Abstract
Chimeric oncoproteins resulting from fusion of MLL to a wide variety of partnering proteins cause biologically distinctive and clinically aggressive acute leukemias. However, the mechanism of MLL-mediated leukemic transformation is not fully understood. Dot1, the only known histone H3 lysine 79 (H3K79) methyltransferase, has been shown to interact with multiple MLL fusion partners including AF9, ENL, AF10, and AF17. In this study, we utilize a conditional Dot1l deletion model to investigate the role of Dot1 in hematopoietic progenitor cell immortalization by MLL fusion proteins. Western blot and mass spectrometry show that Dot1-deficient cells are depleted of the global H3K79 methylation mark. We find that loss of Dot1 activity attenuates cell viability and colony formation potential of cells immortalized by MLL oncoproteins but not by the leukemic oncoprotein E2a-Pbx1. Although this effect is most pronounced for MLL-AF9, we find that Dot1 contributes to the viability of cells immortalized by other MLL oncoproteins that are not known to directly recruit Dot1. Cells immortalized by MLL fusions also show increased apoptosis, suggesting the involvement of Dot1 in survival pathways. In summary, our data point to a pivotal requirement for Dot1 in MLL fusion protein–mediated leukemogenesis and implicate Dot1 as a potential therapeutic target.
Introduction
In addition to their critical function regulating normal gene expression, chromatin-modifying enzymes are also important mediators of neoplastic processes. In this report, we focus on the histone H3 lysine 79 (H3K79) methyltransferase Dot1 and its participation in the immortalization of hematopoietic stem cells by leukemic oncogenes resulting from rearrangements of the mixed lineage leukemia (MLL) gene.
DOT1 (for disruptor of telomere silencing) was originally identified in a genetic screen for high-copy suppressors of telomere silencing in Saccharomyces cerevisiae (1). The Dot1 protein is highly conserved from yeast to mammals (in which it has been designated Dot1L in humans and Dot1l in mice), suggesting an important role in the biology of eukaryotes (2). Mouse Dot1l generates 5 alternatively spliced variants producing isoforms designated Dot1a–e, with Dot1a sharing 84% identity and 88% similarity with human Dot1L (3). Dot1 is a unique histone methyltransferase (HMT) in two respects. First, it is the sole enzyme that methylates lysine residue 79 of histone H3 that is located in the globular domain, not the histone tail, of the protein (4–6). Second, unlike other HMTs, the catalytically active site of Dot1 lacks a SET domain (7).
In mammals, the function of Dot1-mediated H3K79 methylation is not completely understood. H3K79 methylation is coupled to transcriptionally active genes in a number of mammalian cell lines (8), making Dot1L a positive regulator of transcription. Moreover, hypomethylated H3K79 is associated with regions of repressed chromatin (6, 9). On the other hand, Dot1 and H3K79 methylation has been shown to be directly involved in repression of several genes including the epithelial sodium channel gene αENaC and the connective tissue growth factor gene CTGF in the mouse kidney (10, 11), as well as in the developing cerebral cortex where Dot1 represses the transcription factor-encoding Tbr1 gene (12). Thus, the significance of Dot1l methylation of H3K79 seems to be locus dependant.
In the case of αENaC and Tbr1, the activity of Dot1 is positively affected by AF9/MLLT3, which has been shown to bind directly to Dot1 (10, 12). AF9 was first identified as one of the many known fusion partners of the MLL oncoprotein. The t(9;11)(p22;q23) translocation that gives rise to the MLL-AF9 oncoprotein is among the most commonly encountered chromosomal rearrangements in patients with so-called MLL leukemia involving the MLL gene at chromosome band 11q23 (13). The mechanism by which MLL fusion proteins result in leukemic transformation has not been fully elucidated, but in the case of MLL-AF9, aberrant recruitment of Dot1 to MLL-regulated genes has been proposed as one possibility (14). Complicating this model, many other MLL fusion proteins have no structural or functional similarities to AF9 and are unlikely to directly recruit Dot1. Nevertheless, in leukemia cell lines expressing functionally diverse MLL fusion proteins, dimethylated H3K79 is enriched in the promoter regions of genes that are typically overexpressed in MLL leukemias (15). This raises the possibility that Dot1 may be a relevant therapeutic target for MLL leukemias. To explore whether inhibition of Dot1 represents a valid approach to treat MLL leukemias, we tested the effects of genetic ablation of Dot1l function in murine hematopoietic cells immortalized by distinct MLL oncoproteins.
Materials and Methods
Generation of Dot1l f/f and Dot1l f/Δ mouse lines
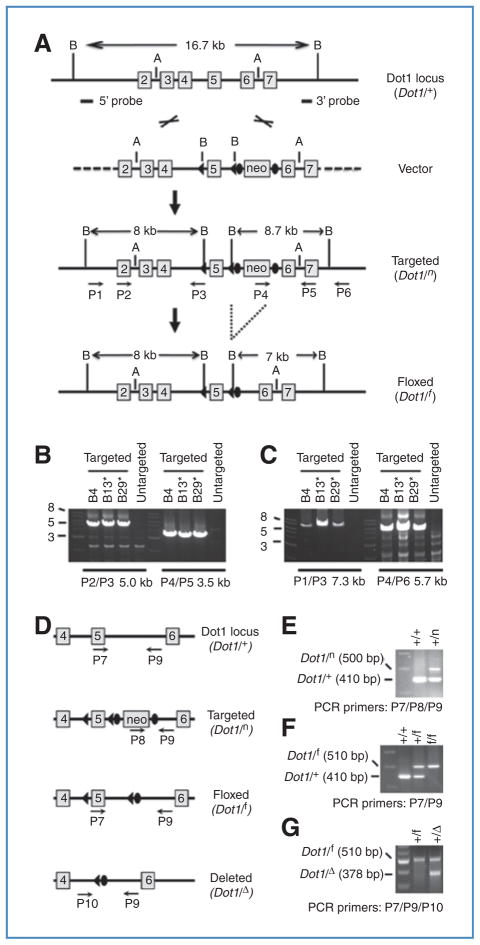
A Dot1l target vector was constructed using a recombineering method (16, 17). Briefly, an approximately 14-kb DNA fragment spanning from intron 1 to intron 7 of the murine Dot1l gene was retrieved from BAC clone RP23-164H17 and converted into a plasmid. A LoxP site and a LoxP-FRT-neo-LoxP-FRT cassette were subsequently introduced into intron 4 and intron 5, respectively, generating the final target vector (WZPL493) for conditional deletion of exon 5. The process involved multiple steps and generated 9 intermediate constructs. The detailed cloning strategy, mouse genotyping, Southern blot analysis, and mating schemes are described in Supplemental Materials and Figs. S1–S3.
