Abstract
Aspergilli have a long history in biotechnology as expression platforms for the production of food ingredients, pharmaceuticals and enzymes. The achievements made during the last years, however, have the potential to revolutionize Aspergillus biotechnology and to assure Aspergillus a dominant place among microbial cell factories. This mini-review will highlight most recent breakthroughs in fundamental and applied Aspergillus research with a focus on new molecular tools, techniques and products. New trends and concepts related to Aspergillus genomics and systems biology will be discussed as well as the challenges that have to be met to integrate omics data with metabolic engineering attempts.
Keywords: Aspergillus, Expression platform, Genomics, Systems biology, Genetic engineering, Organelle engineering
Introduction
There are about 250 named species in the genus Aspergillus (Geiser et al. 2007), including important industrially exploited species (A. niger, A. oryzae, A. awamori, A. sojae, A. terreus) and harmful species being pathogenic to animals and/or plants (e.g. A. fumigatus, A. parasiticus, A. flavus). Due to their extraordinary metabolic versatility, aspergilli are used in biotechnology for the production of a variety of products such as organic acids, pharmaceuticals, proteins and enzymes (for reviews see (Meyer 2008; Lubertozzi and Keasling 2009)). Aspergillus stands out from other microbial cell factories of bacterial or yeast origin due to its ability to tolerate extreme cultivation conditions (Raper and Fennel 1965; Kis-Papo et al. 2003; Machida and Gomi 2010). For example, Aspergillus can be cultivated over a wide range of temperatures (10–50°C), pH (2–11), salinity (0–34%), water activity (0.6–1) and under oligotrophic or nutrient-rich conditions. Hence, Aspergillus can be used for solid-state or submerged fermentations and respective fermentation protocols have been established for large-scale industrial processes. Most importantly, Aspergillus can degrade and utilize diverse biopolymers such as starch, (hemi-)cellulose, pectin, xylan and proteins, allowing it to be cultivated on renewable resources such as plant biomass.
As of 2010, ten genome sequences of the most important industrial and medical Aspergilli are publically accessible, making this genus one of the best to be studied by comparative genome analysis. This resource has spurred multiple research activities, including functional genomics and systems biology attempts aiming at the identification of new leads for strain development and at the understanding of the onset and progression of Aspergillus diseases. The aim of this mini-review is to highlight recent breakthroughs in fundamental and applied Aspergillus research, focusing on new tools, techniques and products and to discuss new trends, concepts and challenges important for future strain development programs in the post-genomic era.
Genetic tools
Although Aspergillus shares with bacterial and yeast cell factories much of their ease of cultivation, the knowledge on how to genetically dissect and engineer industrial Aspergillus strains lagged behind for a long time. However, the past decade of Aspergillus research has been a fascinating period of new discoveries and breakthroughs, which resulted in many new genetic tools and techniques. These include the establishment of (i) efficient genetic transformation systems, (ii) high-throughput gene targeting tools, (iii) expression systems for high level and controlled protein production and (iv) live-imaging techniques for cell biological studies. In the following, some of these development will be touched, however, for more in-depth information, the reader is directed to the following reviews (Meyer 2008; Hickey and Read 2009; Lubertozzi and Keasling 2009; Fleissner and Dersch 2010; Kück and Hoff 2010; Meyer et al. 2010a).
Genetic manipulation
For a long time, genetic manipulation of industrially used Aspergillus strains was hampered due to the lack of efficient genetic transformation systems, limited availability of selection markers and low gene targeting efficiencies (usually around 1–5%). All three problems have been solved during the last decade. Protoplast-mediated transformation became the method of choice to efficiently transform A. niger, A. oryzae, A. sojae and A. terreus, whereas Agrobacterium-mediated transformation is most efficient for A. awamori (reviewed by (Michielse et al. 2005; Meyer 2008)). In addition, versatile selection systems including antibiotic resistance markers (hph, ble, oliC3), auxotrophic markers (pyrG, pyrE, argB, adeA, adeB, niaD, trpC, sC) and nutritional markers (amdS, ptrA) in combination with integrative and autonomously replicating vectors have been developed, offering nowadays highest flexibility for genetic manipulation of industrial Aspergillus strains (Jin et al. 2004; Carvalho et al. 2010a; Fleissner and Dersch 2010; Meyer et al. 2010a). In this context, it is worth emphasizing that the usage of the counter-selectable markers pyrE, pyrG and amdS does fulfill the requirements of the European Union for self-cloning approaches. An interesting alternative option for efficient marker recycling and removal of heterologous DNA sequences makes use of the yeast FLP/FRT recombination system. This system was recently adapted for the penicillin producer Penicillium chrysogenum (Kopke et al. 2010) and is likely to be applicable to Aspergillus as well.
