Abstract
Protein glycosylation on serine/threonine residues with N-acetylglucosamine (O-GlcNAc) is a dynamic, inducible and abundant post-translational modification. It is thought to regulate many cellular processes and there are examples of interplay between O-GlcNAc and protein phosphorylation. In metazoa, a single, highly conserved and essential gene encodes the O-GlcNAc transferase (OGT) that transfers GlcNAc onto substrate proteins using UDP–GlcNAc as the sugar donor. Specific inhibitors of human OGT would be useful tools to probe the role of this post-translational modification in regulating processes in the living cell. Here, we describe the synthesis of novel UDP–GlcNAc/UDP analogues and evaluate their inhibitory properties and structural binding modes in vitro alongside alloxan, a previously reported weak OGT inhibitor. While the novel analogues are not active on living cells, they inhibit the enzyme in the micromolar range and together with the structural data provide useful templates for further optimisation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00726-010-0688-y) contains supplementary material, which is available to authorized users.
Keywords: O-GlcNAc, Post-translational modification, Inhibitor, Signalling, Crystal structure
Introduction
Reversible post-translational modification of many cytoplasmic and nuclear proteins in eukaryotic cells by glycosylation of serine and threonine residues with β-linked N-acetylglucosamine (O-GlcNAc) has been shown to regulate cellular processes as diverse as transcription, translation, insulin sensitivity, protein trafficking and degradation (Torres and Hart 1984; Zachara and Hart 2004; Love and Hanover 2005; Hart et al 2007). Only two enzymes are responsible for the dynamic cycling of O-GlcNAc. The O-GlcNAc transferase (OGT) transfers GlcNAc, using UDP–GlcNAc as the sugar donor, via an inverting mechanism involving as yet unidentified active site residues. The O-GlcNAc hydrolase (OGA) cleaves the glycosidic bond, thus reversing the modification. Dysregulation of O-GlcNAc is thought to play a role in human pathogenesis, such as cancer (Chou and Hart 2001; Liu et al. 2002; Donadio et al. 2008), Alzheimer’s (Griffith and Schmitz 1995; Yao and Coleman 1998; Liu et al. 2004; Wells and Hart 2003; Dias and Hart 2007) and diabetes (McClain et al. 2002; Copeland et al. 2008). Hundreds of cytoplasmic and nucleoplasmic proteins have been shown to be O-GlcNAc modified, although the precise glycosylation sites and functional implications have been determined for only a few of these. Interestingly, examples of crosstalk between protein O-GlcNAcylation and phosphorylation have been recently reported, with the O-GlcNAcylation site being either identical or adjacent to protein phosphorylation sites (Yang et al. 2006). However, the precise molecular mechanisms by which OGT and OGA recognise and act on hundreds of proteins, thereby regulating cellular signalling cascades, remain to be discovered (Hurtado-Guerrero et al. 2008). The OGA enzyme has been characterised in humans, rat, Drosophila and C. elegans (Kelly and Hart 1989; Dong and Hart 1994; Gao et al. 2001; Comtesse et al. 2001; Forsythe et al. 2006). The OGA reaction mechanism has been elucidated and structural insights have been obtained recently from bacterial OGA homologues (Macauley et al. 2005; Rao et al. 2006; Dennis et al. 2006; Ficko-Blean et al. 2009). A wealth of chemical biological tools exist to raise intracellular O-GlcNAc levels in living cells by inhibition of O-GlcNAcase. Until recently the only inhibitors of hOGA were the aspecific compounds PUGNAc (Haltiwanger et al. 1998) and streptozotocin (STZ) (Liu et al. 2000). Lately, several new compounds have been described that selectively and potently inhibit human OGA (Macauley et al. 2005; Dennis et al. 2006; Dorfmueller et al. 2006; Dorfmueller et al. 2009; Stubbs et al. 2006; Yuzwa et al. 2008). These chemical tools are currently enabling studies towards the role of O-GlcNAc in a range of signal transduction pathways, although it is becoming clear that certain cell types are remarkably tolerant of inhibitor-induced hyper-O-GlcNAcylation.
OGT was discovered first in rat (Haltiwanger et al. 1992), subsequently in Arabidopsis thaliana (Jacobsen et al. 1996), humans (Kreppel et al. 1997), C. elegans (Lubas et al. 1997) and more recently in Giardia and Cryptosporidium parvum (Banerjee et al. 2009). The gene that encodes for human OGT (hOGT) resides on the X chromosome and is essential for mammalian development (Shafi et al. 2000). hOGT is ubiquitously expressed, however, at particularly high levels in T cells, B cells and macrophages, whereas lower expression levels are found in pancreatic β-cells and the central nervous system (Hanover et al. 2009).
The first insights into OGT structure have recently been obtained from an apparent bacterial OGT orthologue from Xanthomonas campestris (XcOGT) (Clarke et al. 2008; Martinez-Fleites et al. 2008). Structural complexes with UDP and an UDP–GlcNAc phosphonate analogue revealed features of the active site and three distinct domains: (1) multiple tetratricopeptide repeats (TPRs), (2) a linker region and (3) the catalytic (glycosyltransferase activity) domain, belonging to the GT41 family in the CAZy database (Coutinho et al. 2003). The active site is located between the two lobes of the GT41 domain. While hOGT mutants informed by the structural complexes (Clarke et al. 2008; Martinez-Fleites et al. 2008) have helped to identify several inactive mutants, however, the precise catalytic mechanism of OGT yet remains to be discovered.
The O-GlcNAc modification is a dynamic modification, and thus potent and selective hOGT inhibitors would be beneficial to investigate the role of O-GlcNAc modifications in signalling pathways. The first hOGT inhibitor reported was alloxan, an uracil analogue that is presumed to inhibit hOGT by binding to the uracil binding pocket or alternatively has been proposed to act through a covalent modification of cysteine residues (Konrad et al. 2002). Alloxan, as a small chemical compound, is unlikely to inhibit hOGT specifically in vivo (Zachara et al. 2004). Furthermore, alloxan is a chemically unstable compound with a half-life time of 1.5 min at physiological pH (Lenzen and Munday 1991). Although alloxan inhibits hOGT with an IC50 of 100 μM (Konrad et al. 2002), it is required at millimolar concentrations to decrease cellular levels of O-GlcNAcylation (Liu et al. 2005, 2006, 2007; Noach et al. 2007; Kang et al. 2008). Recently, three novel hOGT inhibitors were identified by high-throughput screening (HTS) (Gross et al. 2005), although the original report did not show that these compounds were able to decrease global cellular O-GlcNAc levels by inhibition of hOGT in living cells. A very recent report showed that one of these inhibitors identified by HTS reduces O-GlcNAc levels in MCF-10A-ErbB2 breast cancer cells when applied at 500 μM (Caldwell et al. 2010).
Here, we report the design and evaluation of a novel glycosyl thiophosphate analogue of UDP–GlcNAc (UDP-S-GlcNAc) and an α,β-methylene bisphosphonate analogue of UDP (C-UDP) as hOGT inhibitors. We have determined inhibition constants of these compounds against hOGT and used X-ray crystallography to demonstrate that the uridine pyrophosphate moiety (or its mimic) occupies the nucleotide sugar binding site in both cases. We have also determined the binding mode of alloxan, a widely used (unspecific) hOGT inhibitor. Together, these compounds and associated data form the basis for the future rational design of more potent substrate analogue OGT inhibitors.