Retrovirus production and myeloid progenitor immortalization assay
The oncogenes MLL-AF9, MLL-GAS7, MLL-AFX, and E2a-Pbx1 were all cloned into MSCV-based retroviral vectors containing a neomycin resistance gene as previously described (18–21). The MLL-GAS7, MLL-AFX, and E2a-Pbx1 constructs were provided by Dr. Michael Cleary (Stanford University, Stanford, CA). The bicistronic MSCV-Cre-IRES-GFP (Cre-GFP) and the MSCVpuro-Cre (Cre-puro) were provided by Dr. Jiwang Zhang (Loyola University Chicago) and the GFP control vector MigR1 was provided by Dr. Warren Pear (University of Pennsylvania Philadelphia, PA). The MSCVpuro control vector was from Clontech.
Eco Phoenix packaging cells were transfected with retro-viral vectors using CalPhos reagent (Clontech). Supernatants were collected 2 and 3 days later, concentrated by centrifugal concentrating filters (Centricon-70; Millipore) and stored at −80°C.
Bone marrow was isolated from the leg bones of Dot1f/f and Dot1f/Δ mice that had been cared for in accordance with an institutionally approved animal use and care committee protocol (University of Texas Medical School at Houston). c-kit+ progenitor cells were enriched using the EasySep kit (StemCell Technologies) and transduced with retrovirus by spinoculation as we have previously described (21). Cells were plated in methylcellulose medium (M3234; StemCell Technologies) supplemented with growth factors: 50-ng/mL stem cell factor (SCF), 10-ng/mL interleukin-3 (IL-3), 10 ng/mL IL-6, and 10-ng/mL granulocyte macrophage-colony stimulating factor (GM-CSF; Invitrogen). Antibiotics were added to the medium for selection of transduced cells [1 mg/mL geneticin (Hyclone)]. For secondary transduction, 105 to 106 oncogene-immortalized cells were transduced with retrovirus by spinoculation and cultured for 2 days in liquid medium before sorting of GFP+ cells by fluorescence-activated cell sorting (BD FACSAria; BD Biosciences). GFP+ cells were plated in either M3234 methylcellulose or liquid medium supplemented with growth factors as above. Colony-forming units (CFU) per 10,000 cells were enumerated 5 to 7 days after plating in methylcellulose. Cells collected after culture in methylcellulose medium were processed by cytospin and stained with Hema 3 (Fisher).
RT-PCR analysis
Total RNA was purified using an RNeasy mini kit (Qiagen), and 1 μg was reverse-transcribed using Superscript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen) according to the manufacturer’s instructions. Ten nanograms of cDNA was used in each real-time PCR reaction, which was carried out in triplicate using Taqman probes and the 7300 Real Time PCR System. TaqMan probes for Hoxa9 (Mm00439364_m1) and GAPDH were purchased from Applied Biosystems and Integrated DNA Technologies, respectively. Expression levels of Hoxa9 relative to that of GAPDH were calculated using a comparative Ct method.
Microscopy and image acquisition
Digital photomicrographs of colonies and individual cells were obtained using a Leica inverted microscope with a Canon PowerShot digital camera and an Olympus AX80 microscope with a Qimaging Retiga 4000R CCD camera, respectively.
Histone extraction, Western blot, and liquid chromatography–mass spectrometry
Ten nanograms of histone extract (see Supplemental Materials) was separated by SDS-4%–20% PAGE and either transferred to a nitrocellulose membrane or stained with Coomassie blue R250. For Western blot, antihistone H3 (Abcam), anti-dimethyl–H3K79 (Millipore), and anti-tri-methyl–H3K79 (Abcam) antibodies were used as primary antibodies. Signals were detected with enhanced chemiluminescence using standard protocols.
For liquid chromatography–mass spectrometry (LC-MS), trypsin-digested samples were analyzed with a Thermo-Fisher LTQ-XL linear ion trap mass spectrometercoupling with an Eksigent nanoLC. Detailed procedures are provided in Supplemental Materials.
Flow cytometric analysis
The PE-Annexin V apoptosis kit (BD) was used to label GFP+-transduced cells cultured in liquid medium for analysis of apoptosis. Flow cytometry was carried out with a FACS-Canto flow cytometer (BD) and the data were analyzed with FlowJo software. In addition, cells maintained in liquid culture were analyzed at serial time points for GFP expression.
