comparison I.
(1.23 MB XLS)
Expression data for genes remaining after genes induced in samples A4, A5, B2, and B3 (see Figure 2H) were subtracted from the genes differentially expressed in comparison I (Table S1). Worksheets for differentially expressed genes, up-regulated genes (Group 1), "early" genes, "transient" genes, and "late" genes are provided.
(0.85 MB XLS)
Expression data for genes remaining (Group 2) after genes induced in samples A4, A5, B2, and B3 identified using less stringent selection criteria were subtracted from the genes differentially expressed in comparison I (Table S1).
(0.08 MB XLS)
Primers used for performing qRT-PCR (A) and generating RNA probes (B).
(0.05 MB PDF)
Acknowledgments
We are grateful to Leslie E. Sieburth, Philip Benfey, Ortrun Mittelsten Scheid, and Daniel Schubert for critical reading of the manuscript. We thank Klaus Kaserer and Bettina Pichlhöfer (Clinical Institute for Pathology, Medical University Vienna) for access to LCM facilities, Vukoslav Komnenovic for excellent histological support, and Bob Zimmermann and Huy Dinh for help with bioinformatics. Wan-Hsing Cheng provided the ABA2:GUS line, Caren Chang the RTE1:GUS line, John Schiefelbein the SCM:GUS line, and Robert Sharrock the ACA8:GUS line.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grant P21258-B03 of the Austrian Science Fund (FWF, http://www.fwf.ac.at/en/index.asp). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc'h A, Carnero E, et al. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 2009;57:626–644. doi: 10.1111/j.1365-313X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- 2.Che P, Lall S, Nettleton D, Howell SH. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 2006;141:620–637. doi: 10.1104/pp.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greb T, Clarenz O, Schäfer E, Müller D, Herrero R, et al. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003;17:1175–1187. doi: 10.1101/gad.260703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sehr EM, Agusti J, Lehner R, Farmer EE, Schwarz M, et al. Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 2010;63:811–822. doi: 10.1111/j.1365-313X.2010.04283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du J, Groover A. Transcriptional regulation of secondary growth and wood formation. J Integr Plant Biol. 2010;52:17–27. doi: 10.1111/j.1744-7909.2010.00901.x. [DOI] [PubMed] [Google Scholar]
- 6.Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, et al. Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 1998;14:425–430. doi: 10.1046/j.1365-313x.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- 7.Altamura MM, Possenti M, Matteucci A, Baima S, Ruberti I, et al. Development of the vascular system in the inflorescence stem of Arabidopsis. New Phyt. 2001;151:381–389. [Google Scholar]
- 8.Sauer M, Balla J, Luschnig C, Wisniewska J, Reinohl V, et al. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 2006;20:2902–2911. doi: 10.1101/gad.390806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elo A, Immanen J, Nieminen K, Helariutta Y. Stem cell function during plant vascular development. Semin Cell Dev Biol. 2009;20:1097–1106. doi: 10.1016/j.semcdb.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Schrader J, Moyle R, Bhalerao R, Hertzberg M, Lundeberg J, et al. Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J. 2004;40:173–187. doi: 10.1111/j.1365-313X.2004.02199.x. [DOI] [PubMed] [Google Scholar]
- 11.Little CHA, MacDonald JE, Olsson O. Involvement of indole-3-acetic acid in fascicular and interfascicular cambial growth and interfascicular extraxylary fiber differentiation in Arabidopsis thaliana inflorescence stems. International Journal of Plant Sciences. 2002;163:519–529. [Google Scholar]
- 12.Schrader J, Baba K, May ST, Palme K, Bennett M, et al. Polar auxin transport in the wood-forming tissues of hybrid aspen is under simultaneous control of developmental and environmental signals. Proc Natl Acad Sci U S A. 2003;100:10096–10101. doi: 10.1073/pnas.1633693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donner TJ, Sherr I, Scarpella E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development. 2009;136:3235–3246. doi: 10.1242/dev.037028. [DOI] [PubMed] [Google Scholar]
- 14.Scarpella E, Marcos D, Friml J, Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20:1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wenzel CL, Schuetz M, Yu Q, Mattsson J. Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J. 2007;49:387–398. doi: 10.1111/j.1365-313X.2006.02977.x. [DOI] [PubMed] [Google Scholar]
- 16.Sachs T. The control of the patterned differentiation of vascular tissues. Adv Bot Res. 1981;9:151–162. [Google Scholar]
- 17.Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, et al. Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 2005;42:618–640. doi: 10.1111/j.1365-313X.2005.02403.x. [DOI] [PubMed] [Google Scholar]
- 18.Ko JH, Han KH. Arabidopsis whole-transcriptome profiling defines the features of coordinated regulations that occur during secondary growth. Plant Mol Biol. 2004;55:433–453. doi: 10.1007/s11103-004-1051-z. [DOI] [PubMed] [Google Scholar]
- 19.Ko JH, Han KH, Park S, Yang J. Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol. 2004;135:1069–1083. doi: 10.1104/pp.104.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh S, Park S, Han KH. Transcriptional regulation of secondary growth in Arabidopsis thaliana. J Exp Bot. 2003;54:2709–2722. doi: 10.1093/jxb/erg304. [DOI] [PubMed] [Google Scholar]