Materials and methods
Protein production and purification
XcOGT wild-type (WT), mutant D471A-XcOGT and hOGT (26-end) were expressed and purified as described previously (Clarke et al. 2008). The proteins were concentrated to 10 mg/ml (XcOGT, D471A-XcOGT) and 10 μM (hOGT), respectively.
Surface plasmon resonance experiments
The molecular interaction between WT/mutant D471A-XcOGT and nucleotide analogues were studied by surface plasmon resonance (SPR), using a BIAcore T100 instrument (BiaCore, Uppsala, Sweden). WT and the D471A mutant-XcOGT were immobilised using standard amine-coupling chemistry in buffer consisting of 10 mM HEPES (pH 7.4) and 150 mM NaCl. The sensor chip surface was activated at 25°C with a 7 min injection of a 1:1 mixture of EDC and NHS. Protein was injected at 0.2 mg/ml in acetate buffer pH 5.5 at a flow rate of 10 μl/min for 1 min to achieve suitable density of approximately 5000 RU. Finally, 1 M ethanolamine–HCl was injected for 7 min to deactivate the surface. One flow cell was used as a reference to subtract nonspecific binding, drift and bulk refractive index. All experiments were performed in 25 mM Tris–HCl (pH 7.5), and 150 mM NaCl at 25°C. Compounds were diluted in the running buffer at a maximum concentration of 1 mM and then threefold diluted to a concentration series ranging from 1 mM to 457 nM. Association was measured for 30 s and dissociation for 60 s at a 30 μl/min flow rate. Injections were carried out in duplicate. Blank injections of running buffer were used during the experiment to control the drift of baseline. As the experimental run time exceeded 15 h, control injections of UDP at 100 μM were included within the run at every eighth sample injection to detect possible changes of activity of immobilised proteins on the surface. Data processing and affinity analysis were performed using Scrubber 2 software (BioLogic Software). Data were corrected for blank reference surface to subtract non-specific binding and for blank buffer injections to subtract possible drift in sensorgrams. Binding of compounds was measured using SPR to WT-XcOGT and mutant D417A. No decrease of response for control injections of UDP (100 μM) was observed during the experimental run, suggesting that the binding capacity of immobilised protein remained the same (data not shown). Data were fitted using equilibrium affinity analysis. Since most of the compound affinities were in millimolar range exceeding the highest concentration of injected compound, the maximum binding capacity (RMAX) could not be reached. Therefore, to calculate RMAX, UDP was used as a control. The affinity of UDP was calculated as 110 μM and reached maximum binding capacity of approximately 36 RU. The responses for the rest of the compounds were normalised according to their molecular weights and fitted using equilibrium fit with RMAX previously calculated for UDP. In this way, binding affinities could be estimated for compounds as weak as 30 mM.
Determination of the XcOGT complex crystal structures
Purified WT-XcOGT protein was crystallised essentially as described previously (Clarke et al. 2008). Vapour diffusion crystallisation experiments were performed by mixing 1 μl of XcOGT protein (10 mg/ml) and 1 μl of mother liquor (0.1 M CHES, pH 9.5 and 25% polyethylene glycol 3350). To obtain the XcOGT–UDP-S-GlcNAc/C-UDP and alloxan complexes, crystals were soaked by addition of 1 μl of 20 mM UDP-S-GlcNAc/C-UDP dissolved in mother liquor or solid alloxan. The crystals were cryoprotected by immersion in mother liquor with increased (40%) polyethylene glycol 3350 concentration. Synchrotron diffraction data were collected and processed using the HKL suite (Pape and Schneider 2004). Refinement of the ligand complexes was initiated from the native XcOGT–UDP–GlcNAc phosphonate structure (2JLB) (Clarke et al. 2008). Further model building with COOT (Emsley and Cowtan 2004) and refinement with REFMAC (Murshudov et al. 1997) yielded the final models with statistics shown in Table 3. In all three complexes, defined |F o|−|F c|, ϕcalc electron density for the compounds was apparent from the early stages of refinement. Compound models were built and refined using coordinates and topologies generated by PRODRG (Schuettelkopf and van Aalten 2004). Comparison of the XcOGT–UDP-S-GlcNAc and XcOGT–alloxan complexes with previously described XcOGT structures (Clarke et al. 2008; Martinez-Fleites et al. 2008) shows that there is more disorder in the N-terminal TPR domains, which were only partially built. This is reflected in higher B-factors (Table 3).
Table 3.
Details of data collection and structure refinement for UDP-S-GlcNAc, C-UDP and alloxan bound to XcOGT
| XcOGT–UDP-S-GlcNAc | XcOGT–C-UDP | XcOGT–alloxan | |
|---|---|---|---|
| Space group | P212121 | P212121 | P212121 |
| Unit cell (Å) | a = 83.7 | a = 84.3 | a = 83.5 |
| b = 100.0 | b = 95.0 | b = 100.0 | |
| c = 155.5 | c = 157.7 | c = 157.2 | |
| Resolution range (Å) | 30.00–2.60 (2.69–2.60) | 30.00–2.40 (2.46–2.40) | 30.00–2.55 (2.64–2.55) |
| # Observed reflections | 181,565 | 170,621 | 171,199 |
| # Unique reflections | 37,400 (3,453) | 47,424 (2,347) | 42,658 (4,136) |
| Redundancy | 4.9 (4.9) | 3.6 (3.2) | 4.0 (3.9) |
| I/σI | 32.5 (2.9) | 13.6 (2.15) | 16.3 (2.1) |
| Completeness (%) | 91.7 (86.4) | 92.9 (70.1) | 97.8 (96.7) |
| R merge | 0.061 (0.691) | 0.067 (0.543) | 0.048 (0.438) |
| R, R free | 23.9, 27.7 | 24.4, 30.4 | 25.4, 29.7 |
| RMSD from ideal geometry | |||
| Bonds (Å) | 0.01 | 0.01 | 0.02 |
| Angles (°) | 1.5 | 1.4 | 1.5 |
| B-factor RMSD (backbone bonds) <B> (Å2) | 0.5 | 0.5 | 0.5 |
| Protein | 73.4 | 45.0 | 75.6 |
| Ligand | 78.7 | 38.2 | 71.7 |
| Solvent | 66.6 | 48.1 | 72.1 |
Values within brackets are for the highest resolution shell. All measured data were included in structure refinement
Inhibition measurements
hOGT assays were carried out at room temperature (RT) in 384-well white optiplates (Perkin Elmer). Each assay was performed in a 20 μl reaction volume containing 50 mM Tris buffer, pH 7.5, 2 mM dithiothreitol (DTT), 3 mM MgCl2, 0.05 mg/ml BSA (Thermo), 0.125 μM [3H]UDP–GlcNAc (0.048 Ci mmol−1) (Sigma/ARC), 1 μM biotinylated DEBtide (Blair et al. manuscript submitted) (GlycoBioChem), 50 nM hOGT and various concentrations of test compounds. Test compounds were placed in the columns 1 and 13 of a 384-well polypropylene plate and then serially diluted in 100% DMSO through half log increments using a JANUS 8-channel Varispan automated workstation (PerkinElmer) to give the compound source plate (100× final assay concentration). From this, 0.25 μl of the compounds were transferred to all wells using a Cartesian Hummingbird (Genomics Solution) before 10 μl of either enzyme/peptide mix or peptide alone (control) was added to the assay plates. The reaction was initiated with 10 μl of UDP–GlcNAc and stopped after 150 min with 40 μl of a stop solution containing 0.2 M phosphoric acid, pH 4.0 and 1.5 M MgCl2 and 2.5 mg/ml PVT SPA beads (GE Healthcare). All reaction mixture additions were carried out using a Thermo Scientific WellMate (Matrix). Plates were sealed and read on a TopCount NXT Microplate Scintillation and Luminescence Counter (Perkin Elmer). Dose–response curves were prepared and analysed using GraFit (Leatherbarrow 2001) (Fig. 1b).