Results
Generation of Dot1l f/f and Dot1l f/Δ mice
To understand the biological functions of Dot1l in MLL oncogene-mediated immortalization and cell survival, mice carrying a floxed Dot1l allele were created. As Dot1l null mice exhibit an embryonic lethal phenotype (22), and also confirmed in this study, we generated a Dot1l conditional target vector using the recombineering method (16, 17). Exon 5 of Dot1l was flanked by two LoxP sites, and an FRT-flanked G418 resistance gene (neo) cassette was inserted between exon 5 and exon 6 (Fig. 1; Supplementary Fig. S1). It is anticipated that excision of exon 5 upon Cre-mediated recombination will create a Dot1l null allele. Deletion of exon 5 leads to loss of the sequence encoding the methyltransferase domain as well as a frameshift of the downstream coding region. The resulting transcript is predicted to generate a truncated protein that contains only residues 1–87 (of a 1,543-amino acid wild-type Dot1a protein). Therefore, Dot1l function and thus cellular histone H3K79 methyltransferase activity should be abolished following Cre expression in the Dot1lf/f mouse. Additional details describing the animals are provided in Supplementary Materials and Figs. S1–S3.
Figure 1.
Generation and characterization of Dot1l f/f and Dot1l f/Δ mice. A, targeting vector was cut with AatII to release an 11-kb insert with 4.9-kb 5′ arm (away from the 5′ LoxP site) and 2.6-kb 3′ arm (away from the 3′ FRT sites) for recombination. Boxes with numbers inside indicate exons. The relative positions of BamHI (B), AatII (A) sites, 5′ and 3′ probes used for Southern blot (see Supplementary Fig. S2), and primers for ES clone genotyping, as well as the predicted sizes of BamHI fragments are shown. B and C, PCR-based genotyping of correctly targeted ES cells. Shown are agarose gel analyses of PCR products using the indicated primers. ES cells were microinjected into MF-1 blastocysts for generation of chimeras. D, diagram showing the relative positions of the primers used for mouse genotyping. E–G, PCR-based mouse genotyping. Shown are agarose gel analyses of PCR products from mice carrying different Dot1l alleles.
Dot1lf/f hematopoietic progenitor cells are immortalized by different MLL oncoproteins
There are more than 50 known MLL fusion partners, and, not surprisingly, a unified model of MLL-induced leukemia has been difficult to formulate. However, experimental evidence suggests that one of at least three general functional properties can be attributed to the majority of the fusion partners. First, several fusion partners that are normally found in the cytoplasm contain dimerization domains and promote homodimerization of their respective MLL fusion proteins. The significance of homodimerization has been confirmed both by blocking the ability of the fusion partners to self-associate and by fusing artificial homodimerization domains to the amino-terminus of MLL (23–25). Another class of fusion partners comprises bone fide transcription factors and transcriptional coactivators including forkhead family members as well as CBP and p300 (26–29). Next is a collection of proteins that assemble to recruit the transcriptional elongation complex P-TEFb. Included in this group of fusion partners is AF4 and its family members, LAF4 and AF5q31, as well as AF9 and its close homologue, ENL (30, 15). As noted above, AF9 (as well as ENL) is also capable of recruiting Dot1l. However, recent evidence indicates that AF9 cannot simultaneously bind P-TEFb and Dot1l, and, importantly, it seems that recruitment of P-TEFb is an essential initiating event in leukemic transformation (15).
We selected representative MLL fusion proteins from each of the 3 classes described above to immortalize c-kit+ hematopoietic precursor cells collected from Dot1l f/f mice. Specifically, MLL-GAS7 functions as a homodimer, MLL-AFX is composed of a forkhead family transcription factor fused to MLL, and MLL-AF9, as previously indicated, recruits a P-TEFb–containing complex. Retroviral transduction of murine hematopoietic precursors with virus expressing these genes has been shown to efficiently immortalize cells in serial replating assays (20, 28, 31). We selected c-kit+ cells harvested from the bone marrow of Dot1lf/f mice and transduced the cells with retrovirus expressing MLL-GAS7, MLL-AFX, or MLL-AF9, or another leukemia-associated oncogene E2a-Pbx1, which is also known to transform myeloid progenitors in vitro (32, 33). All four leukemic oncogenes rendered Dot1l f/f hematopoietic precursor cells from 3 individual animals capable of continued proliferation after 3 successive rounds of replating in methylcellulose-based medium (Supplementary Fig. S4 and data not shown). Therefore, introduction of loxP sites flanking the Dot1l loci had no apparent effect on the ability of these oncogenes to immortalize hematopoietic precursors.
