Abstract
Natural products represent a significant reservoir of unexplored chemical diversity for early-stage drug discovery. The identification of lead compounds of natural origin would benefit from therapeutically relevant bioassays capable of facilitating the isolation of bioactive molecules from multi-constituent extracts. Towards this end, we developed an in vivo bioassay-guided isolation approach for natural product discovery that combines bioactivity screening in zebrafish embryos with rapid fractionation by analytical thin-layer chromatography (TLC) and initial structural elucidation by high-resolution electrospray mass spectrometry (HRESIMS). Bioactivity screening of East African medicinal plant extracts using fli-1:EGFP transgenic zebrafish embryos identified Oxygonum sinuatum and Plectranthus barbatus as inhibiting vascular development. Zebrafish bioassay-guided fractionation identified the active components of these plants as emodin, an inhibitor of the protein kinase CK2, and coleon A lactone, a rare abietane diterpenoid with no previously described bioactivity. Both emodin and coleon A lactone inhibited mammalian endothelial cell proliferation, migration, and tube formation in vitro, as well as angiogenesis in the chick chorioallantoic membrane (CAM) assay. These results suggest that the combination of zebrafish bioassays with analytical chromatography methods is an effective strategy for the rapid identification of bioactive natural products.
Introduction
Small molecules from natural sources are recognized as evolved, privileged structures with greater likelihood than many synthetic compounds to exhibit specific bioactivities [1]. For example, 73% of cancer therapeutics approved to date are either natural products or derivatives thereof [2]. Nevertheless, the use of natural products in drug discovery has significantly declined in the past two decades, due in part to persisting difficulties in the systematic isolation and synthesis of such molecules [3]. One promising strategy to better exploit the therapeutic potential of natural products could be the use of more biomedically relevant assays – ideally in vivo models – for the screening and bioactivity-guided fractionation of plant, fungal and microbial extracts.
Many currently known bioactive natural products were originally identified using in vitro assays (e.g. cytotoxicity in tumor cells) for their activity-guided isolation from extracts. The biological activity of many other natural products was determined only after their initial isolation on the basis of physical characteristics (e.g., preparative chromatography followed by mass spectrometry and NMR spectroscopy analysis). Because of the low throughput of conventional in vivo models such as mice and rats, in addition to the relatively large amounts of compound required for testing in these systems, in vivo assay-guided fractionation is currently not a widely-used approach for the discovery of drug-like natural products.
Novel opportunities for in vivo natural product discovery have arisen through the recent emergence of zebrafish as an effective model system for the identification of disease-relevant genes and bioactive small molecules [4]. Large-scale genetic screens in zebrafish carried out since the early 1990s have led to the identification of therapeutically relevant genes in several indication areas, including cardiovascular, neurological, gastrointestinal, musculoskeletal, and metabolic disorders [5], [6], [7], [8]. In addition, small-molecule screens carried out in zebrafish within the past decade have confirmed the ability of this model system to identify bioactive compounds in a target-independent manner, thereby enabling the discovery of novel mechanisms of action [9], [10], [11], [12], [13].
The primary advantages of zebrafish for drug discovery include their high genetic, physiologic, and pharmacologic similarity with humans, as well as the small size, optical transparency, rapid development, and large numbers of their embryos and larvae, which are the primary system for experimental analysis. Because of their small size (1 to 5 mm), zebrafish embryos and larvae are compatible with microtiter plates for screening (primarily 24- and 96-well plates, but even 384-well plates are possible), thereby requiring only microgram amounts of each extract, fraction, or compound to be tested. Because of the high fecundity of zebrafish, large numbers of embryos and larvae can be produced and analyzed in a more cost-effective manner than, for example, mice and rats. Combined, these features define zebrafish as an ideal in vivo model for the systematic identification of bioactive natural products with therapeutic potential [4].
For an initial evaluation of zebrafish as a platform for natural product discovery, we opted to identify novel inhibitors of angiogenesis. Despite the recent regulatory approval of recombinant antibodies and small molecules targeting the vascular endothelial growth factor (VEGF) pathway, the clinical efficacy of these therapies for various cancers is limited [14]. Also, despite the large number of compounds targeting this pathway [15], many of these have shown limited or insufficient efficacy in clinical trials, or are associated with toxicities such as arterial thromboembolic events [16]. For these reasons, there is still a need for novel anti-angiogenic compounds with different mechanisms of action, some of which might be suitable for use in combinatorial therapy strategies [17].
Numerous in vivo and in vitro assays have been developed since the 1970s for the evaluation of anti-angiogenic molecules [18], yet because of various disadvantages (low throughput, high cost, and the requirement for larger compound amounts in the case of in vivo assays, and limited predictive value in the case of in vitro assays), these are not ideal as front-line assays for natural product discovery (i.e. for high-throughput screening and rapid bioassay-guided fractionation). Zebrafish, however, offer an interesting combination of (1) being an in vivo model and (2) enabling high-throughput, low-volume screening. Within the past decade, zebrafish embryos have become well-established as an in vivo model for the analysis of angiogenesis and vascular development [9], [19], [20], [21], [22], [23]. To test the suitability of zebrafish as an in vivo frontline assay for the bioassay-guided fractionation of complex natural extracts, we therefore combined an embryonic zebrafish angiogenesis assay with analytical chromatography methods, with the goal of rapidly isolating phytochemicals from medicinal plant extracts capable of inhibiting vascular outgrowth in this assay.
Results and Discussion
We chose an angiogenesis assay based on the evaluation of intersegmental vessel (ISV) outgrowth in fli-1:EGFP transgenic embryos [19], which exhibit vasculature-specific expression of enhanced green fluorescent protein (EGFP) in the trunk and tail during embryonic and larval development (Figs. 1a, b). With respect to natural product research, fli-1:EGFP zebrafish have been used to characterize the angiogenic activity of Angelica sinensis (dong quai) [24], as well as the anti-angiogenic activity of solenopsin, an alkaloid isolated from Solenopsis invicta (fire ants) [25]. Similar transgenic lines, with fluorescent reporter proteins expressed under the control of the endothelial cell-specific flk-1/VEGFR2 promoter, have recently enabled an ENU mutagenesis screen to identify genetic determinants of vascular development [26] and a small-molecule screen to identify novel angiogenesis inhibitors [27].