A quantum leap for molecular Aspergillus research was based on a finding made in the fungal model organism Neurospora crassa. Here it was observed that homologous recombination frequencies can increase up to 100%, when the recipient strain is deficient for the non-homologous end joining (NHEJ) pathway (Ninomiya et al. 2004). This finding was confirmed in many aspergilli including A. oryzae, A. sojae and A. niger (Takahashi et al. 2006; Meyer et al. 2007; Mizutani et al. 2008; Carvalho et al. 2010a) and many recipient strains are now available in which the NHEJ pathway is permanently or transiently inactivated (reviewed in (Meyer 2008; Kück and Hoff 2010)). This discovery made genome-wide deletion projects feasible, the first of which was launched in 2007 for N. crassa (www.dartmouth.edu/~neurosporagenome/) followed by the A. nidulans project in 2010 (www.fgsc.net/Aspergillus/KO_Cassettes.htm).
Expression systems
A comprehensive review on different expression system for recombinant protein production in Aspergillus was most recently given (Fleissner and Dersch 2010). About two dozen Aspergillus promoters are available for high-yield production of homologous and heterologous proteins. The promoters used are either constitutively active (e.g. the glyceraldehyde-3-phosphate dehydrogenase promoter PgpdA) or are inducible based on the carbon source present in the fermentation medium (e.g. the glucoamylase promoter PglaA, the alcohol dehydrogenase promoter PalcA, the α-amylase promoter PamyA). However, none of the available promoters allows tight, tunable and carbon source-independent expression control, posing limitations in their use. To overcome this problem, an artificial gene expression system based on the Escherichia coli tetracycline-resistance operon (Tet-On/Tet-Off system) was adopted for use in A. fumigatus (Vogt et al. 2005). The Tet-On system was further refined for use in A. niger and systematically evaluated (Meyer et al. 2010b). The data obtained showed that the Tet-On system is tight under non-induced conditions, is able to respond within minutes after inducer addition and allows tunable gene control in a gene dosage and/or inducer concentration dependent manner. Most importantly, the strength of the A. niger Tet-On system can compete with the gpdA promoter, making it as a very promising tool for future protein overproductions in Aspergillus.
Omics tools
The sequencing of fungal genomes led Aspergillus research into a new era referred to as systems biology. Instead of studying one gene or protein at a time, the complete set of genes, proteins and/or metabolites are studied simultaneously, aiming at a comprehensive understanding of the robustness, viability and productivity of industrial Aspergillus strains and at the identification of factors important for pathogenicity of medical Aspergilli. For each of the omics subdisciplines (transcriptomics, proteomics, metabolomics) a dedicated set of equipment, techniques and software tools has to be developed, a process which is still on-going for many Aspergillus species. For the whole genus, only about 30 omics studies appear yearly in Pubmed (Fig. 1), illustrating that Aspergillus systems biology is still in its infancy. Among these, the following publications on industrially used Aspergilli are worth highlighting: (i) a metabolic network was established for A. niger and A. oryzae based on genomic databases and extensive literature surveys (Andersen et al. 2008; Vongsangnak et al. 2008). (Note that a web-based pathway viewer has recently been launched for A. fumigatus (Tuckwell et al. 2010.) (ii) The starch-, pectin- and inulin-degrading enzyme network of A. niger was identified using a combined database mining and transcriptomic approach (Martens-Uzunova et al. 2006; Yuan et al. 2006, 2008; Martens-Uzunova and Schaap 2009). (iii) An integrated transcriptomics and proteomics approach identified new leads for improved protein secretion of A. niger (Guillemette et al. 2007; Levin et al. 2007; Jacobs et al. 2009). (iv) A transcriptomics and genetic study predicted regulatory networks important for the morphological control of A. niger growth (Meyer et al. 2009). (v) A transcriptomics and metabolomics study disclosed regulatory genes involved in lovastatin biosynthesis in A. terreus (Askenazi et al. 2003). (vi) A transcriptome analysis identified genes important for high-yield solid-phase fermentations in A. oryzae (Tamano et al. 2008).