Loss of Dot1l has profound effects on MLL-AF9–immortalized cells
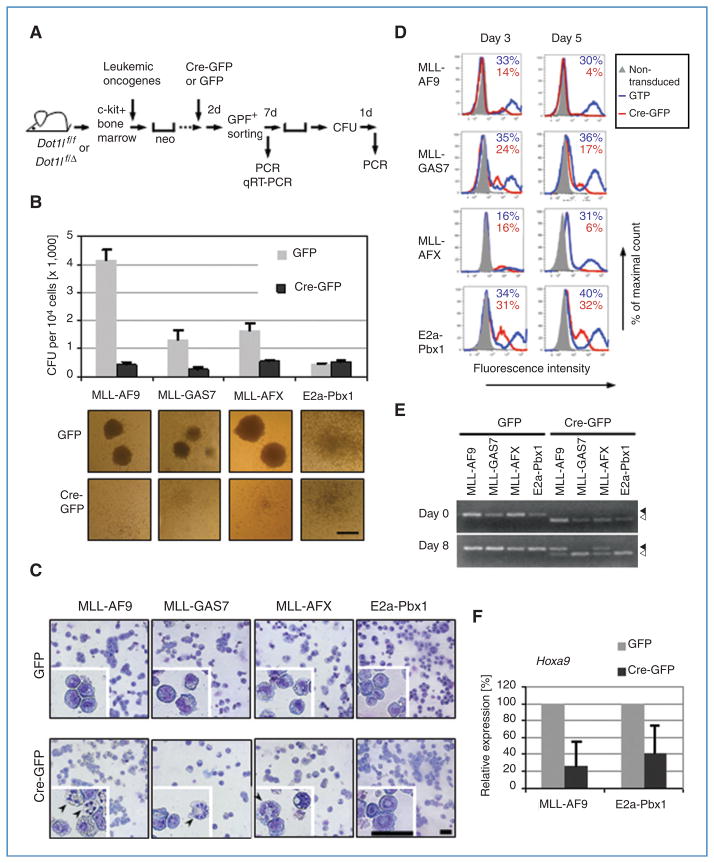
We next evaluated the consequences of Dot1 ablation in these oncogene-immortalized hematopoietic cells. Inactivation of Dot1l was achieved by deleting exon 5 through Cre-mediated recombination. Oncogene-immortalized cells were transduced with a retrovirus expressing a bicistronic Cre recombinase–GFP transcript (Cre-GFP). The GFP-expressing parent vector MigR1 served as a control (GFP). Following transduction, GFP-positive cells were selected by FACS, plated in methylcellulose-based medium containing growth factors, and incubated for one week before colonies were enumerated and genotyped by PCR (Fig. 2A). Figure 2B depicts the number of distinct colonies that developed in both Cre-GFP- and GFP-transduced cells. In the case of MLL-AF9–immortalized cells, introduction of the Cre recombinase led to a dramatic reduction in the number of viable colonies when compared with controls. The same result was observed in a serial replating colony assay in which the excision of Dot1l was mediated by a retrovirus expressing Cre recombinase in conjunction with a puromycin selection gene (Supplementary Fig. S4). In addition, the colonies that did develop from Cre-expressing cells were morphologically distinct. In contrast to the compact, cell-dense colonies of cells transduced with the empty GFP vector, cells transduced with Cre recombinase formed diffuse, ill-defined colonies (Fig. 2B). GFP+ sorted cells were subjected to cytospin and stained with Wright–Giemsa. A large proportion of Dot1l– deleted (Dot1lΔ/Δ) MLL-AF9–immortalized cells showed condensed and fragmented nuclei characteristic of apoptotic cells (Fig. 2C). In addition, in contrast to the small and uniform morphology of immortalized Dot1l f/f cells, Dot1lΔ/Δ cells have large and highly vacuolated cytosol.
Figure 2.
Cre-mediated deletion of Dot1l impairs survival in mouse hematopoietic progenitor cells immortalized by MLL-GAS7, MLL-AFX, and MLL-AF9 but not by E2a-Pbx1. A, experimental scheme to evaluate the effect of Dot1l deletion shows the time points when CFU activity, genotype (by PCR), or Hoxa9 expression (by qRT-PCR) was examined. All experiments were carried out using cells from Dot1l f/f mice unless specified. B, CFUs per 104 cells and representative colony morphologies (20× magnification) of cells immortalized by the indicated fusion oncogenes after Cre-mediated Dot1l deletion. Error bars indicate SD from 3 independent experiments with the exception of E2a-Pbx1, which is from 2 independent experiments. Each independent experiment was conducted in duplicate. Scale bar, 1 mm. C, Wright–Giemsa stain of GFP-sorted cells 6 days after transduction. Representative cells are enlarged in insets to show morphologic details. Magnification is 400 × and scale bars are 40 μm. Arrowheads indicate cells with condensed, fragmented nuclei. D, percentage of GFP+ cells at 3 or 5 days after GFP or Cre-GFP transduction in cells expressing indicated oncogens. Nontransduced cells were used as controls (gray fill) to determine GFP positivity. Cells from Dot1f/Δ mice were used in all samples with the exception of the MLL-AFX–immortalized cells, which originated from Dot1lf/f mice. The percentage of GFP+ cells is shown on each histogram. E, Dot1l genomic status was examined by PCR at days 0 and 8 in methylcellulose culture. Arrowhead, floxed allele at 510 bp; open arrowhead, deleted allele at 378 bp. F, relative expression levels of Hoxa9 after Cre-mediated Dot1l deletion. Expression levels are normalized to GAPDH and expressed relative to GFP-transduced cells (set to 100%). Error bars indicate the SD of analyses carried out in triplicate.
In the case of Dot1l f/f cells immortalized by MLL-GAS7 and MLL-AFX, introduction of Cre recombinase also resulted in diminished colony numbers and changes in colony morphology similar to MLL-AF9–immortalized cells; however, the overall effect of loss of Dot1l in these cells was less pronounced (Fig. 2B; Supplementary Fig. S4). In concordance with the colony counts, the proportion of morphologically apoptotic cells in Dot1l Δ/Δ cells was decreased in the case of MLL-GAS7 or MLL-AFX compared with that of MLL-AF9 (Fig. 2C). Strikingly, Cre-mediated gene excision of Dot1lf/f had no apparent effect on colony number or morphology in cells immortalized by E2a-Pbx1 (Fig. 2B and C; Supplementary Fig. S4).