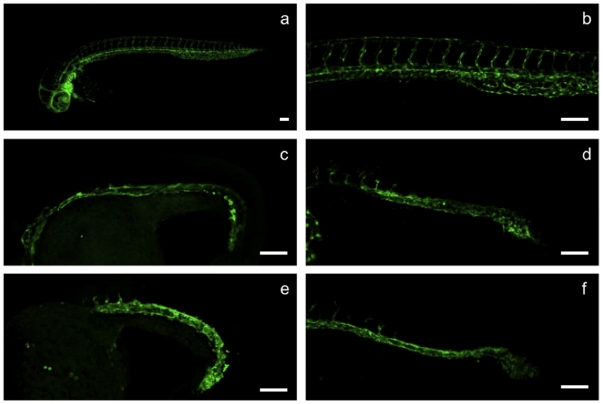
Figure 1. Crude methanolic extracts and isolated compounds inhibit vascular outgrowth in fli-1:EGFP transgenic embryos.
All embryos are 40 hpf, with anterior to the left, scale bar = 100 µm. a, b, DMSO-treated control; c, embryo treated with 200 µg/ml O. sinuatum extract; d, embryo treated with 10 µg/ml P. barbatus extract; e, embryo treated with 8 µM emodin; f, embryo treated with 2 µM coleon AL.
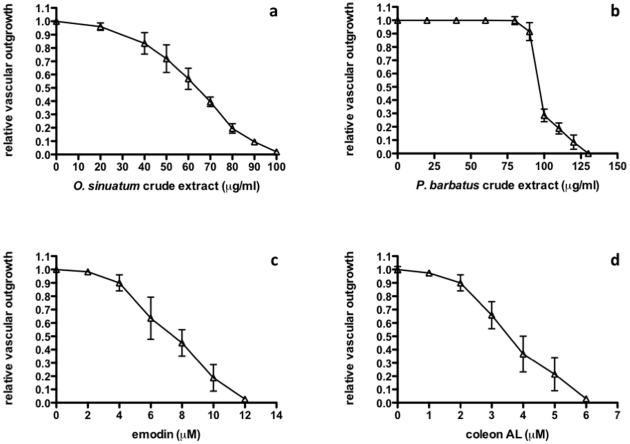
To test the utility of this zebrafish assay for natural product discovery, we screened crude methanolic extracts from over 80 East African medicinal plants. Two extracts, from Oxygonum sinuatum (Meisn.) Dammer (Polygonaceae) and Plectranthus barbatus Andrews (Lamiaceae), inhibited ISV outgrowth in fli-1:EGFP embryos in a dose-dependent manner (Figs. 1c, d and 2a, b).
Figure 2. Crude methanolic extracts and isolated compounds exhibit concentration-dependent effects on vascular outgrowth in zebrafish embryos.
Relative vascular outgrowth scores are shown (see Materials & Methods). a, O. sinuatum crude extract; b, P. barbatus crude extract; c, emodin; d, coleon AL.
In terms of known bioactivities for these plants, O. sinuatum has been documented as an ethnobotanical treatment in Kenya for several unrelated disorders [28]. No phytochemical analysis of this plant has been reported to date. P. barbatus (previously also known as Coleus forskohlii Briq.) is widely used in traditional medicine in Africa and Latin American to treat a range of human ailments [29]. This species is also well-known as the primary source of forskolin, a labdane diterpenoid and activator of cAMP signaling [30], [31]. Intruigingly, although forskolin has been shown to inhibit angiogenesis in the chick chorioallantoic membrane assay [32] and in vitro [33], it is also known to upregulate VEGF expression [34], making its overall effect on angiogenesis in vivo difficult to predict. We determined that forskolin does not inhibit angiogenesis in zebrafish (data not shown) and since it is isolated from P. barbatus roots (an extract from leaves being the subject of this analysis), we concluded that the anti-angiogenic activity seen in zebrafish embryos for the P. barbatus extract is likely due to the bioactivity of another compound.
We next sought to isolate from O. sinuatum and P. barbatus extracts the principle components responsible for their anti-angiogenic effects. Both crude methanolic extracts were fractionated via thin-layer chromatography (TLC), using toluene/ethyl formate/formic acid (5:4:1) as the solvent. A single analytical-scale TLC plate (20×20 cm) was used to separate 10 mg of each extract, and was subsequently divided into 10-15 horizontal strips based on the presence of UV254 -absorbing and UV365-emitting components (Figs. 3 a, b). The silica was removed from these strips and extracted with methanol, after which the eluted constituents were subjected to bioactivity analysis in zebrafish, followed by high-resolution electrospray ionization mass spectroscopy (HRESIMS) for those exhibiting anti-angiogenic activity. For both O. sinuatum and P. barbatus, single TLC fractions were identified in this manner which phenocopied the anti-angiogenic activity shown by the crude extracts.