Fig. 1.
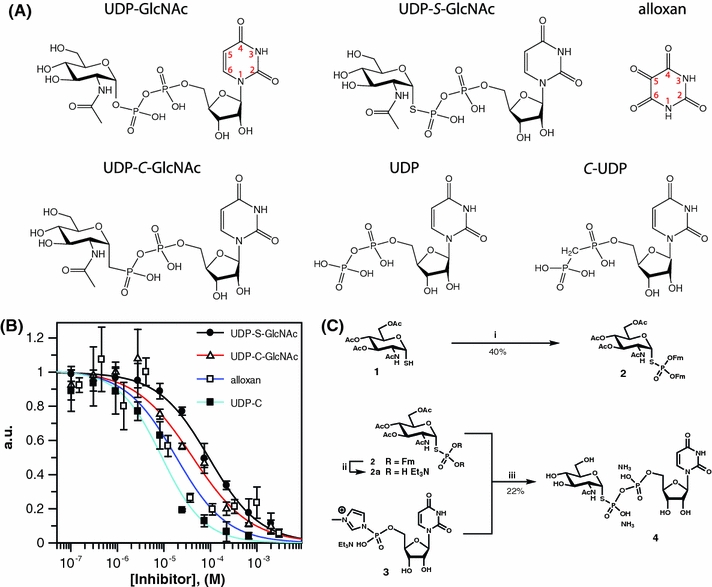
Design, synthesis and inhibitory properties of hOGT substrate/product analogues. a Chemical structures of UDP–GlcNAc, the substrate of hOGT, the substrate analogues UDP-C-GlcNAc and UDP-S-GlcNAc, the hOGT product UDP and the analogue C-UDP and alloxan. The atoms in the uracil moiety are numbered. Alloxan ring atoms are numbered according to the uracil numbering system. b Dose–response curves of the inhibitors with hOGT. The transfer of GlcNAc from UDP–GlcNAc onto a peptide [DEBtide (Blair et al. manuscript submitted)] by hOGT was assayed in presence of increasing concentrations of UDP-S-GlcNAc, UDP-C-GlcNAc, alloxan and C-UDP. The remaining activity is plotted against the inhibitor concentration. Data were fitted using the standard IC50 equation in the GraFit program (Leatherbarrow 2001). c Reagents and conditions for UDP-S-GlcNAc synthesis: (i) Bis(9H-fluoren-9-ylmethyl)-diisopropylamidophosphite, 1H-tetrazole, MeCN, RT, 30 min; then dimethyldioxirane, −40°C to −20°C, 30 min; (ii) DCM:Et3N (4:1), RT, 24 h; (iii) (a) 2a + 3, MeCN, RT, 16 h; (b) MeOH:H2O:Et3N (5:2:1), RT, 16 h; followed by size exclusion chromatography
Synthesis of UDP-S-GlcNAc
Compound 2 (Fig. 1c): To a solution of 2-acetamido-2-deoxy-3,4,6-tri-O,O,O-acetyl-1-thio-α-d-glucopyranose (Knapp and Myers 2001) (0.393 g, 1.08 mmol) and bis(9H-fluoren-9-ylmethyl)-diisopropylamidophosphite (Bialy and Waldmann 2004) (0.846 g, 1.6 mmol) in MeCN (4 ml), a stock 0.45 M solution of 1H-tetrazole (7.1 ml, 3.2 mmol) was added at RT. After 30 min, the reaction mixture was cooled down to −40°C and a solution of dimethyldioxirane (19 ml; 0.1 M, 1.9 mmol) was added in one portion. The reaction mixture was further stirred for 30 min while the temperature was allowed to reach −20°C. The reaction was removed from the cooling bath and concentrated. The residue was dissolved in DCM and washed with water, dried and concentrated. The residue was purified by flash chromatography in Tol-Me2CO 5–30% to give 0.344 g of the target product 2 (0.43 mmol; 40%) (supplementary materials).
Compound 4 (Fig. 1c): A solution of the protected thiophosphate 2 (0.12 g, 0.15 mmol) in a mixture of DCM (3 ml) and triethylamine (0.75 ml) was kept at RT for 24 h. The reaction mixture was concentrated and dried in vacuum to give crude glycosyl thiophosphate (2a), (supplementary materials).
To a stirred slurry of uridine 5′-monophosphate bis-triethylammonium salt (0.076 g, 0.144 mmol) in a mixture of anhydrous MeCN (0.6 ml), N, N-dimethylaniline (0.070 ml, 0.56 mmol) and Et3N (0.04 ml, 0.288 mmol), a solution of trifluoroacetic anhydride (0.12 ml, 0.864 mmol) in MeCN (0.2 ml) was added via capillary at 0°C. The reaction mixture was stirred for a few minutes at RT, while clear yellowish solution was formed and concentrated to remove excess of trifluoroacetic anhydride. The yellow oily residue was re-dissolved in MeCN (0.5 ml) and cooled to 0°C. A solution of N-methylimidazole (0.057 ml, 0.72 mmol) and Et3N (0.12 ml, 0.864 mmol) in anhydrous MeCN (0.2 ml) was then added to the above solution via capillary. The reaction mixture was further stirred for 15 min at RT. The prepared solution of UMP-N-methylimidazolide (3) was added via capillary to a flask containing a solution of the crude product 2a in MeCN (0.8 ml) at 0°C. The resulting turbid yellow solution was further stirred overnight at RT. The reaction was quenched by addition of cold Et3NHHCO3 buffer (1 ml) and stirred for 20 min. The reaction was concentrated; the residue was dissolved in a mixture of MeOH:H2O:Et3N (5:2:1, 8 ml) and kept at RT for 16 h. The reaction mixture was concentrated, diluted with water, and extracted with DCM. The layers were separated and the aqueous phase was additionally extracted with DCM. The aqueous layer was concentrated to give yellowish oily residue. This was purified by size exclusion chromatography (Bio-Gel P2 fine; column 2.6 × 100 cm; flow rate 0.4 ml/min; elution with 0.25 M NH4HCO3). The fractions containing the product were pooled and evaporated. The residue was re-dissolved in water and freeze dried to give 0.0216 g (0.012 mmol, 22%) of the target product 4 (95% purity by 1H NMR) as fluffy white solid (supplementary materials).