A separate set of Cre-GFP or GFP-transduced cells were maintained in liquid culture and analyzed by flow cytometry. Cells immortalized by MLL-AF9, MLL-GAS7, and MLL-AFX revealed a reduction in GFP+ cells from day 3 to day 5 after Cre-GFP transduction whereas cells transduced with the GFP control vector maintained substantial GFP+ populations (Fig. 2D; Supplementary Fig. S5). For example, among MLL-AF9–immortalized cells, GFP+ cells decreased from 14% at day 3 to 4% at day 5 after GFP-Cre transduction. These fractions were only slightly decreased from 33% to 30% in GFP-transduced control cells. Cre-dependent reduction of GFP+ cells was not observed in E2a-Pbx1–immortalized cells as evidenced by the similar percentage of GFP+ cells in both groups. This indicates that many of the MLL oncogene–immortalized cells are lost upon Cre-mediated excision of Dot1l.
We then verified the status of the Dot1l loci in the surviving cells. After transduction with GFP-Cre or GFP control vector, cells were GFP-sorted and either harvested immediately (designated day 0) or following culture in methylcellulose (designated day 8). Genomic DNA was isolated and subjected to PCR amplification using primer sets (P7/P9 or P9/P10) specific for either the floxed allele or the excised Dot1l allele (Dot1lΔ; Fig. 1D; Supplemental Materials). Figure 2E reveals that immediately following transduction (day 0), only the excised allele of Dot1l can be detected in sorted Cre-GFP–transduced cells. However, the same GFP-sorted cells grown in methylcellulose for 8 days show varying amounts of the excised allele. Importantly, in cells immortalized by MLL-AF9 and by MLL-AFX in which Cre-GFP was expressed, a significant number of the surviving cells retain the floxed allele consistent with a strong selective pressure against loss of Dot1l under these conditions. In fact, the floxed Dot1l allele predominated over the excised allele in surviving MLL-AF9–immortalized cells. In contrast, the floxed allele could not be detected in cells immortalized either by MLL-GAS7 or by E2a-Pbx1 following Cre expression indicating complete or near-complete excision of Dot1l. These data suggest that many of the MLL-AF9- and MLL-AFX–immortalized cells that survive 8 days after introduction of Cre recombinase retain at least 1 functional (floxed) allele of Dot1l. On the other hand, cells immortalized by MLL-GAS7 and E2a-Pbx1 survive even after loss of all detectable functional Dot1l. In the case of MLL-GAS7–immortalized cells, however, survival is nevertheless compromised by Dot1l deletion as shown in Figs. 2B–D and Supplementary Figs. S4 and S5.
The promoter region of one of the most widely studied MLL target genes, HOXA9, is hypermethylated at H3K79, and over-expression of HOXA9 is a hallmark of MLL leukemias (15). We measured transcript abundance of Hoxa9 in cells immortalized by MLL-AF9 and E2a-Pbx1 to determine whether loss of Dot1 function downregulated expression of this gene. Immortalized cells were transduced with GFP-Cre or the GFP control vector, and GFP+ cells were selected by FACS. Figure 2F depicts the results of this analysis. Hoxa9 expression is substantially diminished in MLL-AF9–immortalized cells following Dot1l excision and loss of Hoxa9 expression. However, Hoxa9 is expressed at lower levels in similarly treated cells immortalized by E2a-Pbx1 suggesting that Dot1 contributes to the regulation of this gene under general circumstances. Nevertheless, the reduction in Hoxa9 expression may be of greater biological significance in cells that require this gene for leukemic transformation. As described below, ChIP analysis of methylated H3K79 is not possible as this modification is absent in cells lacking Dot1.
Dot1l deletion abolishes histone H3 lysine 79 methylation
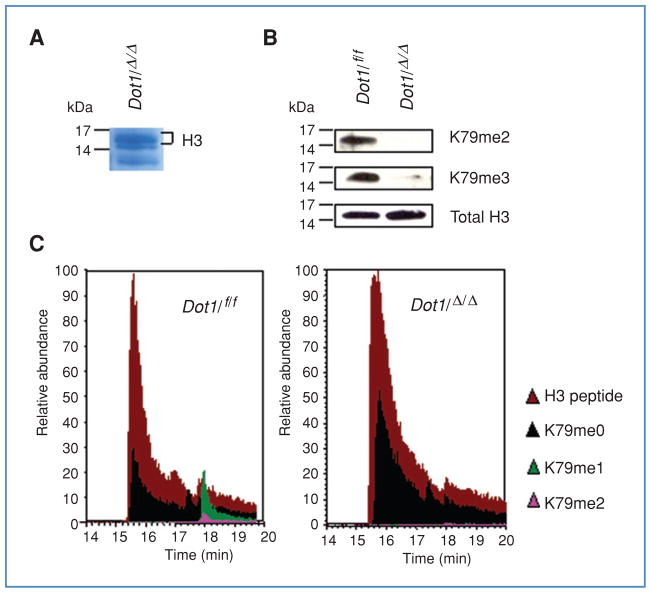
Dot1 is the only known histone modifying enzyme that methylates histone H3 at the lysine 79 residue. In yeast, more than 90% of nucleosomes are methylated by Dot1 (34). Given the ubiquitous expression of Dot1l and the embryonic lethal phenotype of Dot1l-deficient mice, we investigated whether viable immortalized cells in which Dot1l had been experimentally excised lacked all H3K79 methyltransferase activity. We observed that E2a-Pbx1–immortalized cells with deletion of Dot1l survived under continuous culture conditions. Therefore, a single colony of E2a-Pbx1–immortalized cells with PCR-confirmed Dot1l Δ/Δ genotype was expanded in liquid culture and analyzed for H3K79 methylation. First, histones were extracted, separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies to total histone H3, dimethyl H3K79, and trimethyl H3K79. As the available antibody against monomethyl H3K79 (ab2886; Abcam) nonspecifically recognizes histone H3, its application to detect monomethyl H3K79 was omitted (22). Figure 3B shows that both Dot1l f/f- and Dot1l Δ/Δ-immortalized cells have equal amounts of histone H3. Dimethyl H3K79 and trimethyl H3K79 are easily detected in the Dot1l f/f cells immortalized by E2a-Pbx1; however, these protein modifications could not be detected by Western blot in Dot1l Δ/Δ cells (Fig. 3B).