- 21.Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 2005;138:803–818. doi: 10.1104/pp.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, et al. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell. 2004;16:2278–2292. doi: 10.1105/tpc.104.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groover AT, Mansfield SD, DiFazio SP, Dupper G, Fontana JR, et al. The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol Biol. 2006;61:917–932. doi: 10.1007/s11103-006-0059-y. [DOI] [PubMed] [Google Scholar]
- 24.Fisher K, Turner S. PXY, a Receptor-like Kinase Essential for Maintaining Polarity during Plant Vascular-Tissue Development. Curr Biol. 2007;17:1061–1066. doi: 10.1016/j.cub.2007.05.049. [DOI] [PubMed] [Google Scholar]
- 25.Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci U S A. 2008;105:15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etchells JP, Turner SR. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development. 2010;137:767–774. doi: 10.1242/dev.044941. [DOI] [PubMed] [Google Scholar]
- 27.De Smet I, Voss U, Jurgens G, Beeckman T. Receptor-like kinases shape the plant. Nat Cell Biol. 2009;11:1166–1173. doi: 10.1038/ncb1009-1166. [DOI] [PubMed] [Google Scholar]
- 28.Ji J, Strable J, Shimizu R, Koenig D, Sinha N, et al. WOX4 promotes procambial development. Plant Physiol. 2009;152:1346–1356. doi: 10.1104/pp.109.149641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirakawa Y, Kondo Y, Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 32.Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003;132:27–35. doi: 10.1104/pp.102.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agusti J, Merelo P, Cercos M, Tadeo FR, Talon M. Comparative transcriptional survey between laser-microdissected cells from laminar abscission zone and petiolar cortical tissue during ethylene-promoted abscission in citrus leaves. BMC Plant Biol. 2009;9:127. doi: 10.1186/1471-2229-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet. 2009;41:258–263. doi: 10.1038/ng.282. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Madi S, Borsuk L, Nettleton D, Elshire RJ, et al. Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet. 2007;3:e101. doi: 10.1371/journal.pgen.0030101. doi: 10.1371/journal.pgen.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 37.Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci U S A. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatfield SP, Stirnberg P, Forde BG, Leyser O. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 2000;24:159–169. doi: 10.1046/j.1365-313x.2000.00862.x. [DOI] [PubMed] [Google Scholar]
- 39.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 40.Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y. APL regulates vascular tissue identity in Arabidopsis. Nature. 2003;426:181–186. doi: 10.1038/nature02100. [DOI] [PubMed] [Google Scholar]
- 41.Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsson J, Karlberg A, Antti H, Lopez-Vernaza M, Mellerowicz E, et al. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell. 2008;20:843–855. doi: 10.1105/tpc.107.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scarpella E, Francis P, Berleth T. Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development. 2004;131:3445–3455. doi: 10.1242/dev.01182. [DOI] [PubMed] [Google Scholar]
- 44.Devic M. The importance of being essential: EMBRYO-DEFECTIVE genes in Arabidopsis. C R Biol. 2008;331:726–736. doi: 10.1016/j.crvi.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 45.Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science. 2005;307:1111–1113. doi: 10.1126/science.1105373. [DOI] [PubMed] [Google Scholar]
- 46.George L, Romanowsky SM, Harper JF, Sharrock RA. The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol. 2008;146:716–728. doi: 10.1104/pp.107.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, et al. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol. 2007;143:745–758. doi: 10.1104/pp.106.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong CH, Rivarola M, Resnick JS, Maggin BD, Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 2008;53:275–286. doi: 10.1111/j.1365-313X.2007.03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Elo A, Helariutta Y. Arabidopsis as a model for wood formation. Curr Opin Biotechnol Epub ahead of print. 2010 doi: 10.1016/j.copbio.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Groover AT. What genes make a tree a tree? Trends Plant Sci. 2005;10:210–214. doi: 10.1016/j.tplants.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, et al. Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 2009;5:e1000621. doi: 10.1371/journal.pgen.1000621. doi: 10.1371/journal.pgen.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sauer M, Friml J. In vitro culture of Arabidopsis embryos. Methods Mol Biol. 2008;427:71–76. doi: 10.1007/978-1-59745-273-1_5. [DOI] [PubMed] [Google Scholar]
- 53.Snow R. Activation of cambial growth by pure hormones. New Phyt. 1935;34:347–360. [Google Scholar]
- 54.Björklund S, Antti H, Uddestrand I, Moritz T, Sundberg B. Cross-talk between gibberellin and auxin in development of Populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 2007;52:499–511. doi: 10.1111/j.1365-313X.2007.03250.x. [DOI] [PubMed] [Google Scholar]