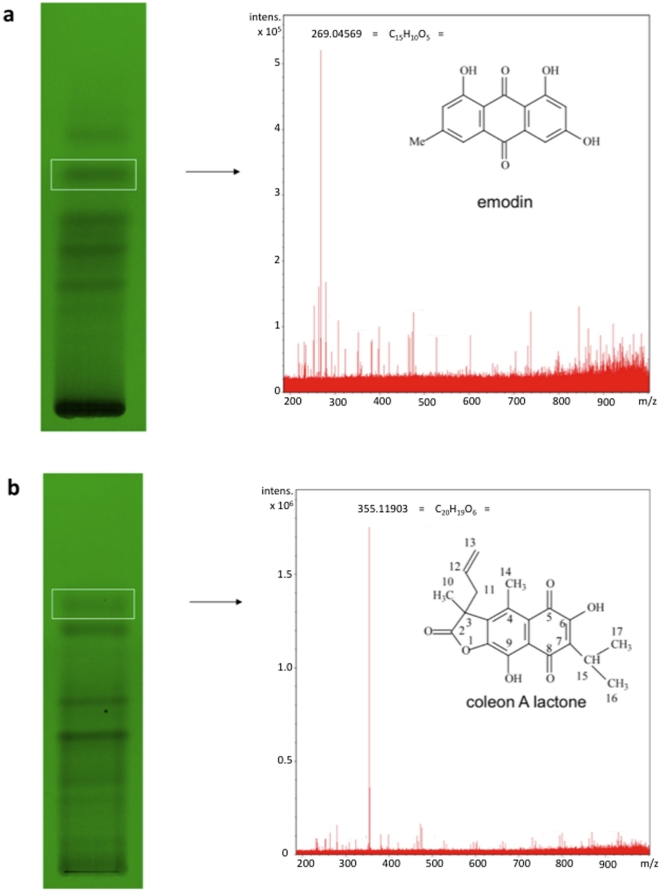
Figure 3. TLC fractionation and HRESIMS analysis identify emodin and coleon AL as bioactive constituents.
Thin-layer chromatograms, HRESIMS spectra, and predicted structures of isolated, bioactive compounds. a, O. sinuatum; b, P. barbatus.
O. sinuatum yielded a single bioactive compound whose molecular formula was determined to be C15H10O5 based on the deprotonated molecular ion at m/z 269.0457 identified by HRESIMS analysis (negative mode), suggesting 6-methyl-1,3,8-trihydroxyanthraquinone (emodin) – a known product of several other Polygonaceae species. Emodin has recently been reported as an inhibitor of angiogenesis in vitro [35] and in vivo [36], and has been shown to be an inhibitor of the protein kinases Lck [37], HER-2 [38], and CK2 [39]. More recently, emodin was shown to inhibit angiogenesis in vitro at least in part by restricting the phosphorylation of VEGFR2 [40]. In addition; CK2 has been found to directly phosphorylate Akt [41], one of several downstream elements of VEGF signaling, and this modification has been shown to be essential for nuclear retention of FOXO1, an important cytoplasmic inhibitor of angiogenesis [42]. Semi-synthetic emodin revealed an MS spectrum identical to the bioactive compound isolated from O. sinuatum (data not shown) and, importantly, phenocopied both this compound and the crude extract (Fig. 1e, 2c), thereby confirming emodin as the primary constituent responsible for this plant's bioactivity. Furthermore, emodin and other anthraquinones synthesized by Rheum species (rhubarb root, or Dahuang) have recently also been shown to inhibit vascular outgrowth in zebrafish using a histochemical assay to visualize blood vessels [43].
P. barbatus yielded a bioactive molecule with an apparent Mw of 355.1190 based on HRESIMS analysis and the predicted molecular formula C20H19O6, suggesting coleon A lactone, a known product [44] of another Lamiaceae species but with no previously described bioactivity. Following isolation by preparative-scale chromatography, the identity of coleon A lactone (hereinafter referred to as coleon AL) was confirmed by NMR (Table 1).
Table 1. NMR spectroscopy analysis confirms identity of coleon AL.
| a. 1H NMR data in CDCl3 | ||
| Ref. 44 | experimental | |
| 1.31 (6H, d, J = 7.1 Hz) | CH3-16/17 | 1.306 (6H, d, J = 7.0 Hz) |
| 1.65 (3H, s) | CH3-10 | 1.662 (3H, s) |
| 2.69 (3H, s) | CH3-14 | 1.689 (3H, s) |
| 2.83 (2H, d, J = 6.0 Hz) | H-11AH-11B | 2.811 (1H, dd, 2J = 14.0 Hz, 3J = 7.5 Hz)2.868 (1H, dd, 2J = 14.0 Hz, 3J = 7.3 Hz) |
| 3.38 (1H, pent, J = 7.1 Hz) | H-15 | 3.378 (1H, sept, J = 7.0 Hz) |
| 4.90 – 5.60 (3H, m) | H-13AH-13BH-12 | 4.955 (1H, dd, J = 9.9 Hz, J = 1.1 Hz)5.054 (1H, dd, J = 17.0 Hz, J = 1.1 Hz)5.326 (1H, ddt, J = 17.0/9.9/7.4 Hz) |
| 7.96 (1H, s) | 9-OH | 8.01 (1H, s) |
| 13.25 (1H, s) | 6-OH | 13.35 (1H, s) |
a, Comparison of 1H-NMR chemical shift assignments from Ref. 44 and our experimental data, with more resolution for protons H-11A, 11B, 13A, 13B and 12; b, 13C NMR experimental chemical shift assignments, with 3 carbonyl peaks at 177–190 ppm, and olefinic peaks at 117–154 ppm corresponding to the aromatic fused-ring moeities.