Figure 3.
Dot1l deletion abolishes histone H3K79 methylation.
A, Coomassie-stained SDS-PAGE gel of histone extract with histone H3 band indicated. B, histone H3K79 methylation status in E2a-Pbx1–immortalized cells obtained from Dot1lf/f or Dot1lΔ/Δ backgrounds was examined by Western blot using antibodies specific for dimethyl (K79me2) or trimethyl (K79me3) marks. The total histone H3 serves as a loading control. C, SDS-PAGE–purified histone H3 from E2a-Pbx1–immortalized cells with Dot1lf/f or Dot1lf/Δ genotype was subjected to LC-MS analysis. The relative abundance of the H3 peptide was manually set to 100. H3 peptide, YRPGTVALR; K79me0, K79me1, and K79me3 represent the unmodified (EIAQDFK), monomethyl and dimethyl (EIAQDFKTDLR) H3K79 peptides, respectively.
Second, histones extracted from Dot1l f/f- and Dot1l Δ/Δ-immortalized cells were separated by SDS-PAGE, excised from a Coomassie-stained gel (Fig. 3A), and subjected to mass spectrometry. The peptide fragment EIAQDFKTDLR resulting from trypsin digestion of H3 contains lysine 79 and was detected in samples from both Dot1l f/f and Dot1l Δ/Δ cells (Fig. 3C). Under our experimental conditions, the unmodified peptide EIAQDFKTDLR is further cleaved into two fragments, EIAQDFK and TDLR. In Dot1l f/f cells, mono- and dimethylated peptides were found whereas in Dot1l Δ/Δ cells only the unmodified EIAQDFK peptide was detectable. It is likely that no trimethylated peptide was identified by mass spectrometry due to the current detection limit of mass spectrometry and the low abundance of the trimethylated species, which has previously been shown to represent only 0.1% of the total H3K79 peptides (Fig. 3C; ref. 22). Nevertheless, using 2 independent analyses, there was no evidence of H3K79 methylation in Dot1l Δ/Δ cells immortalized by E2a-Pbx1. These findings indicate that the gene product of the excised Dot1l allele lacks methyltransferase activity. Furthermore, Dot1l is not absolutely required for cell survival at least in the setting of hematopoietic cells immortalized by E2a-Pbx1.
Loss of Dot1l triggers apoptosis in MLL-AF9-, MLL-GAS7-, and MLL-AFX–immortalized cells
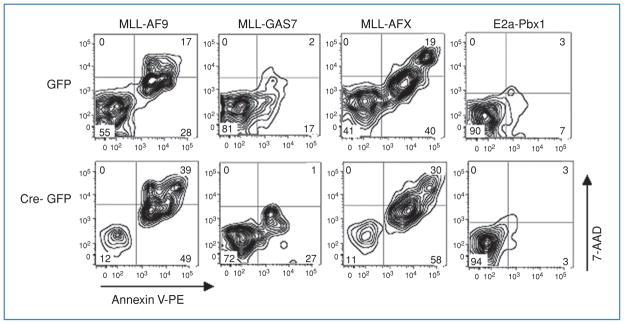
Colony-forming assays indicate that loss of Dot1l in cells immortalized by MLL-GAS7, MLL-AFX, and MLL-AF9 impairs survival. Flow cytometric analysis of cells transduced with Cre-GFP showed a significant reduction in GFP+ cells following Cre-mediated recombination in cells immortalized by any of these three oncogenes. However, the percentage of GFP+ cells was unaffected in the case of immortalization by E2a-Pbx1 (Fig. 2D; Supplementary Fig. S5). Microscopic analysis of Dot1lΔ/Δ cells, in particular those immortalized by MLL-AF9, showed a number of cells with condensed, fragmented nuclei that are characteristic of apoptotic cells (Fig. 2C). Embryonic stem cells derived from Dot1lΔ/Δ murine blastocysts can be maintained in cell culture but do exhibit an increased rate of apoptosis (22). We tested whether there were differences in the frequency of apoptotic in immortalized progenitor cells transduced by GFP-Cre compared with controls. For this, cells were labeled with Annexin-V and 7-amino-actinomycin D (7-AAD) and analyzed by flow cytometry. Figure 4 depicts representative plots of the four different oncogene-immortalized hematopoietic cells. The percentage of Annexin-V-positive, 7-AAD-negative cells that are indicative of apoptosis is significantly increased among cells immortalized by MLL-GAS7, MLL-AFX, and MLL-AF9 five days following transduction with Cre-GFP compared with GFP-transduced cells. Again, no meaningful difference among similarly treated cells immortalized by E2a-Pbx1 was found. Thus, reduced colony numbers and loss of GFP+ cells can at least partly be attributed to apoptosis when Dot1l function is lost in cells immortalized by these MLL oncogenes.
Figure 4.
Increased Annexin V labeling in Dot1l deleted cells immortalized by MLL-AF9, MLL-GAS7, or MLL-AFX but not by E2a-Pbx1.
Immortalized hematopoietic cells expressing the indicated oncogenes were transduced with GFP or Cre-GFP, labeled with Annexin V-PE/7-AAD, and analyzed by flow cytometry 5 days after transduction. The Annexin V-positive/7-AAD-negative cells in the lower right quadrant and the Annexin V–positive/7-AAD-positive cells in the upper right quadrant represent early apoptotic and late apoptotic/necrotic cells, respectively. GFP+ cells are presented as dual parameter contour plots and the percentage of cells in each quadrant is indicated.