Fig. 1.
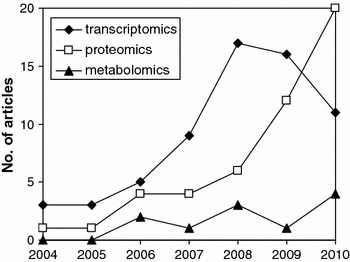
Number of Aspergillus omics articles published over the last years in Pubmed (by October 2010)
Key for systems biology attempts is the availability of well-annotated genome sequences, standardized analysis tools (e.g. microarray platforms, RNA sequencing technologies, high-throughput chromatographic separations and mass spectrometry tools) and suitably configured databases for data storage and network reconstruction. Most recently, the open access online tool ‘BioMet toolbox’ has been launched (www.sysbio.se/BioMet/) which allows integration and systematic analysis of omics data from A. niger and A. oryzae (Cvijovic et al. 2010). A major challenge in systems biology approaches is the fact that the data gained has varying levels of reliability. Data-sets can be error prone due to technical irreproducibility, e.g. cultivations of Aspergillus performed in shake flasks are not reproducible due to continuous changes in pH, temperature, oxygen availability and eventually in morphology. In contrast, cultivations in lab-scale bioreactors allow on-line control on these parameters and thus ensure highest reproducibility (Jorgensen et al. 2010; Meyer et al. 2010a). Besides technical problems, biological variations caused by heterogeneous gene expression in Aspergillus populations (Vinck et al. 2005; Levin et al. 2007) will introduce errors as well. Hence, the application of statistical procedures and bioinformatics pipelines for handling and modeling errors as well as internal controls are crucial to gain legitimate interpretations out from high-throughput omics data.
Strain developments
A. niger, A. oryzae and A. awamori are well-established expression platforms for the production of secreted proteins and enzymes (Punt et al. 2002). However, their productivities are limited due to several reasons. First, the protein of interest can become degraded by secreted host proteases, the number of which was estimated to be 200 in A. niger (Pel et al. 2007). To tailor the host protease profile, several approaches have been followed, including deletion or silencing of selected protease genes in A. oryzae or A. awamori (Moralejo et al. 2002; Yoon et al. 2009) and deletion of the protease-specific transcription factor PrtT in A. niger and A. oryzae (Punt et al. 2008). Second, low yields are often observed when proteins of non-fungal origin have to be expressed. It is thought that this results from intracellular misfolding of the target proteins, an event, which initiates compensatory responses in Aspergillus such as ERAD (ER-associated degradation; sensing and removal of misfolded proteins via the proteasome) and UPR (unfolded protein response; degradation or refolding of unfolded proteins). To get insights into the genetic repertoire securing protein secretion, ERAD and UPR, transcriptomics and functional genomics approaches have recently been undertaken in A. niger (Guillemette et al. 2007; Jorgensen et al. 2009; Carvalho et al. 2010b) and A. oryzae (Wang et al. 2010), to name but a few. In order to tackle intracellular degradation of the target protein, the ‘carrier approach’ is one option, where the protein of interest is genetically fused to an abundantly secreted host protein (Punt et al. 2002). An additional promising approach is the optimisation of the codon usage of the gene of interest according to the preferred codon-usage of the expression host (Roubos and van Peij 2008). Finally, protein engineering approaches aiming at increased protein stabilities hold promise as well (Kumita et al. 2006).