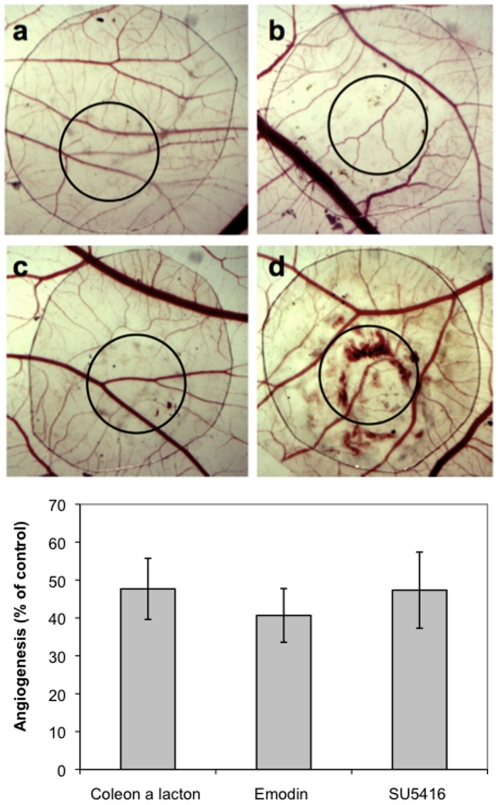
Zebrafish have only recently been utilized for the systematic identification of bioactive small molecules [12], so the predictive power of zebrafish assays for drug discovery will only become clear as molecules found using this platform are advanced into the clinic. In any case, to further evaluate the therapeutic potential of natural products identified using embryonic or larval zebrafish models, bioactive compounds should subsequently be validated using a series of additional in vitro and in vivo assays. To this end, in vitro anti-angiogenesis assays were carried out to further characterize the anti-angiogenic activity of the bioactive natural products isolated in this study, revealing both emodin and coleon AL to inhibit the proliferation, migration and tube formation of mammalian endothelial cells (Table 2, Fig. 4, 5). In addition, both compounds inhibited blood vessel formation in the chick chorioallantoic membrane (CAM) assay (Fig. 6). Emodin and coleon AL inhibited the proliferation of mouse aortic endothelial cells (MAECs) with an IC50 similar to that of the vascular endothelial growth factor (VEGF) receptor inhibitor SU5416, a synthetic indoline derivative [45], and inhibited the proliferation of bovine aortic endothelial cells (BAECs) with an IC50 similar to that of the PI3K inhibitors wortmannin, a fungal furanosteroid [46], and LY294002, a synthetic chromone derivative [47]. In the CAM assay, we determined the anti-angiogenic activity of emodin and coleon AL to be similar to that of SU5416.
Table 2. Emodin and coleon AL inhibit endothelial cell proliferation in vitro.
| IC50 (µM) | ||
| MAEC | BAEC | |
| emodin | 32±11 | 39±9 |
| coleon AL | 30±5 | 31±13 |
| wortmannin | 55±5 | 29±12 |
| LY294002 | 7±2 | 3±1 |
| SU5416 | 29±2 | 8±2 |
IC50 values (± SD) for emodin and coleon A lactone are shown for the inhibition of proliferation of both bovine aortic endothelial cells (BAEC) and mouse aortic endothelial cells (MAEC). For comparison, effects of 3 other anti-angiogenic compounds are shown (wortmannin, LY294002, and SU5416).
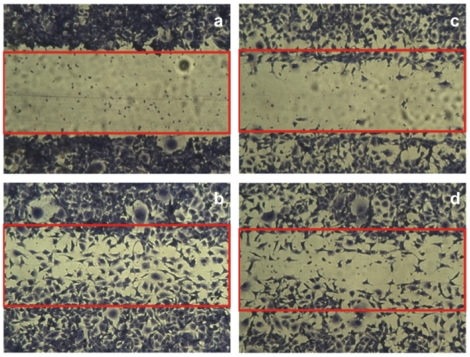
Figure 4. Emodin and coleon AL inhibit endothelial cell migration in vitro.
a, control (immediate); b, control; c, 30 µM emodin; d, 30 µM coleon AL.
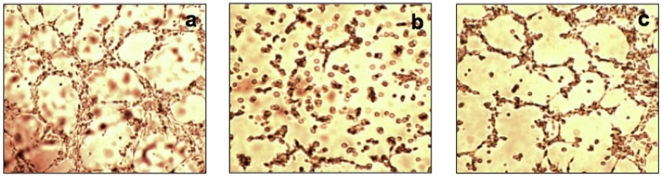
Figure 5. Emodin and coleon AL inhibit endothelial tube formation in vitro.
a, DMSO-treated control; b, 10 µM emodin; c, 10 µM coleon AL.
Figure 6. Emodin and coleon AL inhibit angiogenesis in the chick chorioallantoic membrane (CAM) assay.
a, negative control; b, positive control (50 nmol SU5416); c, 50 nmol emodin; d, 50 nmol coleon AL.
As a next step, in vivo angiogenesis assays should be carried out for these compounds in mammalian models, including mouse tumor assays [18]. Additional in vitro assays should also be performed to determine the activity of anti-angiogenic compounds under hypoxic conditions, an important condition for possible application in antitumor therapy [48].
Zebrafish were first proposed as a model for in vivo drug discovery in 1957 by Jones and Huffmann of the Oklahoma Research Foundation [49], and soon thereafter were used for the first time to analyze the bioactivities of natural products [50]. Only more recently, however, have zebrafish been widely used for the function-based identification of disease-relevant genes and bioactive compounds [12]. An important advantage of using zebrafish embryos and larvae for the identification of bioactive molecules is that they are living organisms, thereby enabling the rapid in vivo evaluation of compounds not only in terms of their pharmacological activity but also of their possible toxicity (particularly cardio-, neuro-, and hepatotoxicity) early in the drug discovery process [51], [52]. Within the past decade, zebrafish have furthermore emerged as a powerful model for chemical genetics, both with respect to the in vivo dissection of signaling pathways [20], [53] and to the elucidation of the mechanism of action of bioactive natural products [54], [55]. In the area of natural product discovery, one recent report describes the application of a histochemical assay in zebrafish to guide the isolation of anti-angiogenic terpenoids from Tripterygium wilfordii, an anti-inflammatory Chinese medicinal plant, using preparative chromatography methods [56].
Here, we demonstrate the utility of zebrafish bioassay-guided fractionation by analytical chromatography techniques, and further establish zebrafish as an in vivo platform for the discovery of bioactive natural products. Based on these initial results, it appears possible that zebrafish can help address a critical bottleneck in natural product discovery by enabling the rapid, in vivo and microgram-scale bioactivity analysis, bioassay-guided fractionation, and dereplication of complex natural extracts. While data described here were obtained using zebrafish bioassay-guided TLC fractionation, additional advantages for accelerating natural product discovery will be realized through the combination of zebrafish bioassays with more advanced HPLC techniques – in particular, those enabling microfractionation [57], [58]. With a wide array of biomedically relevant assays now established in zebrafish [4], the advantages of this in vivo system for natural product discovery should facilitate the systematic identification of a new generation of bioactive natural products with potential utility in drug discovery.