UDP-C-GlcNAc and C-UDP analogues were synthesised as described previously (Clarke et al. 2008; Borodkin et al., manuscript submitted). Alloxan was purchased from Sigma–Aldrich.
Results and discussion
Design and synthesis of substrate/product analogues as potential OGT inhibitors
In the absence of potent, cell permeable OGT inhibitors we decided to explore substrate analogues as potential inhibitor scaffolds. We aimed to investigate UDP–GlcNAc analogues as hOGT inhibitors, in which the glycosidic oxygen would be replaced with non-hydrolysible isosteric linkages conferring stability towards enzymatic and chemical hydrolysis. Recent studies have described the synthesis and evaluation of a UDP-GlcNAc glycosyl C-phosphonate analogue (UDP-C-GlcNAc, Fig. 1a) against O-GlcNAc transferases of different origin (Clarke et al. 2008; Chang et al. 2006). Interestingly, UDP-C-GlcNAc did not show any inhibitory activity against hOGT (Hajduch et al. 2008), potentially indicating an essential role of the glycosidic oxygen atom in substrate binding. It is possible that the lack of hydrogen bonds between the isosteric, albeit non-isopolar, methylene group of the glycosyl C-phosphonate explains why UDP-C-GlcNAc is a weak hOGT inhibitor.
In a search for novel non-hydrolysable UDP–GlcNAc congeners with enhanced biological activity profiles, a thioglycosyl (thiophosphate) analogue of UDP–GlcNAc (UDP-S-GlcNAc, Fig. 1a) was envisaged as a possible target. Replacement of the glycosyl oxygen with the sulphur atom would give rise to an analogue, which despite not being isosteric (van der Waals radius of sulphur is 1.85 Å compared to 1.44 Å for oxygen) would still bear more resemblance to the natural O-linked substrate than the glycosyl C-phosphonate analogue. This non-hydrolysable substrate analogue might further reveal insight into the binding mode of UDP–GlcNAc, since the previously reported XcOGT–UDP-C-GlcNAc complex (Clarke et al. 2008) revealed a substrate binding site which is not fully compatible with catalysis of larger nucleotide–sugar analogues that were reported previously (Gross et al. 2005; Laughlin and Bertozzi 2009).
No thiophosphate analogues of nucleotide diphosphate sugars have so far been synthesised, except for an anomeric sulphur analogue of CMP-Neu5Ac (Cohen and Halcomb 2000). This unique compound was found to be surprisingly hydrolytically stable compared to the natural O-linked substrate and two orders of magnitude poorer substrate for α-2,3-sialyl transferase than the natural O-linked substrate (Cohen and Halcomb 2000). No evidence of a perceptible inhibition of the sialyl transferase activity with this compound was reported.
We envisioned that the UDP-S-GlcNAc (Fig. 1a) could be prepared by coupling of suitably protected GlcNAc glycosyl thiophosphate and activated 5′-uridine monophosphate by analogy with the common practice of pyrophosphate bond construction in the chemical synthesis of nucleoside diphosphate sugars. Initially, GlcNAc diallylthiophosphate (not shown) was prepared from the α-configured GlcNAc glycosyl thiol 1 (Fig. 1c) as a potential precursor of GlcNAc glycosyl thiophosphate according to the published procedure (Knapp and Myers 2001). However, all attempts to liberate thiophosphate functionality by removing allyl groups from this compound using tetrakis (triphenylphosphine) palladium catalyst in the presence of various allyl cation scavengers were utterly unsuccessful, leading predominantly to the formation of dephosphorylated products. Seeking for an alternative solution, we suggested that change of the thiophosphate protecting groups to 9-fluorenylmethyl would allow the deprotection in mild anhydrous conditions. Thus, the glycosyl thiol 1 (Fig. 1c) was S-phosphitylated with bis(9H-fluoren-9-ylmethyl)-diisopropylamidophosphite (Bialy and Waldmann 2004) followed by oxidation with dimethyldioxirane to give the GlcNAc bis-9-fluorenylmethyl glycosyl thiophosphate 2 (Fig. 1c) in acceptable yield (31P; δ 23.36). Removal of the protecting groups by treatment with Et3N in DCM for 24 h produced the requisite glycosyl thiophosphate 2a (Fig. 1c) as judged by 31P NMR spectrum, showing the presence of the single phosphorous containing product (31P; δ 13.06). The crude glycosyl thiophosphate was then reacted with UMP-N-methylimidazolide 3 (Fig. 1c; Marlow and Kiessling 2001) to furnish the target molecule in moderate yield after deprotection and purification by gel filtration chromatography. Formation of the pyrophosphate bond was evident from the presence of typical pair of doublets δ 5.10 and −11.97 (J P,P′ = 29 Hz) in the 31P spectrum of compound 4.
To investigate the inhibition of UDP–GlcNAc and UDP analogues, kinetic studies were carried out using recombinant hOGT protein. A synthetic peptide (DEBtide) was used as the acceptor substrate. The incorporation of [3H]-GlcNAc from [3H]-UDP–GlcNAc was monitored using a newly developed radioactive assay (Blair et al. manuscript submitted). The K m for UDP–GlcNAc was determined to be 1 μM, in agreement to the K m determined previously (Haltiwanger et al. 1992). In our hands, hOGT displays product inhibition with UDP (IC50 of 1.8 μM, Fig. 1a). We therefore also decided to explore the inhibition profile of a non-hydrolysible α,β-methylene bisphosphonate analogue with the pyrophosphate oxygen replaced by a methylene group (C-UDP).
Synthesis of C-UDP was performed using published procedures (Borodkin et al., manuscript submitted).
UDP-S-GlcNAc—a sub-millimolar inhibitor of hOGT and substrate binding probe
Using the glycosyltransferase assay with a model acceptor peptide, UDP-S-GlcNAc appears to inhibit hOGT with an IC50 of 93 μM (Fig. 1b; Table 1), similar to the previously described weak hOGT inhibitor UDP-C-GlcNAc (Clarke et al. 2008; Hajduch et al. 2008) (IC50 = 41 μM, Fig. 1b; Table 1). Human OGT has so far resisted expression and purification to levels required for structural studies and biophysical measurement of ligand affinity. To further study the binding mode of the novel UDP–GlcNAc analogues, we therefore used the apparent hOGT homologue from Xanthomonas campestris pv. campestris (XcOGT) for which structural data and protein crystals suitable for soaking studies are available (Clarke et al. 2008; Martinez-Fleites et al. 2008). SPR experiments with recombinant and purified XcOGT protein revealed that UDP has a binding affinity (K d) of 112 μM for XcOGT (Table 2), which should be compared to inhibition with IC50 = 1.8 μM of hOGT activity (Tables 1, 2). The micromolar hOGT inhibitors UDP-S-GlcNAc and UDP-C-GlcNAc both bind XcOGT weakly, with a K d > 1 mM (Table 2). UDP–GlcNAc itself binds to XcOGT weakly with a K d of approximately 15 mM (Table 2). We next investigated whether UDP-S-GlcNAc is a substrate for hOGT or XcOGT. Kinetic experiments using recombinant hOGT/XcOGT and protein acceptor substrates in conjunction with a general O-GlcNAc antibody did not show any evidence for O-GlcNAcylation. Furthermore, a colorimetric assay with the thio-reactive reagent DTNB [5,5′-dithio-bis(2-nitrobenzoic acid)], and a fluorescent assay with N-(1-pyrene)maleimide did not shown any hydrolysis of UDP-S-GlcNAc by hOGT or XcOGT.