Discussion
The large diversity of MLL fusion partners poses a challenge to developing a unified model of the pathobiology of MLL leukemia. Despite this, gene expression signatures of MLL leukemias are remarkably similar and can be reliably used to distinguish leukemias with MLL rearrangements from other subtypes (35). Mistargeting of Dot1 by the MLL-AF10 onco-protein was described by Zhang and colleagues. They proposed that Dot1-catalyzed chromatin modifications of loci normally regulated by wild-type MLL contribute to leukemogenesis (36). The importance of Dot1 in MLL leukemias was given additional weight when it was found that the MLL fusion partners AF9, ENL, and AF17 are components of a novel Dot1L-containing complex (DotCom; ref. 37). Our previous studies have shown that Dot1l (mouse isoform a) directly interacts with AF9 and AF17 in multiple assays (10, 38). More evidence that Dot1 may play a central role in many MLL leukemias has been provided by other groups as well. Hematopoietic precursor cells are not immortalized when transduced with a mutant MLL-ENL that encodes an ENL moiety that does not bind Dot1. Furthermore, H3K79 methylation is abundant in the promoter regions of genes that are over-expressed in cells transformed by MLL-ENL as well as MLL-AF4 (39–41).
To directly test the requirement for Dot1 in an experimental model of MLL leukemia, we have generated a new Dot1 conditional knockout mouse (Dot1l f/f) with a floxed exon 5. As a consequence of Cre-mediated Dot1l deletion, the majority, if not all of the Dot1a function, including the methyl-transferase activity and the AF9/AF17-binding domain, which maps to residues 479–659 of Dot1a (10, 38), is eliminated. Although there are several other isoforms of Dot1l in the mouse, the function of Dot1b should also be completely disrupted because its translation start site is found in exon 5 (3). The translation start sites for Dot1c-e have not been determined, but the transcripts would lack exon 5 and the resulting isoforms would not possess H3K79 methyltransferase activity. Our results indicate that this is, in fact, the case. There is no detectable H3K79 methyltransferase activity in Dot1lΔ/Δ cells.
Zhang and colleagues were the first to experimentally show the requirement for Dot1 in at least one type of MLL fusion-mediated leukemia (36). Expression of a dominant-negative allele of Dot1L inhibited the proliferation of mouse hematopoietic precursor cells immortalized by MLL-AF10. This artificial mutant expresses a full-length Dot1L protein in which the catalytic site has been inactivated. In contrast to our finding that Dot1l deletion diminishes the proliferation of cells immortalized by MLL-AFX, the dominant-negative Dot1L allele did not affect the plating efficiency of cells immortalized by this oncoprotein. This suggests that Dot1 may have other functions in addition to its methyltransferase activity that are retained in the dominant-negative protein but which are lost in the truncated protein encoded by the Dot1l Δ allele. This putative activity, in turn, may be important for the survival of MLL-AFX–immortalized cells.
Despite evidence that supports a role for Dot1, others find that assembly of a so-called AEP complex (for AF4 and ENL family proteins in complex with P-TEFb) is also an essential characteristic of many MLL fusion proteins including MLL-AF4, MLL-AF5q31, MLL-ENL, and MLL-AF9 (15). In this model, normal gene expression is perturbed by at least 2 events. First, the MLL oncoprotein stimulates transcriptional elongation by recruiting P-TEFb and the AEP complex. Next, Dot1 is recruited to the promoter region where H3K79 methylation maintains a transcriptional memory (15, 42). Our findings support this model insofar as they highlight the importance of Dot1 function in MLL leukemias. It is noteworthy that three distinct MLL oncogenes are affected by the loss of Dot1 function, including oncogenes that are unlikely to act through an AEP-initiated process. However, Dot1 activity seems to be most important in the case of MLL-AF9 for which recruitment of an AEP complex seems to be critical for leukemogenesis. The results presented here also show that Dot1 is not a requirement for cellular immortalization by all leukemia-associated oncogenes. This is shown by the continued growth and proliferation of cells immortalized by E2a-Pbx1 following Dot1l excision. Thus, at least under some circumstances, cells can survive indefinitely in the absence of Dot1.
In sum, we conclude that Dot1-mediated chromatin modifications are essential for the continued survival of MLL leukemias. Specific inhibitors of the methyltransferase activity of Dot1 have yet to be described, and given the importance of the enzyme, may have significant systemic toxicity. Nonetheless, if toxicity can be limited, pharmacologic inhibition of Dot1 may provide a useful adjunct for the treatment of MLL leukemias.
Supplementary Material
Acknowledgments
Drs. Jeffrey J. Lin and Zhijing Zhang provided important technical assistance. We thank Dr. Neal G. Copeland for providing reagents of the recombineering system. We greatly appreciate the assistance of Drs. Yinhuai Chen and Tina Grisham at the Gene Targeted Mouse Service Core, Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati, for helping generate Dot1l+/n mice. We thank Drs. Michael L. Cleary and Warren Pear for sharing retroviral constructs and Dr. Jiwang Zhang for providing DNA constructs as well as valuable suggestions. The LSUHSC Proteomics Core Facility is partially supported by the LSUHSC-School of Medicine and the Louisiana Cancer Research Consortium-Stanley S. Scott Cancer Center.