A third, long-lasting and still unsolved problem is related to the morphology of Aspergillus. As with all filamentous fungi, either freely dispersed filaments or highly compact pellets can be formed during submerged fermentations, both of which do limit process productivity (Papagianni 2004; Grimm et al. 2005). So far, the formation of these macroscopic morphologies can only descriptively be monitored but not controlled. To overcome this constraint and to optimize the morphology by rational genetic engineering, transcriptomics and functional genomics studies have recently been initiated aiming at the reconstruction of the morphogenetic machinery of A. niger (Meyer et al. 2008, 2009).
New products in the post-genomic era
The availability of Aspergillus genome sequences has triggered an area of research called ‘genome mining’ which assumes that Aspergillus can be a source of new products such as therapeutics or drugs. Indeed, genome analyses disclosed more genes involved in secondary metabolite (SM) synthesis than anticipated. The respective pathway genes are conveniently arranged in gene clusters (30–40 clusters are typically found in Aspergilli), and have been predicted to be involved in the synthesis of various polyketides or non-ribosomal peptides (Brakhage and Schroeckh 2010). To facilitate the systematic mining and mapping of these clusters, the web-based tool SMURF (Secondary Metabolite Unknown Regions Finder; www.jcvi.org/smurf/) has recently been launched as open access bioinformatics tool (Khaldi et al. 2010). However, most SM clusters are silent or cryptic, i.e. are not expressed under laboratory or industrial conditions (Pel et al. 2007). To activate expression of silent clusters and to identify the metabolite(s) produced, different strategies based on molecular, epigenetics and cultivation methods have been undertaken in Aspergillus and recently reviewed (Chiang et al. 2009; Brakhage and Schroeckh 2010). Among these, four approaches stand out. First, it was reported that controlled activation of a pathway-specific transcription factor in the model A. nidulans activated a silent metabolic pathway, as a result of which two novel SM were identified (Bergmann et al. 2007). Second, targeted manipulation of the chromatin state, i.e. removal of a chromatin silencing gene, activated expression of at least two other gene clusters (Bok et al. 2009). Thirdly, it was shown that an intimate physical interaction during fungal-bacterial co-cultivations is the prerequisite for selective activation of another SM cluster (Schroeckh et al. 2009). And finally, cultivation at very low specific growth rates, which almost reached zero-growth conditions, induced expression of SM clusters in A. niger (Jorgensen et al. 2010). Hence, multiple approaches can be followed or combined to fully harness the SM potential of Aspergilli.
Lubertozzi and Keasling (2009) list in their comprehensive review many more potential natural Aspergillus products with commercial relevance, an impressive list that emphasises the great potential of Aspergillus to become a multi-purpose expression platform. Besides polyketides and non-ribosomal peptides as potential therapeutic agents, isoprenoids are interesting as new neutraceuticals or aroma compounds, lipopeptides and AFP-derived proteins as new antifungals and poly-unsaturated fatty acids or lipids as food additives or fuel feedstocks. Moreover, the future product spectrum of industrial Aspergillus strains is not restricted to its natural genetic repertoire. The new genetic engineering toolkit for Aspergillus and the advent of synthetic biology open new avenues to many more compounds of (non)fungal origin to be expressed and produced in Aspergillus.
New concepts and perspectives
To fully explore and exploit Aspergillus as a multi-purpose expression platform, conceptually new approaches will be followed, some of which have the potential to revolutionize Aspergillus biotechnology. It is likely that newly engineered production strains will be completely devoid of unwanted SM gene clusters and used as chassis for the production of food and feed ingredients, enzymes, pharmaceuticals or even fuel feedstocks. The gene(s) or gene cluster(s) of interest will be of Aspergillus or (non)fungal origin and polished according to the Aspergillus codon-bias. Expression constructs will be plugged into selected genomic loci allowing highest control over gene expression. The gene(s) of interest will be expressed under the control of constitutive or inducible promoters, depending on the toxicity of the final product. The product repertoire, the timing of product formation and productivity will be optimised using different cultivation strategies. For example, fermentations under zero-growth conditions can uncouple growth from product formation and have the potential to improve SM production (Jorgensen et al. 2010).