Table 1.
Inhibition data of the hOGT substrate/product analogues
| Compound | IC50 [μM]a | BEIb |
|---|---|---|
| UDP | 1.8 ± 1.0 | 15 |
| C-UDP | 9.0 ± 1.0 | 13 |
| UDP-C-GlcNAc | 41 ± 7 | 7 |
| UDP-S-GlcNAc | 93 ± 15 | 7 |
| Alloxan | 18 ± 1 | 34 |
aThe dose–response experiments were performed with a UDP-GlcNAc concentration tenfold below the K m
bBinding efficiency index (Abad-Zapatero and Metz 2005), BEI = −log(K i)/M, with M being the mass in kDa and K i is derived from IC50 data using the Cheng and Prusoff equation with [S] = 125 nM and K m = 1 μM. Cheng–Prusoff equation: K i = IC50/1 + ([S]/K m)
Table 2.
Dissociation constant (K d) of compounds for XcOGT-WT from SPR experiments
| Compound | K d (mM) | |
|---|---|---|
| WT-XcOGT | D471A-XcOGT | |
| UDP | 0.110 ± 0.001 | n.b. |
| C-UDP | 0.530 ± 0.006 | n.b. |
| UDP-GlcNAc | 15.1 ± 0.5 | n.b. |
| UDP-S-GlcNAc | >1 | >1 |
| UDP-C-GlcNAc | >1 | >1 |
| Alloxan | >1 | n.d. |
n.b. = no binding
n.d. = not determined
To investigate the binding mode of the UDP-S-GlcNAc substrate analogue, XcOGT protein crystals were soaked with the compound. Collection of synchrotron diffraction data and subsequent refinement yielded a final model with statistics shown in Table 3. UDP-S-GlcNAc binds in the active site of XcOGT (Fig. 2a). The uracil moiety occupies a pocket formed by Lys441 to His444, whilst stacking with the side chain of Tyr447 (Fig. 2a). The uracil moiety is involved in a total of three hydrogen bonds with the protein, with the backbone of Leu442/Pro443 and the His444 side chain (Fig. 2a). The ribose moiety of UDP-S-GlcNAc makes a hydrogen bond with Asp471 (Fig. 2), a side chain that is conserved in OGTs from organisms across the evolutionary spectrum. Recent studies reported that a mutant of the equivalent position in hOGT (Asp925) is no longer able to O-GlcNAcylate an acceptor substrate (Clarke et al. 2008; Martinez-Fleites et al. 2008). SPR measurements show that the D471A-XcOGT mutant no longer binds UDP–GlcNAc (Table 2).
Fig. 2.
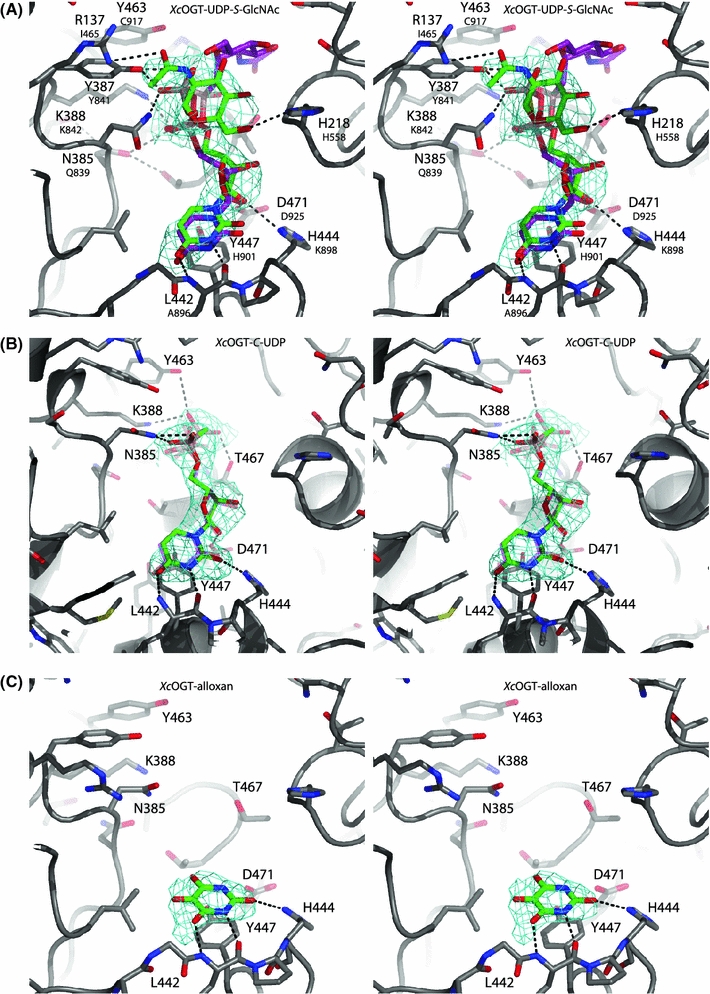
X-ray crystallographic determination of inhibitor binding modes. a Stereo figure of XcOGT in complex with UDP-S-GlcNAc and superposition of the previously described XcOGT–UDP-C-GlcNAc complex [PDB entry 2JLB (Clarke et al. 2008)]. XcOGT active site residues are shown in sticks with grey carbon, red oxygen, blue nitrogen atoms. UDP-S-GlcNAc and UDP-C-GlcNAc are shown with green and purple carbon, respectively. Hydrogen bonds for the XcOGT–UDP-S-GlcNAc complex are indicated by black dashed lines. Unbiased |F o|−|F c|, ϕcalc electron density map (2.75σ) is shown as cyan chickenwire. b Stereo figure of XcOGT in complex with C-UDP compared to the UDP complex [PDB entry 2VSN (Martinez-Fleites et al. 2008)]. XcOGT active site residues are shown in sticks with grey carbon, red oxygen, blue nitrogen atoms (transparent sticks for XcOGT–UDP), ligands shown in green carbon atoms. Active site residues involved in hydrogen bonds are labelled. Hydrogen bonds are indicated by black dashed lines. Unbiased |F o|−|F c|, ϕcalc electron density map (2.25σ) is shown as cyan chickenwire. c Stereo figure of XcOGT in complex with alloxan. XcOGT active site are shown in sticks with green carbon, red oxygen, blue nitrogen atoms in the active site of XcOGT (sticks with grey carbon atoms). Hydrogen bonds are indicated by black dashed lines. Unbiased |Fo|−|Fc|, ϕcalc electron density map (2.25σ) is shown as cyan chickenwire. Black dashed lines showing hydrogen bonds for XcOGT–alloxan complex
Surprisingly, a structural comparison with the previously described UDP-C-GlcNAc complex (Clarke et al. 2008) shows that while the uridine moiety occupies a similar position in both complexes (maximum atom shift = 0.8 Å), the sugar and phosphate moieties occupy different binding sites (Fig. 2a). The UDP-S-GlcNAc phosphate mimics occupy a groove, interacting with the side chains of Lys388 (Lys842 in hOGT) and Asn385 [Gln839 in hOGT, mutation of which inactivates the enzyme (Clarke et al. 2008; Martinez-Fleites et al. 2008)]. The sugar moiety occupies a position above the pyrophosphate, hydrogen bonding with His218 and Arg137 (Fig. 2a). In contrast to the previously described UDP-C-GlcNAc–XcOGT crystallographic complex (Clarke et al. 2008), this binding mode is compatible with the reported tolerance of hOGT for larger substitutions on the N-acetyl group (Gross et al. 2005; Laughlin and Bertozzi 2009), and also exposes the β-face of the anomeric carbon for nucleophilic attack (Fig. 2a). Loss of activity in hOGT His558 mutants (His218 in XcOGT) has been proposed to reflect a role as a general base in the reaction mechanism (Martinez-Fleites et al. 2008), which would be incompatible with the UDP-S-GlcNAc conformation described here (Fig. 2a). Similarly, Cys917 (Tyr463 in XcOGT) has been proposed to be the catalytic base to activate the peptide substrate, but is not in proximity to the GlcNAc anomeric carbon in the UDP-S-GlcNAc complex. Interestingly, the complex shows that the hydroxyl group of Tyr387 is positioned within 4.5 Å from the anomeric carbon. This Tyr387 is conserved amongst XcOGT and hOGT (Fig. 2a; Clarke et al. 2008) and is in close proximity to Lys388, which could prime the tyrosine to act as a base to activate the acceptor substrate. However, the identity of the hOGT catalytic machinery remains to be firmly established.