Materials and Methods
Zebrafish
The fli-1:EGFP transgenic line [19] was obtained from the Zebrafish International Resource Center at the University of Oregon (Eugene, Oregon, USA). Zebrafish husbandry, embryo collection, and embryo and larvae maintenance were performed as described earlier [59], [60]. Zebrafish assays were standardly performed in 24-well microtiter plates using 10 embryos per well in 1 ml of 0.3× Danieau's medium (17 mM NaCl, 2 mM KCl, 0.12 mM MgSO4, 1.8 mM Ca(NO3)2 and 1.5 mM HEPES, pH 7.6). Embryos were exposed to extracts and compounds at 16 hours post-fertilization (hpf) – approximately 8 hours prior to the initiation of intersegmental vessel (ISV) outgrowth – and scored for relative vascular outgrowth at 40 hpf. Extracts and compounds were solubilized in dimethyl sulfoxide (DMSO, Agros Organics, Geel, Belgium), and were added to the medium up to a maximum DMSO concentration of 1%. The extent of outgrowth of intersegmental vessels (ISVs) was determined using a scoring method that takes into account both the approximate number of outgrowing vessels (100, 75, 50, 25, or 0%) and the average degree to which these vessels have extended into the trunk from the dorsal aorta/posterior cardinal vein (DA/PCV) (100, 75, 50, 25, or 0%). These two values are multiplied to give the relative vascular outgrowth (RVO) score.
Extracts
Plant samples were collected from different locations in Tanzania and their respective voucher specimens deposited at the Department of Pharmacognosy, Faculty of Pharmacy of the Muhimbili University of Health and Allied Sciences (MUHAS), Dar es Salaam, Tanzania. For each plant sample, plant materials were dried at room temperature and ground. The dry, powdery plant samples were exhaustively extracted with methanol by maceration. Dry methanolic extracts were obtained after removing the solvent by evaporation under reduced pressure. Prior to testing, aliquots of each dry methanolic extract were suspended in 100% DMSO; these stock solutions were then kept at −20 °C.
Compounds
Semi-synthetic emodin was obtained from Janssen Chimica (Geel, Belgium). SU5416 and wortmannin were obtained from Sigma-Alrich (Bornem, Belgium) and LY294002 was obtained from Cayman Chemical (Talinn, Estonia). Coleon A lactone (coleon AL) was isolated from leaves of Plectranthus barbatus collected in Handeni, Tanga Region, Tanzania. Leaves were dried at ambient temperature under sunlight, homogenized, and extracted 3 times with chloroform (1 l chloroform per 100 g leaves). This chloroform extract was concentrated 100:1 on a rotary evaporator and subjected to chromatographic separation on a LaFlash chromatography apparatus from VWR (West Chester, Pennsylvania, USA) using VWR SuperVarioFlash silica cartridges (30 g, Si60, pore size 15-40 µM), with chloroform:acetic acid 200:1 as the solvent. Coleon AL was isolated as the second major peak showing absorbance at 254 nm.
Thin-layer chromatography
TLC plates were obtained from Macherey-Nagel (Düren, Germany). For the experiments described here, 20×20 cm aluminum plates coated with TLC silica gel 60 (layer thickness, 0.2 mm) containing a UV254 fluorescence indicator were used (ALUGRAM SIL G/UV254, Machery-Nagel product number 818133). Plates were loaded manually, using a finely tapered micropipette tip, with 10 mg of crude extract (50 mg/ml in methanol), dried for 15 seconds with a hair dryer at low heat, and placed in an enclosed, upright 25×25×10 cm glass chamber containing 100 ml toluene/ethyl formate/formic acid 5:4:1 (pre-equilibrated for at least 15 minutes, and with 3 vertical faces covered with solvent-saturated filter paper).
High-resolution electrospray ionization mass spectrometry
Electrospray ionization (ESI) mass spectra were recorded in positive and negative mode on an orthogonal acceleration quadrupole time-of-flight mass spectrometer (Q-Tof 2, Micromass, Manchester, UK). The electrospray needle voltage was set to 3000 V or −2850 V for the positive and negative mode respectively. Fragment ion spectra were obtained by selecting the precursor ion in the quadrupole and collisional activation with argon gas in the collision cell. Accurate mass measurements were performed at a resolution of 9000 using the protonated leucine-enkephaline (YGGFL) ion as lock mass.
NMR spectroscopy
1H and 13C NMR spectra were recorded on a Bruker (Fällanden, Switzerland) Avance II 500 spectrometer operating at 500.130 MHz for 1H and at 125.758 MHz for 13C, and using a gradient-equipped inverse 5 mm triple probe with π/2 pulses of 6.5, and 14.5 µs respectively. The standard Bruker Topspin 2.1 software under Windows XP was used throughout. All experiments were performed at 22 °C in deuterochloroform solution with the solvent peak as internal standard set at 7.27 ppm (1H) or 77.0 (13C) vs. TMS respectively. First-order analysis was applied throughout, and first-order multiplets or apparent first-order multiplets were denoted as follows: s = singlet, d = doublet, dd = double doublet, t = triplet. J-values were extracted directly from the splittings in the spectrum, and are not optimised. Spectral assignments were based not only on the usual chemical shift rules and coupling patterns, but especially on routine 2D-correlations such as COSY45- (homonuclear H,H J-correlations), GHSQC- (single bond C,H 1J-correlations) and GHMBC-experiments (multiple bond C,H 3J-correlations). The data for coleon AL are summarized in Fig. 4 and compared with previously reported values [44].