In addition, vesicle and organelle engineering approaches offer exciting opportunities. For example, intracellular proteins can become secreted via ‘peroxicretion’, an artificial route where proteins become targeted to peroxisomes which themselves have genetically been decorated with v-SNARES. They ensure transport of the peroxisomes to the plasma membrane, where the protein cargo becomes released into the medium (Sagt et al. 2009). Moreover, SM production can be improved by optimizing the volume fraction of organelles in which (part of) SM biosynthesis takes place. A proof-of-concept was recently shown for penicillin synthesis in Penicillium chrysogenum, where artificially induced proliferation of peroxisomes resulted in increased penicillin titres (Kiel et al. 2005; Meijer et al. 2010). Most interestingly, natural product formation in Aspergillus can also be dependent on the circadian rhythm—not only in the case for SM as known for a long time (Brakhage and Schroeckh 2010), but likely also in the case of secreted enzymes as shown for cellulase production in the industrial fungus Trichoderma reesei (Castellanos et al. 2010). Hence, understanding circadian regulatory mechanisms might also give new leads for genetic engineering strategies.
What is pivotal to successfully merge Aspergillus biotechnology, systems biology tools and metabolic engineering attempts? Controlled cultivation and harvesting conditions for Aspergillus as well as standardised methodological and bioinformatics tools are crucial to deduce accurate and meaningful conclusions out from transcriptome, proteome and metabolome data. Genome-wide metabolic models and metabolic flux analyses are key for the identification of pathway limitations and predicting beneficial metabolic engineering strategies. However, to fully describe and engineer cellular phenomena in Aspergillus, the implementation of cell biological studies such as in vivo live imaging are required. Only the understanding of compartmentalized product biosynthesis as well as transport and traffic phenomena will uncover the link between morphology, secretion and product formation in Aspergillus and will eventually give significant leads for redesigning this versatile expression host.
Acknowledgments
The authors would like to acknowledge China’s Scholarship Council (CSC) for supporting the research stay of BW in our lab. This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Andersen MR, Nielsen ML, Nielsen J. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol Syst Biol. 2008;4:178. doi: 10.1038/msb.2008.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askenazi M, Driggers EM, Holtzman DA, Norman TC, Iverson S, Zimmer DP, Boers ME, Blomquist PR, Martinez EJ, Monreal AW, Feibelman TP, Mayorga ME, Maxon ME, Sykes K, Tobin JV, Cordero E, Salama SR, Trueheart J, Royer JC, Madden KT. Integrating transcriptional and metabolite profiles to direct the engineering of lovastatin-producing fungal strains. Nat Biotechnol. 2003;21:150–156. doi: 10.1038/nbt781. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA, Schroeckh V (2010) Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet Biol. Epub ahead of print [DOI] [PubMed]
- Carvalho ND, Arentshorst M, Jin Kwon M, Meyer V, Ram AF. Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl Microbiol Biotechnol. 2010;87:1463–1473. doi: 10.1007/s00253-010-2588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho ND, Arentshorst M, Kooistra R, Stam H, Sagt CM, van den Hondel CA, Ram AF (2010b) Effects of a defective ERAD pathway on growth and heterologous protein production in Aspergillus niger. Appl Microbiol Biotechnol. Epub ahead of print [DOI] [PMC free article] [PubMed]