The α,β-methylene bisphosphonate UDP is a micromolar hOGT inhibitor
We have found that the α,β-methylene bisphosphonate UDP analogue (C-UDP) appears to inhibit hOGT with an IC50 of 9 μM (Table 1; Fig. 1b). The binding affinity of C-UDP to XcOGT was investigated by SPR experiments, revealing a K d of 530 μM (Table 2). This binding constant is approximately fivefold weaker than for UDP, but in agreement with the ranking observed for hOGT inhibition (Tables 1, 2).
We determined the crystal structure of the XcOGT–C-UDP complex to investigate the molecular basis of the weaker hOGT inhibition and XcOGT binding compared to UDP (Fig. 2b). The uracil ring of C-UDP interacts with the XcOGT protein via the same three hydrogen bonds and π–π stacking interactions as observed for the UDP-S-GlcNAc complex vide supra (Fig. 2a, b). The key hydrogen bond of the ribose with Asp471 is also conserved. In agreement with this, SPR measurements with the XcOGT-D471A mutant revealed that neither UDP nor C-UDP bind to the mutant enzyme (Table 2). Interestingly, however, the α,β-phosphonates are shifted (maximum atomic shift = 1.4 Å) compared to the previously published XcOGT–UDP complex (Martinez-Fleites et al. 2008). While Asn385 and Lys388 still hydrogen bond the phosphonates (albeit with swapped interactors compared to the XcOGT–UDP-S-GlcNAc complex; Fig. 2a), no hydrogen bonds between the β-phosphonate and Thr468/Lys388 are present (4.5 and 3.7 Å, respectively) as a result of the methylene-induced positional shift of the β-phosphonate. However, an additional hydrogen bond is introduced between Tyr468 and the β-phosphonate. This could explain the lower inhibition constant of C-UDP in comparison to UDP (Table 1).
Alloxan mimics the uracil binding mode in the XcOGT structure
The first hOGT inhibitor described was the uracil derivative alloxan (Konrad et al. 2002; Fig. 1a). It was proposed that alloxan inhibits hOGT through covalent modification of cysteine residues (Konrad et al. 2002). In our assay, alloxan is a low micromolar inhibitor of hOGT (IC50 of 18 μM) compared to the previously reported IC50 of 100 μM (Konrad et al. 2002). It is revealing to express the average contribution per atom to the ΔG of binding [the so-called binding efficiency index (BEI) (Abad-Zapatero and Metz 2005)] for the hOGT inhibitors (Table 1). Interestingly, even though the small compound alloxan is a weaker inhibitor of hOGT than UDP and C-UDP, however, in absolute terms, in terms of ligand efficiency alloxan is a more efficient binder (BEI of 34) than UDP and C-UDP (BEI of 15 and 13, respectively). Thus, these data indicate that alloxan, although chemically unstable, may be a very efficient fragment to probe the hOGT uracil-binding site, perhaps as a moiety or as part of a large inhibitor molecule. To allow such inhibitors to be developed, we attempted to obtain an XcOGT–alloxan complex. XcOGT crystals were soaked with alloxan and data were collected to 2.55 Å (Table 3). Well-defined electron density for alloxan is located in the uracil pocket, with the compound mimicking the previously described XcOGT–uracil interactions (Fig. 2c). The C2 and C4/C6-carbonyl groups (numbering system according to Fig. 1a) accept hydrogen bonds from the side chain nitrogen of His444 and the backbone nitrogen atom of Leu442, respectively (Fig. 2c). A third hydrogen bond is formed between the alloxan N1/N3 and the backbone carbonyl of Leu442. π–π stacking interactions are observed with the aromatic side chain of Tyr447 (His901 in hOGT). Previous work has described an ‘active site lid’ (Clarke et al. 2008) that controls access to the donor binding site—this loop is observed to be in the ‘closed’ conformation in the alloxan complex. There is no evidence to support the recent hypothesis that alloxan covalently modifies active site cysteine residues (Konrad et al. 2002). Unexpectedly, SPR experiments carried out with XcOGT revealed a weak binding constant of alloxan to XcOGT (K d > 1 mM). It is possible that alloxan degraded to alloxanic acid over the course of the experiment, but it should also be noted that the two substrate analogues reported here showed significantly weaker binding to XcOGT than their observed inhibition of hOGT.
Concluding remarks
In the search for novel hOGT inhibitors, we have focussed our work on the synthesis and evaluation of novel hOGT substrate and product analogues (Fig. 1). The inhibitory potency of these compounds revealed that they are all sub-millimolar inhibitors of hOGT (Table 1). Structural data obtained from UDP-S-GlcNAc soaking experiments with XcOGT crystals revealed a competitive binding mode, yet different from the previously published UDP-C-GlcNAc complex (Clarke et al. 2008). The sugar moiety projects above the diphosphate group mimic positioning the thioglycosidic bond within hydrogen bonding distance of Asn385, which might play a role in catalysis [mutation of the equivalent Gln839 in hOGT generates an inactive species (Clarke et al. 2008; Martinez-Fleites et al. 2008)]. This could be similar to the role of Gln189 in LgtC, a retaining galactosyltransferase from Neisseria meningitidis (Tvaroska 2004). On the basis of structural, biochemical and molecular modelling, Gln189 in LgtC was predicted to be involved in hydrogen bond formation with the donor and the acceptor in the transition state (Tvaroska 2004). However, the identity of the catalytic base in hOGT remains to be firmly established.