Imaging
Zebrafish were screened for GFP fluorescence using an Axiovert 40 CFL microscope from Zeiss (Oberkochen, Germany) equipped with an MBQ 52 AC fluorescence lamp from LEJ (Jena, Germany). Micrographs of zebrafish embryos were taken on a Stemi 2000 stereo microscope from Zeiss equipped with a DP200 CMOS digital camera and using DpxView Pro EE EF software, both from Deltapix (Maalov, Denmark). Confocal fluorescence micrographs of zebrafish embryos were acquired using a Nikon A1R confocal unit mounted on a Ti2000 inverted microscope (Nikon, Tokyo, Japan). The microscope was equipped with 4× (0.2 N.A.) and 10× (0.45 N.A.) objective lenses, and fluorescence was revealed using a 488 nm laser line (CVI Melles Griot, Albuquerque, NM, USA). For imaging, zebrafish embryos were anesthetized using 0.1 mg/ml ethyl 3-aminobenzoate methanesulfonate (Sigma) in 0.3× Danieau's solution.
Cell cultures
Mouse aortic endothelial cells (MAEC) and bovine aortic endothelial cells (BAEC) were kindly provided by Prof. M. Presta (Brescia, Italy). The cells were grown in Dulbecco's modified minimum essential medium (DMEM, Life Technologies, Rockville, MD, USA) supplemented with 10 mM Hepes (Life Technologies) and 10% fetal calf serum (FCS, Harlan Sera-Lab, Loughborough, UK).
Cell proliferation assays
Cells (MAEC or BAEC) were seeded in 48-well plates at 10,000 cells per cm2. After 16 h, the cells were incubated in fresh medium in the presence of different concentrations of the test compounds (i.e. 100 - 20 - 4 - 1 - 0.2 µM). On day 5, cells were trypsinized and counted by a Coulter counter (Analis, Belgium). The compound concentration that inhibits cell growth by 50 % (i.e. IC50) was calculated based on cell counts in control cultures.
Cell migration assay
Wounds were created in confluent MAE cell monolayers with a 1.0-mm wide micropipette tip. Then, cells were incubated in fresh medium containing 10% FCS in the presence of the test compounds. After 8 h, the wounds were photographed, and endothelial cells invading the wound were quantified by computerized analysis of the digitalized images.
Tube formation assay
Wells of a 96-well plate were coated with 60 µl matrigel (10 mg/ml, BD Biosciences, Heidelberg, Germany) at 4 °C. After gelatinization at 37 °C during 30 min, BAEC (60,000 cells) were seeded on top of the matrigel in 200 µl DMEM containing 1% FCS and the test compounds. After 6 hours of incubation, the cell structures were photographed at 100×magnification. Tube formation was quantified by counting the number of branching points.
Chorioallantoic membrane (CAM) assay
The in vivo CAM angiogenesis model was performed as described with slight modifications [61]. Fertilized chicken eggs were incubated for 3 days at 37 °C when 3 ml of albumen was removed (to detach the shell from the developing CAM) and a window was opened on the eggshell exposing the CAM. The window was covered with cellophane tape and the eggs were returned to the incubator until day 9 when the compounds were applied. The compounds were placed on sterile plastic discs (Ø 8 mm), which were allowed to dry under sterile conditions. A solution of cortisone acetate (100 µg/disc, Sigma-Aldrich, St. Louis, MO, USA) was added to all discs in order to prevent an inflammatory response. A loaded and dried control disc was placed on the CAM approximately 1 cm away from the disc containing the test compound(s). Next, the windows were covered and the eggs further incubated until day 11 when the area around the discs was cut-off and photographed. Then, 2 concentric circles were positioned on the digitalized pictures and all vessels intersecting these circles were counted. A two-tailed paired Student's t-test was performed to assess the significance of the obtained results.
Acknowledgments
We thank Eef Meyen and Elke Simons (Rega Institute) for dedicated technical assistance. We gratefully acknowledge the support of the Imaging Core Facility (Tomas Luyten, Ludwig Missiaen) and the Aquaculture Core Facility (Sophie Louwette, Frédéric Hendrickx) of the Biomedical Sciences Group at K.U.Leuven. We thank the Zebrafish International Resource Center (University of Oregon) and Brant Weinstein (NICHD, NIH) for providing mating pairs of the fli-1:EGFP transgenic zebrafish line. We also thank Peter Rüedi (Institut für Organische Chemie, Universität Zürich) for useful discussions and kind gifts of pharmacological reagents.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded in part by the Industrieel Onderzoeksfonds (IOF-HB/06/018) of K.U. Leuven, the Centers of Excellence of the K.U. Leuven (EF-05/15) and the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (G. 0486.08). Sandra Liekens is a postdoctoral researcher of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4:206. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 3.Clardy J, Walsh C. Lessons from natural molecules. Nature. 2004;432:829. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- 4.Crawford AD, Esguerra CV, de Witte PAM. Fishing for drugs from nature: zebrafish as a technology platform for natural product discovery. Planta Med. 2008;74:1. doi: 10.1055/s-2008-1034374. [DOI] [PubMed] [Google Scholar]
- 5.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 7.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101:12792. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2:956. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- 9.Serbedzija GN, Flynn E, Willett CE. Zebrafish angiogenesis: a new model for drug screening. Angiogenesis. 1999;3:353. doi: 10.1023/a:1026598300052. [DOI] [PubMed] [Google Scholar]