Grant Support
This work was supported by the National Institutes of Health (CA 105049 to N. Zeleznik-Le, CA 098459 to C.S. Hemenway, and DK 080236 to W. Zhang), the American Heart Association (Beginning Grant-in-Aid 0865271F to W. Zhang), the American Society of Nephrology (Carl W. Gottschalk Research Scholar Award to W. Zhang), and the Leukemia and Lymphoma Society (7327-07 to C.S. Hemenway).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
The authors have no conflicts of interest to disclose
References
- 1.Singer MS, Kahana A, Wolf AJ, et al. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–32. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawada K, Yang Z, Horton JR, Collins RE, Zhang X, Cheng X. Structure of the conserved core of the yeast. Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J Biol Chem. 2004;279:43296–306. doi: 10.1074/jbc.M405902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Hayashizaki Y, Kone BC. Structure and regulation of the mDot1 gene, a mouse histone H3 methyltransferase. Biochem J. 2004;377(Pt 3):641–51. doi: 10.1042/BJ20030839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng HH, Feng Q, Wang H, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–27. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacoste N, Utley RT, Hunter JM, Poirier GG, Côte J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–4. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 6.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–56. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 7.Feng Q, Wang H, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–8. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 8.Steger DJ, Lefterova MI, Ying L, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–39. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci USA. 2003;100:1820–5. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–68. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Z, Kong Q, Kone BC. CREB trans-activation of disruptor of telomeric silencing-1 mediates forskolin inhibition of CTGF transcription in mesangial cells. Am J Physiol Renal Physiol. 2010;298:F617–24. doi: 10.1152/ajprenal.00636.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Büttner N, Johnsen SA, Kügler S, Vogel T. Af9/Mllt3 interferes with Tbr1 expression through epigenetic modification of histone H3K79 during development of the cerebral cortex. Proc Natl Acad Sci USA. 2010;107:7042–7. doi: 10.1073/pnas.0912041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balgobind BV, Raimondi SC, Harbott J, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. 2009;114:2489–96. doi: 10.1182/blood-2009-04-215152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marschalek R. Mixed lineage leukemia: roles in human malignancies and potential therapy. FEBS J. 2010;277:1822–31. doi: 10.1111/j.1742-4658.2010.07608.x. [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monica K, LeBrun DP, Dedera DA, Brown R, Cleary ML. Transformation properties of the E2a-Pbx1 chimeric oncoprotein: fusion with E2a is essential, but the Pbx1 homeodomain is dispensable. Mol Cell Biol. 1994;14:8304–14. doi: 10.1128/mcb.14.12.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So CW, Cleary ML. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol Cell Biol. 2002;22:6542–52. doi: 10.1128/MCB.22.18.6542-6552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.So CW, Karsunky H, Passegué E, Cozzio A, Weissman IL, Cleary ML. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 2003;3:161–71. doi: 10.1016/s1535-6108(03)00019-9. [DOI] [PubMed] [Google Scholar]
- 21.Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omon-kowska M, et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010;17:62–8. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones B, Su H, Bhat A, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin ME, Milne TA, Bloyer S, et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4:197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 24.So CW, Lin M, Ayton PM, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4:99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 25.Liedtke M, Ayton PM, Somervaille TC, Smith KS, Cleary ML. Self-association mediated by the Ras association 1 domain of AF6 activates the oncogenic potential of MLL-AF6. Blood. 2010;116:63–70. doi: 10.1182/blood-2009-09-243386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ida K, Kitabayashi I, Taki T, et al. Adenoviral E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13) Blood. 1997;90:4699–704. [PubMed] [Google Scholar]
- 27.Sobulo OM, Borrow J, Tomek R, et al. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t (11;16)(q23;p13.3) Proc Natl Acad Sci USA. 1997;94:8732–7. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.So CW, Cleary ML. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol Cell Biol. 2002;22:6542–52. doi: 10.1128/MCB.22.18.6542-6552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.So CW, Cleary ML. Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood. 2003;101:633–9. doi: 10.1182/blood-2002-06-1785. [DOI] [PubMed] [Google Scholar]
- 30.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 31.Popovic R, Riesbeck LE, Velu CS, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–22. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang CP, de Vivo I, Cleary ML. The Hox cooperativity motif of the chimeric oncoprotein E2a-Pbx1 is necessary and sufficient for oncogenesis. Mol Cell Biol. 1997;17:81–8. doi: 10.1128/mcb.17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamps MP, Wright DD. Oncoprotein E2A-Pbx1 immortalizes a myeloid progenitor in primary marrow cultures without abrogating its factor-dependence. Oncogene. 1994;9:3159–66. [PubMed] [Google Scholar]
- 34.van Leeuwen F, van Steensel B. Histone modifications: from genome-wide maps to functional insights. Genome Biol. 2005;6:113. doi: 10.1186/gb-2005-6-6-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsutsumi S, Taketani T, Nishimura K, Ge X, Taki T, Sugita K, et al. Two distinct gene expression signatures in pediatric acute lymphoblastic leukemia with MLL rearrangements. Cancer Res. 2003;63:4882–7. [PubMed] [Google Scholar]
- 36.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–78. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–89. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reisenauer MR, Anderson M, Huang L, Zhang Z, Zhou Q, Kone BC, et al. AF17 competes with AF9 for binding to Dot1a to up-regulate transcription of epithelial Na+ channel alpha. J Biol Chem. 2009;284:35659–69. doi: 10.1074/jbc.M109.038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–54. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–68. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller D, García-Cuéllar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin JJ, Hemenway CS. Hsp90 directly modulates the spatial distribution of AF9/MLLT3 and affects target gene expression. J Biol Chem. 2010;285:11966–73. doi: 10.1074/jbc.M110.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.