To further investigate the reduced inhibitory effects of the potent UDP analogue, we determined the crystallographic complex of XcOGT–C-UDP. A hydrogen bond network, similar to the previously reported XcOGT–UDP complex, was observed, however, conformational changes for the β-phosphonate result in elimination of two hydrogen bonds which could explain the lower inhibition constant of C-UDP in comparison to UDP (Table 1). Structural and kinetic studies with alloxan, an unspecific hOGT inhibitor, have shown that this small molecule is an efficient fragment to inhibit hOGT. Chemical modifications of this compound (i.e. of the C5/C6-carbonyl groups) can be explored with the help of the binding mode revealed by the structural data presented here.
The novel substrate/product analogue hOGT inhibitors were also tested in cell-based assays, but failed to induce an observable decrease in global O-GlcNAcylation levels. Despite C-UDP being a potent hOGT inhibitor, this nucleotide analogue is a hydrophilic and negatively charged compound that might not be cell permeable. Thus, it remains to be explored whether modified (e.g. esterified) derivatives of C-UDP would be more cell permeable. Finally, it should be noted that all compounds considered in this study may be equally potent inhibitors of other glycosyltransferases using UDP–GlcNAc as the donor (e.g. in N-linked glycosylation), or enzymes involved in sugar nucleotide biosynthesis. It is anticipated that the structural data for the compounds reported here could aid the rational design of more potent and selective substrate/product analogue OGT inhibitors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank the European Synchrotron Radiation Facility, Grenoble, for the time at beam line BM14 and ID14-1. This work was supported by a Wellcome Trust Senior Fellowship and a Lister Institute for Preventive Medicine Research Prize. HCD is supported by the College of Life Sciences Alumni Studentship. The coordinates and structure factors have been deposited with the PDB (PDB entry 2xgm, 2xgs, 2xgo).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abad-Zapatero C, Metz JT. Ligand efficiency indices as guideposts for drug discovery. Drug Discov Today. 2005;10(7):464–469. doi: 10.1016/S1359-6446(05)03386-6. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Robbins PW, Samuelson J. Molecular characterization of nucleocytosolic O-GlcNAc transferases of Giardia lamblia and Cryptosporidium parvum. Glycobiology. 2009;19(4):331–336. doi: 10.1093/glycob/cwn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialy L, Waldmann H. Total synthesis and biological evaluation of the protein phosphatase 2A inhibitor cytostatin and analogues. Chemistry. 2004;10(11):2759–2780. doi: 10.1002/chem.200305543. [DOI] [PubMed] [Google Scholar]
- Blair D, Borodkin V, Gray L, van Aalten D (manuscript submitted)
- Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, Vosseller K, Reginato MJ. Nutrient sensor it O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29:2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- Chang R, Vo T, Finney N. Synthesis of the C1-phosphonate analog of UDP–GlcNAc. Carbohydr Res. 2006;341:1998–2004. doi: 10.1016/j.carres.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Chou TY, Hart GW. O-linked N-acetylglucosamine and cancer: messages from the glycosylation of c-Myc. Adv Exp Med Biol. 2001;491:413–418. doi: 10.1007/978-1-4615-1267-7_26. [DOI] [PubMed] [Google Scholar]
- Clarke AJ, Hurtado-Guerrero R, Pathak S, Schuttelkopf AW, Borodkin V, Shepherd SM, Ibrahim AFM, van Aalten DMF. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 2008;27(20):2780–2788. doi: 10.1038/emboj.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SB, Halcomb RL. Synthesis and characterization of an anomeric sulfur analogue of CMP–sialic acid. J Org Chem. 2000;65(19):6145–6152. doi: 10.1021/jo000646+. [DOI] [PubMed] [Google Scholar]
- Comtesse N, Maldener E, Meese E. Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a β-N-acetylglucosaminidase. Biochem Biophys Res Commun. 2001;283(3):634–640. doi: 10.1006/bbrc.2001.4815. [DOI] [PubMed] [Google Scholar]
- Copeland RJ, Bullen Jr JW, Hart GW. Crosstalk between glconacylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am J Physiol Endocrinol Metab. 2008;295:17–28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho P, Deleury E, Davies G, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, Black GN, Vocadlo DJ, Davies GJ. Structure and mechanism of a bacterial β-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol. 2006;13:365–371. doi: 10.1038/nsmb1079. [DOI] [PubMed] [Google Scholar]
- Dias W, Hart G. O-GlcNAc modification in diabetes and Alzheimer’s disease. Mol Biosyst. 2007;3:766–772. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- Donadio AC, Lobo C, Tosina M, de la Rosa V, Martin-Rufian M, Campos-Sandoval JA, Mates JM, Marquez J, Alonso FJ, Segura JA. Antisense glutaminase inhibition modifies the O-GlcNAc pattern and flux through the hexosamine pathway in breast cancer cells. J Cell Biochem. 2008;103(3):800–811. doi: 10.1002/jcb.21449. [DOI] [PubMed] [Google Scholar]
- Dong DLY, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-β-d-glucosaminidase from rat spleen cytosol. J Biol Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- Dorfmueller HC, Borodkin VS, Schimpl M, Shepherd SM, Shpiro NA, van Aalten DMF. GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-GlcNAcylation levels. J Am Chem Soc. 2006;128:16484–16485. doi: 10.1021/ja066743n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfmueller HC, Borodkin VS, Schimpl M, van Aalten DMF. GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation. Biochem J. 2009;420(2):221–227. doi: 10.1042/BJ20090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ficko-Blean E, Gregg KJ, Adams JJ, Hehemann J-H, Czjzek M, Smith SP, Boraston AB. Portrait of an enzyme, a complete structural analysis of a multimodular beta-N-acetylglucosaminidase from Clostridium perfringens. J Biol Chem. 2009;284(15):9876–9884. doi: 10.1074/jbc.M808954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe ME, Love DC, Lazarus BD, Kim EJ, Prinz WA, Ashwell G, Krause MW, Hanover JA. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: OGA-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer. Proc Natl Acad Sci USA. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins—cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Griffith LS, Schmitz B. O-linked N-acetylglucosamine is upregulated in Alzheimer brains. Biochem Biophys Res Commun. 1995;213(2):424–431. doi: 10.1006/bbrc.1995.2149. [DOI] [PubMed] [Google Scholar]
- Gross BJ, Kraybill BC, Walker S. Discovery of O-GlcNAc transferase inhibitors. J Am Chem Soc. 2005;127:14588–14589. doi: 10.1021/ja0555217. [DOI] [PubMed] [Google Scholar]
- Hajduch J, Nam G, Kim E, Fraehlich R, Hanover J, Kirk K. A convenient synthesis of the C-1-phosphonate analogue of UDP–GlcNAc and its evaluation as an inhibitor of O-linked GlcNAc transferase (OGT) Carbohydr Res. 2008;343:189–195. doi: 10.1016/j.carres.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger R, Blomberg M, Hart G. Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase. J Biol Chem. 1992;267:9005–9013. [PubMed] [Google Scholar]