- 10.Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci USA. 2000;97:1296. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sachidanandan C, Yeh JR, Peterson QP, Peterson RT. Identification of a novel retinoid by small molecule screening with zebrafish embryos. PLoS ONE. 2008;3:e1947. doi: 10.1371/journal.pone.0001947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nature Rev Drug Discov. 2005;4:35. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 13.Berger J, Currie P. The role of zebrafish in chemical genetics. Curr Med Chem. 2007;14:2413. doi: 10.2174/092986707781745532. [DOI] [PubMed] [Google Scholar]
- 14.Cao Y, Zhong W, Sun Y. Improvement of antiangiogenic cancer therapy by understanding the mechanisms of angiogenic factor interplay and drug resistance. Semin Cancer Biol. 2009;19:338. doi: 10.1016/j.semcancer.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Ivy SP, Wick JY, Kaufman BM. An overview of small-molecule inhibitors of VEGFR signaling. Nat Rev Clin Oncol. 2009;6:569. doi: 10.1038/nrclinonc.2009.130. [DOI] [PubMed] [Google Scholar]
- 16.Choueiri TK, Schutz FA, Je Y, Rosenberg JE, Bellmunt J. Risk of arterial thromboembolic events with sunitinib and sorafenib: a systematic review and meta-analysis of clinical trials. J Clin Oncol. 2010;28:2280. doi: 10.1200/JCO.2009.27.2757. [DOI] [PubMed] [Google Scholar]
- 17.Carmeliet P, De Smet F, Loges S, Mazzone M. Branching morphogenesis and antiangiogenesis candidates: tip cells lead the way. Nat Rev Clin Oncol. 2009;6:315. doi: 10.1038/nrclinonc.2009.64. [DOI] [PubMed] [Google Scholar]
- 18.Staton CA, Reed MW, Brown NJ. A critical analysis of current in vitro and in vivo angiogenesis assays. Int J Exp Pathol. 2009;90:195–221. doi: 10.1111/j.1365-2613.2008.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 20.Chan J, Bayliss PE, Wood JM, Roberts TM. Dissection of angiogenic signaling in zebrafish using a chemical genetic approach. Cancer Cell. 2002;1:257. doi: 10.1016/s1535-6108(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 21.Nicoli S, Ribatti D, Cotelli F, Presta M. Mammalian tumor xenografts induce neovascularization in zebrafish embryos. Cancer Res. 2007;67:2927. doi: 10.1158/0008-5472.CAN-06-4268. [DOI] [PubMed] [Google Scholar]
- 22.Kidd KR, Weinstein BM. Fishing for novel angiogenic therapies. Br J Pharmacol. 2003;140:585. doi: 10.1038/sj.bjp.0705496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cha YR, Weinstein BM. Visualization and experimental analysis of blood vessel formation using transgenic zebrafish. Birth Defects Res C Embryo Today. 2007;81:286. doi: 10.1002/bdrc.20103. [DOI] [PubMed] [Google Scholar]
- 24.Lam HW, Lin HC, Lao SC, Gao JL, Hong SJ, et al. The angiogenic effects of Angelica sinensis extract on HUVEC in vitro and zebrafish in vivo. J Cell Biochem. 2008;103:195. doi: 10.1002/jcb.21403. [DOI] [PubMed] [Google Scholar]
- 25.Arbiser JL, Kau T, Konar M, Narra K, Ramchandran R, et al. Solenopsin, the alkaloidal component of the fire ant (Solenopsis invicta), is a naturally occurring inhibitor of phosphatidylinositol-3-kinase signaling and angiogenesis. Blood. 2007;109:560. doi: 10.1182/blood-2006-06-029934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin SW, Herzog W, Santoro MM, Mitchell TS, Frantsve J, et al. A transgene-assisted genetic screen identifies essential regulators of vascular development in vertebrate embryos. Dev Biol. 2007;307:29. doi: 10.1016/j.ydbio.2007.03.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran TC, Sneed B, Haider J, Blavo D, White A, et al. Automated, quantitative screening assay for antiangiogenic compounds using transgenic zebrafish. Cancer Res. 2007;67:11386. doi: 10.1158/0008-5472.CAN-07-3126. [DOI] [PubMed] [Google Scholar]
- 28.Watt JM, Breyer-Brandwijk MG. Edinburgh: E & S Livingstone; 1962. The medicinal and poisonous plants of southern and eastern Africa. 2nd ed. [Google Scholar]
- 29.Lukhoba CW, Simmonds MS, Paton AJ. Plectranthus: a review of ethnobotanical uses. J Ethnopharmacol. 2006;103:1. doi: 10.1016/j.jep.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Seamon KB, Daly JW. Forskolin: a unique diterpene activator of cyclic AMP-generating systems. J Cyclic Nucleotide Res. 1981;7:201. [PubMed] [Google Scholar]
- 31.Ammon HP, Müller AB. Forskolin: from an ayurvedic remedy to a modern agent. Planta Med. 1985;6:473. doi: 10.1055/s-2007-969566. [DOI] [PubMed] [Google Scholar]
- 32.Tsopanoglou NE, Haralabopoulos GC, Maragoudakis ME. Opposing effects on modulation of angiogenesis by protein kinase C and cAMP-mediated pathways. J Vasc Res. 1994;31:195. doi: 10.1159/000159044. [DOI] [PubMed] [Google Scholar]
- 33.del Valle-Pérez B, Martínez-Estrada OM, Vilaró S, Ventura F, Viñals F. cAMP inhibits TGFbeta1-induced in vitro angiogenesis. FEBS Lett. 2004;569:105. doi: 10.1016/j.febslet.2004.05.058. [DOI] [PubMed] [Google Scholar]
- 34.Pueyo ME, Chen Y, D'Angelo G, Michel JB. Regulation of vascular endothelial growth factor expression by cAMP in rat aortic smooth muscle cells. Exp Cell Res. 1998;238:354. doi: 10.1006/excr.1997.3864. [DOI] [PubMed] [Google Scholar]
- 35.Wang XH, Wu SY, Zhen YS. Inhibitory effects of emodin on angiogenesis. Yao Xue Xue Bao (Acta Pharm Sin) 2004;39:254. [PubMed] [Google Scholar]