- 55.Laux T, Mayer KF, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 56.Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stahl Y, Wink RH, Ingram GC, Simon R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19:909–914. doi: 10.1016/j.cub.2009.03.060. [DOI] [PubMed] [Google Scholar]
- 58.Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 59.Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, et al. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nature Genetics. 2008;40:1489–1492. doi: 10.1038/ng.253. [DOI] [PubMed] [Google Scholar]
- 60.Kwak SH Schiefelbein J. A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr Biol. 2008;18:1949–1954. doi: 10.1016/j.cub.2008.10.064. [DOI] [PubMed] [Google Scholar]
- 61.DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, et al. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhu Y, Wang Y, Li R, Song X, Wang Q, et al. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 2010;61:223–233. doi: 10.1111/j.1365-313X.2009.04049.x. [DOI] [PubMed] [Google Scholar]
- 63.Guo Y, Han L, Hymes M, Denver R, Clark SE. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010;23:889–900. doi: 10.1111/j.1365-313X.2010.04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010;152:166–176. doi: 10.1104/pp.109.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Solomon BD. Biofuels and sustainability. Ann N Y Acad Sci. 2010;1185:119–134. doi: 10.1111/j.1749-6632.2009.05279.x. [DOI] [PubMed] [Google Scholar]
- 66.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Research. 2009;37:D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 69.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 70.Kimura M. Cambridge: Camb.Univ.Press; 1983. The neutral Theory of Molecular Evolution.75 [Google Scholar]
- 71.Hwang I, Chen HC, Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–515. doi: 10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akama K, Junker V, Beier H. Identification of two catalytic subunits of tRNA splicing endonuclease from Arabidopsis thaliana. Gene. 2000;257:177–185. doi: 10.1016/s0378-1119(00)00408-x. [DOI] [PubMed] [Google Scholar]
- 73.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression data for all genes spotted on the ATH1 array in comparison I.
(9.56 MB TXT)
Expression data for all genes spotted on the ATH1 array in comparison II.
(9.56 MB TXT)
Expression data for all genes spotted on the ATH1 array in comparison III.
(9.56 MB TXT)
Dose-response analysis of CIS-incubation. (A–F) Stem fragments were incubated for 5 days on split-plates containing 0 (A), 0.03 (B), 0.1 (C), 0.3 (D), 1 (E) and 3 (F) µg/ml NAA in the apical half of the plate. Expansion of the IC-derived tissue is indicated by brackets. An enhancement of the effect up to 1 µg/ml was observed. The positions of primary bundles are labeled by asterisks. Size bar in (A): 100 µm, same magnification in (A–F).
(6.38 MB TIF)
LCM sampling of tissues. (A and B) The starch sheath/dividing zone was labeled (A) and dissected by the laser beam (B). (C and D) Subsequently, the surrounding tissue was labeled (C) and dissected (D). Positions of primary vascular bundles are labeled by asterisks.
(5.51 MB TIF)
Classification of genes found in group 1 into (A) "early" (309), (B) "late" (276), and (C) "transient" (287) genes. See Table S2 for gene identifications.
(7.40 MB TIF)
Validation of microarray-based detection of mRNA abundance and analysis of WOX4 mRNA abundance in LCM-derived samples by qRT-PCR. (A) ACA8 (AT5G57110), (B) AT2G41230, (C) AT2G47180, (D) AT1G02850, (E) MOL1 (AT5G51350), (F) PXY (AT5G61480), (G) RTE (AT2G26070), (H) RUL1 (AT5G05160), (I) SCM (AT1G11130), (J) AT5G54690, (K) WOX4 (AT1G46480). Values obtained by qRT-PCR are in grey, values obtained by microarray analysis are in black. For sample labeling, see Figure 1A and Figure 2H.
(5.38 MB TIF)
Schematic representation of MOL1 (A) and RUL1 (C) gene and protein structure and gene expression in corresponding mutants. The upper panels show the genomic organization, with the transcribed region in gray and white, and exons in white; the lower panels show protein domains as elucidated using CLC Main Workbench 5.5 software. T-DNA insertions are indicated by triangles. (B and D) Transcript levels of MOL1 and RUL1 in the corresponding mutants as revealed by qRT-PCR. Primer positions used for the RT-PCR are indicated by small arrows above the schemes in (A and C). In addition to mol1-1, we analyzed the T-DNA insertion line SAIL_79_A04 as a potential second mutant allele for MOL1. However, we were unable to confirm that the line carries the T-DNA insertion.
(1.51 MB TIF)
Expression data for genes identified as being differentially expressed in comparison I.
(1.23 MB XLS)
Expression data for genes remaining after genes induced in samples A4, A5, B2, and B3 (see Figure 2H) were subtracted from the genes differentially expressed in comparison I (Table S1). Worksheets for differentially expressed genes, up-regulated genes (Group 1), "early" genes, "transient" genes, and "late" genes are provided.
(0.85 MB XLS)
Expression data for genes remaining (Group 2) after genes induced in samples A4, A5, B2, and B3 identified using less stringent selection criteria were subtracted from the genes differentially expressed in comparison I (Table S1).
(0.08 MB XLS)
Primers used for performing qRT-PCR (A) and generating RNA probes (B).
(0.05 MB PDF)