- Castellanos F, Schmoll M, Martinez P, Tisch D, Kubicek CP, Herrera-Estrella A, Esquivel-Naranjo EU. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei. Fungal Genet Biol. 2010;47:468–476. doi: 10.1016/j.fgb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Chiang YM, Lee KH, Sanchez JF, Keller NP, Wang CC. Unlocking fungal cryptic natural products. Nat Prod Commun. 2009;4:1505–1510. [PMC free article] [PubMed] [Google Scholar]
- Cvijovic M, Olivares-Hernandez R, Agren R, Dahr N, Vongsangnak W, Nookaew I, Patil KR, Nielsen J. BioMet Toolbox: genome-wide analysis of metabolism. Nucleic Acids Res. 2010;38(Suppl):W144–W149. doi: 10.1093/nar/gkq404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner A, Dersch P. Expression and export: recombinant protein production systems for Aspergillus. Appl Microbiol Biotechnol. 2010;87:1255–1270. doi: 10.1007/s00253-010-2672-6. [DOI] [PubMed] [Google Scholar]
- Geiser DM, Klich MA, Frisvad JC, Peterson SW, Varga J, Samson RA. The current status of species recognition and identification in Aspergillus. Stud Mycol. 2007;59:1–10. doi: 10.3114/sim.2007.59.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm LH, Kelly S, Krull R, Hempel DC. Morphology and productivity of filamentous fungi. Appl Microbiol Biotechnol. 2005;69:375–384. doi: 10.1007/s00253-005-0213-5. [DOI] [PubMed] [Google Scholar]
- Guillemette T, van Peij NN, Goosen T, Lanthaler K, Robson GD, van den Hondel CA, Stam H, Archer DB. Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics. 2007;8:158. doi: 10.1186/1471-2164-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey PC, Read ND. Imaging living cells of Aspergillus in vitro. Med Mycol. 2009;47(Suppl 1):S110–S119. doi: 10.1080/13693780802546541. [DOI] [PubMed] [Google Scholar]
- Jacobs DI, Olsthoorn MM, Maillet I, Akeroyd M, Breestraat S, Donkers S, van der Hoeven RA, van den Hondel CA, Kooistra R, Lapointe T, Menke H, Meulenberg R, Misset M, Muller WH, van Peij NN, Ram A, Rodriguez S, Roelofs MS, Roubos JA, van Tilborg MW, Verkleij AJ, Pel HJ, Stam H, Sagt CM. Effective lead selection for improved protein production in Aspergillus niger based on integrated genomics. Fungal Genet Biol. 2009;46(Suppl 1):S141–S152. doi: 10.1016/j.fgb.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett. 2004;239:79–85. doi: 10.1016/j.femsle.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Jorgensen TR, Goosen T, Hondel CA, Ram AF, Iversen JJ. Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway. BMC Genomics. 2009;10:44. doi: 10.1186/1471-2164-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen TR, Nitsche BM, Lamers GE, Arentshorst M, van den Hondel CA, Ram AF. Transcriptomic insights into the physiology of Aspergillus niger approaching a specific growth rate of zero. Appl Environ Microbiol. 2010;76:5344–5355. doi: 10.1128/AEM.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel JA, van der Klei IJ, van den Berg MA, Bovenberg RA, Veenhuis M. Overproduction of a single protein, Pc-Pex11p, results in 2-fold enhanced penicillin production by Penicillium chrysogenum. Fungal Genet Biol. 2005;42:154–164. doi: 10.1016/j.fgb.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Kis-Papo T, Oren A, Wasser SP, Nevo E. Survival of filamentous fungi in hypersaline Dead Sea water. Microb Ecol. 2003;45:183–190. doi: 10.1007/s00248-002-3006-8. [DOI] [PubMed] [Google Scholar]
- Kopke K, Hoff B, Kuck U. Application of the Saccharomyces cerevisiae FLP/FRT recombination system in filamentous fungi for marker recycling and construction of knockout strains devoid of heterologous genes. Appl Environ Microbiol. 2010;76:4664–4674. doi: 10.1128/AEM.00670-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück U, Hoff B. New tools for the genetic manipulation of filamentous fungi. Appl Microbiol Biotechnol. 2010;86:51–62. doi: 10.1007/s00253-009-2416-7. [DOI] [PubMed] [Google Scholar]