- 36.Ljubimov AV, Caballero S, Aoki AM, Pinna LA, Grant MB, et al. Involvement of protein kinase CK2 in angiogenesis and retinal neovascularization. Invest Ophthalmol Vis Sci. 2004;45:4583. doi: 10.1167/iovs.04-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayasuriya H, Koonchanok NM, Geahlen RL, McLaughlin JL, Chang CJ. Emodin, a protein tyrosine kinase inhibitor from Polygonum cuspidatum. J Nat Prod. 1992;55:696. doi: 10.1021/np50083a026. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Chang CJ, Bacus SS, Hung MC. Suppressed transformation and induced differentiation of HER-2/neu-overexpressing breast cancer cells by emodin. Cancer Res. 1995;55:3890. [PubMed] [Google Scholar]
- 39.Yim H, Lee YH, Lee CH, Lee SK. Emodin, an anthraquinone derivative isolated from the rhizomes of Rheum palmatum, selectively inhibits the activity of casein kinase II as a competitive inhibitor. Planta Med. 1999;65:9. doi: 10.1055/s-1999-13953. [DOI] [PubMed] [Google Scholar]
- 40.Kwak HJ, Park MJ, Park CM, Moon SI, Yoo DH, et al. Emodin inhibits vascular endothelial growth factor-A-induced angiogenesis by blocking receptor-2 (KDR/Flk-1) phosphorylation. Int J Cancer. 2006;118:2711. doi: 10.1002/ijc.21641. [DOI] [PubMed] [Google Scholar]
- 41.Di Maira G, Salvi M, Arrigoni G, Marin O, Sarno S, et al. Protein kinase CK2 phosphorylates and upregulates Akt/PKB. Cell Death Differ. 2005;12:668. doi: 10.1038/sj.cdd.4401604. [DOI] [PubMed] [Google Scholar]
- 42.Potente M, Urbich C, Sasaki K, Hofmann WK, Heeschen C, et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115:2382. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He ZH, He MF, Ma SC, But PP. Anti-angiogenic effects of rhubarb and its anthraquinone derivatives. J Ethnopharmacol. 2009;121:313. doi: 10.1016/j.jep.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Künzle JM, Rüedi P, Eugster CH. Isolierung und Strukturaufklärung von 36 Diterpenoiden aus Trichomen von Plectranthus edulis (VATKE) T.T. AYE. Helv Chim Acta. 1987;70:1911. [Google Scholar]
- 45.Fong TA, Shawver LK, Sun L, Tang C, App H, et al. SU5416 is a potent and selective inhibitor of the vascular endothelial growth factor receptor (Flk-1/KDR) that inhibits tyrosine kinase catalysis, tumor vascularization, and growth of multiple tumor types. Cancer Res. 1999;59:99. [PubMed] [Google Scholar]
- 46.Wiesinger D, Gubler HU, Haefliger W, Hauser D. Antiinflammatory activity of the new mould metabolite 11-desacetoxy-wortmannin and of some of its derivatives. Experientia. 1974;30:135. doi: 10.1007/BF01927691. [DOI] [PubMed] [Google Scholar]
- 47.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem. 1994;269:5241. [PubMed] [Google Scholar]
- 48.Rapisarda A, Melillo G. Role of the hypoxic tumor microenvironment in the resistance to anti-angiogenic therapies. Drug Resist Updat. 2009;12:74. doi: 10.1016/j.drup.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones RW, Huffmann MN. Fish embryos as bio-assay material in testing chemicals for effects on cell division and differentiation. Trans Amer Microsc Soc. 1957;76:177. [Google Scholar]
- 50.Jones RW, Stout MG, Reich H, Huffmann MN. Cytotoxic activities of certain flavonoids against zebra-fish embryos. Cancer Chemother Rep. 1964;34:19. [PubMed] [Google Scholar]
- 51.McGrath P, Li CQ. Zebrafish: a predictive model for assessing drug-induced toxicity. Drug Discov Today. 2008;13:394. doi: 10.1016/j.drudis.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Eimon PM, Rubinstein AL. The use of in vivo zebrafish assays in drug toxicity screening. Expert Opin Drug Metab Toxicol. 2009;5:393. doi: 10.1517/17425250902882128. [DOI] [PubMed] [Google Scholar]
- 53.Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotech. 2004;22:595. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Yeh JR, Mara A, Ju R, Hines JF, et al. A chemical and genetic approach to the mode of action of fumagillin. Chem Biol. 2006;13:1001. doi: 10.1016/j.chembiol.2006.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luesch H, Chanda SK, Raya RM, DeJesus PD, Orth AP, et al. A functional genomics approach to the mode of action of apratoxin A. Nat Chem Biol. 2006;2:158. doi: 10.1038/nchembio769. [DOI] [PubMed] [Google Scholar]
- 56.He MF, Liu L, Ge W, Shaw PC, Jiang R, et al. Antiangiogenic activity of Tripterygium wilfordii and its terpenoids. J Ethnopharmacol. 2009;121:61. doi: 10.1016/j.jep.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 57.Wennberg T, Kreander K, Lähdevuori M, Vuorela H, Vuorela P. Primary screening of natural products using micro-fractionation combined with a bioassay. J Liq Chromatogr Relat Technol. 2004;27:2571. [Google Scholar]
- 58.Wolfender JL, Queiroz EF, Hostettmann K. Phytochemistry in the microgram domain – an LC-NMR perspective. Magn Reson Chem. 2005;43:697. doi: 10.1002/mrc.1631. [DOI] [PubMed] [Google Scholar]
- 59.Westerfield M. Eugene: University of Oregon Press; 1994. The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio). [Google Scholar]
- 60.Nüsslein-Volhard C, Dahm R. Oxford: Oxford University Press; 2002. Zebrafish: a practical approach. [Google Scholar]
- 61.Liekens S, Hernández AI, Ribatti D, De Clercq E, Camarasa MJ, et al. The nucleoside derivative 5′-O-trityl-inosine (KIN59) suppresses thymidine phosphorylase-triggered angiogenesis via a noncompetitive mechanism of action. J Biol Chem. 2004;279:29598. doi: 10.1074/jbc.M402602200. [DOI] [PubMed] [Google Scholar]