- Haltiwanger RS, Grove K, Philipsberg GA. Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O-GlcNAc-β-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate. J Biol Chem. 1998;273:3611–3617. doi: 10.1074/jbc.273.6.3611. [DOI] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2009;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- Hurtado-Guerrero R, Dorfmueller HC, van Aalten DMF. Molecular mechanisms of O-GlcNAcylation. Curr Opin Struct Biol. 2008;18(5):551–557. doi: 10.1016/j.sbi.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA. 1996;93(17):9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E-S, Han D, Park J, Kwak TK, Oh M-A, Lee S-A, Choi S, Park ZY, Kim Y, Lee JW. O-GlcNAc modulation at Akt1 Ser473 correlates with apoptosis of murine pancreatic beta cells. Exp Cell Res. 2008;314(11–12):2238–2248. doi: 10.1016/j.yexcr.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Hart GW. Glycosylation of chromosomal proteins: localization of O-linked N-acetylglucosamine in Drosophila chromatin. Cell. 1989;57(2):243–251. doi: 10.1016/0092-8674(89)90962-8. [DOI] [PubMed] [Google Scholar]
- Knapp S, Myers D. Alpha-GlcNAc thioconjugates. J Org Chem. 2001;66:3636–3638. doi: 10.1021/jo010088e. [DOI] [PubMed] [Google Scholar]
- Konrad RJ, Zhang FX, Hale JE, Knierman MD, Becker GW, Kudlow JE. Alloxan is an inhibitor of the enzyme O-linked N-acetylglucosamine transferase. Biochem Biophys Res Commun. 2002;293:207–212. doi: 10.1016/S0006-291X(02)00200-0. [DOI] [PubMed] [Google Scholar]
- Kreppel L, Blomberg M, Hart G. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- Laughlin ST, Bertozzi CR. In vivo imaging of caenorhabditis elegans glycans. ACS Chem Biol. 2009;4(12):1068–1072. doi: 10.1021/cb900254y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherbarrow RJ. GraFit. Version 5. Horley: Erithacus Software Ltd; 2001. [Google Scholar]
- Lenzen S, Munday R. Thiol-group reactivity, hydrophilicity and stability of alloxan, its reduction products and its N-methyl derivatives and a comparison with ninhydrin. Biochem Pharmacol. 1991;42(7):1385–1391. doi: 10.1016/0006-2952(91)90449-F. [DOI] [PubMed] [Google Scholar]
- Liu K, Paterson AJ, Chin E, Kudlow JE. Glucose stimulates protein modification by O-linked GlcNAc in pancreatic β-cells: linkage of O-linked GlcNAc to β-cell death. Proc Natl Acad Sci USA. 2000;97:2820–2825. doi: 10.1073/pnas.97.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Paterson AJ, Konrad RJ, Parlow AF, Jimi S, Roh M, Chin EJ, Kudlow JE. Streptozotocin, an O-GlcNAcase inhibitor, blunts insulin and growth hormone secretion. Mol Cell Endocrinol. 2002;194(1–2):135–146. doi: 10.1016/S0303-7207(02)00155-7. [DOI] [PubMed] [Google Scholar]
- Liu K, Paterson A, Zhang F, McAndrew J, Fukuchi K, Wyss J, Peng L, Hu Y, Kudlow J. Accumulation of protein O-GlcNAc modification inhibits proteasomes in the brain and coincides with neuronal apoptosis in brain areas with high O-GlcNAc metabolism. J Neurochem. 2004;89:1044–1055. doi: 10.1111/j.1471-4159.2004.02389.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Yu Y, Fan YZ, Chang H, Liu HM, Cui Y, Chen Q, Wang R. Cardiovascular effects of endomorphins in alloxan-induced diabetic rats. Peptides. 2005;26(4):607–614. doi: 10.1016/j.peptides.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Liu J, Pang Y, Chang T, Bounelis P, Chatham JC, Marchase RB. Increased hexosamine biosynthesis and protein O-GlcNAc levels associated with myocardial protection against calcium paradox and ischemia. J Mol Cell Cardiol. 2006;40(2):303–312. doi: 10.1016/j.yjmcc.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Liu J, Marchase RB, Chatham JC. Glutamine-induced protection of isolated rat heart from ischemia/reperfusion injury is mediated via the hexosamine biosynthesis pathway and increased protein O-GlcNAc levels. J Mol Cell Cardiol. 2007;42(1):177–185. doi: 10.1016/j.yjmcc.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005;312:1–14. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- Lubas W, Frank D, Krause M, Hanover J. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J Biol Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. O-GlcNAcase uses substrate-assisted catalysis–kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005;280:25313–25322. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- Marlow A, Kiessling L. Improved chemical synthesis of UDP–galactofuranose. Org Lett. 2001;3:2517–2519. doi: 10.1021/ol016170d. [DOI] [PubMed] [Google Scholar]
- Martinez-Fleites C, Macauley M, He Y, Shen D, Vocadlo D, Davies G. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat Struct Mol Biol. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- McClain DA, Lubas WA, Cooksey RC, Hazel M, Parker GJ, Love DC, Hanover JA. Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc Natl Acad Sci USA. 2002;99:10695–10699. doi: 10.1073/pnas.152346899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect F Struct Biol Cryst Commun. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Noach N, Segev Y, Levi I, Segal S, Priel E. Modification of topoisomerase I activity by glucose and by O-GlcNAcylation of the enzyme protein. Glycobiology. 2007;17(12):1357–1364. doi: 10.1093/glycob/cwm105. [DOI] [PubMed] [Google Scholar]
- Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. doi: 10.1107/S0021889804018047. [DOI] [Google Scholar]
- Rao FV, Dorfmueller HC, Villa F, Allwood M, Eggleston IM, van Aalten DMF. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006;25:1569–1578. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Cryst. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Shafi R, Lyer SPN, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the x chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs KA, Zhang N, Vocadlo DJ. A divergent synthesis of 2-acyl derivatives of PUGNAc yields selective inhibitors of O-GlcNAcase. Org Biomol Chem. 2006;4:839–845. doi: 10.1039/b516273d. [DOI] [PubMed] [Google Scholar]
- Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes - evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- Tvaroska I. Molecular modeling insights into the catalytic mechanism of the retaining galactosyltransferase LgtC. Carbohydr Res. 2004;339(5):1007–1014. doi: 10.1016/j.carres.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Wells L, Hart GW. O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 2003;546:154–158. doi: 10.1016/S0014-5793(03)00641-0. [DOI] [PubMed] [Google Scholar]
- Yang W, Kim J, Nam H, Ju J, Kim H, Kim Y, Cho J. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- Yao P, Coleman P. Reduction of O-linked N-acetylglucosamine-modified assembly protein-3 in Alzheimer’s disease. J Neurosci. 1998;18(7):2399–2411. doi: 10.1523/JNEUROSCI.18-07-02399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa S, Macauley M, Heinonen J, Shan X, Dennis R, He Y, Whitworth G, Stubbs K, McEachern E, Davies G, Vocadlo D. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- Zachara NE, Hart GW. O-GlcNAc modification: a nutritional sensor that modulates proteasome function. Trends Cell Biol. 2004;14:218–221. doi: 10.1016/j.tcb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Zachara NE, Cheung WD, Hart GW. Nucleocytoplasmic glycosylation, O-GlcNAc: identification and site mapping. Methods Mol Biol. 2004;284:175–194. doi: 10.1385/1-59259-816-1:175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