- Kumita JR, Johnson RJ, Alcocer MJ, Dumoulin M, Holmqvist F, McCammon MG, Robinson CV, Archer DB, Dobson CM. Impact of the native-state stability of human lysozyme variants on protein secretion by Pichia pastoris. FEBS J. 2006;273:711–720. doi: 10.1111/j.1742-4658.2005.05099.x. [DOI] [PubMed] [Google Scholar]
- Levin AM, de Vries RP, Conesa A, de Bekker C, Talon M, Menke HH, van Peij NN, Wosten HA. Spatial differentiation in the vegetative mycelium of Aspergillus niger. Eukaryot Cell. 2007;6:2311–2322. doi: 10.1128/EC.00244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubertozzi D, Keasling JD. Developing Aspergillus as a host for heterologous expression. Biotechnol Adv. 2009;27:53–75. doi: 10.1016/j.biotechadv.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Machida M, Gomi K (eds) (2010) Aspergillus: molecular biology and genomics. Caister Academic Press
- Martens-Uzunova ES, Schaap PJ. Assessment of the pectin degrading enzyme network of Aspergillus niger by functional genomics. Fungal Genet Biol. 2009;46(Suppl 1):S170–S179. doi: 10.1016/j.fgb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Martens-Uzunova ES, Zandleven JS, Benen JA, Awad H, Kools HJ, Beldman G, Voragen AG, Van den Berg JA, Schaap PJ. A new group of exo-acting family 28 glycoside hydrolases of Aspergillus niger that are involved in pectin degradation. Biochem J. 2006;400:43–52. doi: 10.1042/BJ20060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer WH, Gidijala L, Fekken S, Kiel JA, van den Berg MA, Lascaris R, Bovenberg RA, van der Klei IJ. Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum. Appl Environ Microbiol. 2010;76:5702–5709. doi: 10.1128/AEM.02327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer V. Genetic engineering of filamentous fungi—progress, obstacles and future trends. Biotechnol Adv. 2008;26:177–185. doi: 10.1016/j.biotechadv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Meyer V, Arentshorst M, El-Ghezal A, Drews AC, Kooistra R, van den Hondel CA, Ram AF. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J Biotechnol. 2007;128:770–775. doi: 10.1016/j.jbiotec.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Meyer V, Arentshorst M, van den Hondel CA, Ram AF. The polarisome component SpaA localises to hyphal tips of Aspergillus niger and is important for polar growth. Fungal Genet Biol. 2008;45:152–164. doi: 10.1016/j.fgb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Meyer V, Arentshorst M, Flitter SJ, Nitsche BM, Kwon MJ, Reynaga-Pena CG, Bartnicki-Garcia S, van den Hondel CA, Ram AF. Reconstruction of signaling networks regulating fungal morphogenesis by transcriptomics. Eukaryot Cell. 2009;8:1677–1691. doi: 10.1128/EC.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer V, Ram AF, Punt PJ. Genetics, genetic manipulation, and approaches to strain improvement of filamentous fungi. In: Demain AL, Davis J, editors. Manual of industrial microbiology and biotechnology. New York: Wiley; 2010. pp. 318–329. [Google Scholar]
- Meyer V, Wanka F, van Gent J, Arentshorst M, van den Hondel CA, Ram AF (2010b) Fungal gene expression on demand: An inducible, tunable and metabolism-independent expression system for Aspergillus niger. submitted [DOI] [PMC free article] [PubMed]
- Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet. 2005;48:1–17. doi: 10.1007/s00294-005-0578-0. [DOI] [PubMed] [Google Scholar]
- Mizutani O, Kudo Y, Saito A, Matsuura T, Inoue H, Abe K, Gomi K. A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet Biol. 2008;45:878–889. doi: 10.1016/j.fgb.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Moralejo FJ, Cardoza RE, Gutierrez S, Lombrana M, Fierro F, Martin JF. Silencing of the aspergillopepsin B (pepB) gene of Aspergillus awamori by antisense RNA expression or protease removal by gene disruption results in a large increase in thaumatin production. Appl Environ Microbiol. 2002;68:3550–3559. doi: 10.1128/AEM.68.7.3550-3559.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagianni M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol Adv. 2004;22:189–259. doi: 10.1016/j.biotechadv.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d’Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wosten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Punt PJ, van Biezen N, Conesa A, Albers A, Mangnus J, van den Hondel C. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 2002;20:200–206. doi: 10.1016/S0167-7799(02)01933-9. [DOI] [PubMed] [Google Scholar]
- Punt PJ, Schuren FH, Lehmbeck J, Christensen T, Hjort C, van den Hondel CA. Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet Biol. 2008;45:1591–1599. doi: 10.1016/j.fgb.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Raper KD, Fennel DI, editors. The genus Aspergillus. Baltimore: Williams & Williams; 1965. [Google Scholar]
- Roubos JA, van Peij NN. A method for achieving improved polypeptide expression. WO/2008/000632. Netherlands: DSM; 2008. [Google Scholar]
- Sagt CM, ten Haaft PJ, Minneboo IM, Hartog MP, Damveld RA, van der Laan JM, Akeroyd M, Wenzel TJ, Luesken FA, Veenhuis M, van der Klei I, de Winde JH. Peroxicretion: a novel secretion pathway in the eukaryotic cell. BMC Biotechnol. 2009;9:48. doi: 10.1186/1472-6750-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Masuda T, Koyama Y. Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol Genet Genomics. 2006;275:460–470. doi: 10.1007/s00438-006-0104-1. [DOI] [PubMed] [Google Scholar]
- Tamano K, Sano M, Yamane N, Terabayashi Y, Toda T, Sunagawa M, Koike H, Hatamoto O, Umitsuki G, Takahashi T, Koyama Y, Asai R, Abe K, Machida M. Transcriptional regulation of genes on the non-syntenic blocks of Aspergillus oryzae and its functional relationship to solid-state cultivation. Fungal Genet Biol. 2008;45:139–151. doi: 10.1016/j.fgb.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Tuckwell D, Denning DW, Bowyer P (2010) A public resource for metabolic pathway mapping of Aspergillus fumigatus Af293. Med Mycol. Epub ahead of print [DOI] [PubMed]
- Vinck A, Terlou M, Pestman WR, Martens EP, Ram AF, van den Hondel CA, Wosten HA. Hyphal differentiation in the exploring mycelium of Aspergillus niger. Mol Microbiol. 2005;58:693–699. doi: 10.1111/j.1365-2958.2005.04869.x. [DOI] [PubMed] [Google Scholar]
- Vogt K, Bhabhra R, Rhodes JC, Askew DS. Doxycycline-regulated gene expression in the opportunistic fungal pathogen Aspergillus fumigatus. BMC Microbiol. 2005;5:1. doi: 10.1186/1471-2180-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongsangnak W, Olsen P, Hansen K, Krogsgaard S, Nielsen J. Improved annotation through genome-scale metabolic modeling of Aspergillus oryzae. BMC Genomics. 2008;9:245. doi: 10.1186/1471-2164-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Guo G, Wang C, Lin Y, Wang X, Zhao M, Guo Y, He M, Zhang Y, Pan L. Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic Acids Res. 2010;38:5075–5087. doi: 10.1093/nar/gkq256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Kimura S, Maruyama J, Kitamoto K. Construction of quintuple protease gene disruptant for heterologous protein production in Aspergillus oryzae. Appl Microbiol Biotechnol. 2009;82:691–701. doi: 10.1007/s00253-008-1815-5. [DOI] [PubMed] [Google Scholar]
- Yuan XL, Goosen C, Kools H, van der Maarel MJ, van den Hondel CA, Dijkhuizen L, Ram AF. Database mining and transcriptional analysis of genes encoding inulin-modifying enzymes of Aspergillus niger. Microbiology. 2006;152:3061–3073. doi: 10.1099/mic.0.29051-0. [DOI] [PubMed] [Google Scholar]
- Yuan XL, van der Kaaij RM, van den Hondel CA, Punt PJ, van der Maarel MJ, Dijkhuizen L, Ram AF. Aspergillus niger genome-wide analysis reveals a large number of novel alpha-glucan acting enzymes with unexpected expression profiles. Mol Genet Genomics. 2008;279:545–561. doi: 10.1007/s00438-008-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


