Abstract
Purpose
This article provides a review of the transcriptomic expression profiling studies that have been performed on meningiomas so far. We discuss some future prospects and challenges ahead in the field of gene expression profiling.
Methods
We performed a systematic search in the PubMed and EMBASE databases in May 2010 using the following search terms alone or in combination: “meningioma”, “microarray analysis”, “oligonucleotide array sequence analysis”, or “gene expression profiling”. Only original research articles in English that had used RNA hybridized to high-resolution microarray chips to generate gene expression profiles were included.
Results
We identified 13 articles matching the inclusion criteria. All studies had been performed during the last decade.
Conclusions
The main results of the studies can be grouped in three categories: (1) several groups have identified meningioma-specific genes and genes associated with the three WHO grades, and the main histological subtypes of grade I meningiomas; (2) one publication has shown that the general transcription profile of samples of all WHO grades differs in vivo and in vitro; (3) one report provides evidence that microarray technology can be used in an automated fashion to classify tumors. Due to lack of consensus on how microarray data are presented, possible general trends found across the studies are difficult to extract. This could obstruct the discovery of important genes and pathways universally involved in meningioma biology.
Keywords: Meningioma, Microarray analysis, Oligonucleotide array sequence analysis, Gene expression profiling
Introduction
Medicine has been transformed by the genomic revolution, and classical population risk assessment and empirical treatment challenged by molecular classification and the concept of personalized therapy [1]. With high-throughput molecular genetic analyses, genes and pathways associated with, e.g., clinical progression, response to irradiation or drugs, and environmental exposure, can be discovered [2]. Thus, new biomarkers and therapeutic targets may be developed. During the last decade, microarray technology has been implemented in molecular biological laboratories world-wide, and numerous studies have been performed in a wide range of human tissues and conditions.
Microarray chips of the size of histology slides including all known human genes are currently manufactured with high precision. The main methods for the production of such chips are based on robotic spotting of cDNA probes or oligonucleotides to a surface [3] (Fig. 1). By labeling the RNA in the samples at study and hybridization to the corresponding gene probes on the microarray chip it is subsequently possible to measure the abundance of every gene expressed at time of tissue sampling. The measured signals are then converted to numerical values and interpreted with bioinformatic tools to produce the gene expression profile.
Fig. 1.
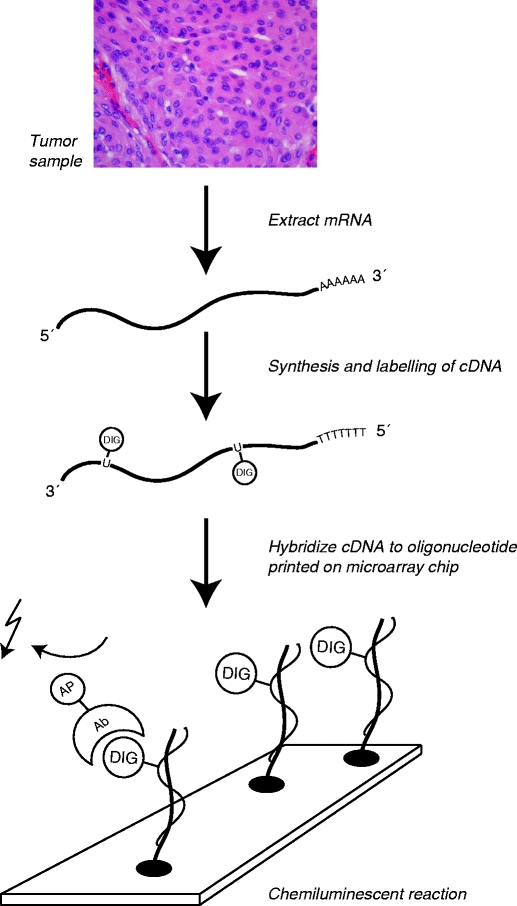
Principle behind microarray analysis (Human Genome Microarray, Applied Biosystems). mRNA extracted from a biological sample is labeled and converted into cDNA. On a microarray slide (“chip”), thousands of oligonucleotides representing individual genes are positioned in predefined positions. If the corresponding cDNA is present, it will hybridize to the oligonucleotide. By a chemiluminescent reaction, the presence of cDNA can be measured
Meningiomas (Fig. 2), being the second most common intracranial neoplasm (20–30% of all cases) [4], have been extensively studied in the past. An important contribution to the understanding of the pathogenesis was the identification of the NF2 gene located on chromosome 22q12.2 [5–8]. Loss of one allele of this gene is causing the autosomal dominant syndrome neurofibromatosis type 2, in which bilateral vestibular schwannomas are pathognomonic, and multiple meningiomas often develop. From research on sporadic (non-NF2-related) meningiomas it has been found that loss of heterozygosity is found in 40–70% of cases and mutations in the remaining allele present in 60% [9–12]. Thus, a main mechanism of meningioma initiation follows the pattern of Knudsons two-hit hypothesis: first, a risk allele is deleted, and then a mutation in the remaining allele containing a tumor suppressor gene initiates neoplastic growth [13]. However, it follows that this mechanism is not uniformly causing meningiomas, and that other genes or pathways are contributing to the tumorigenesis. Hence, gene expression profiling studies have the potential to discover novel genes and signaling pathways with a role in meningioma biology.
Fig. 2.
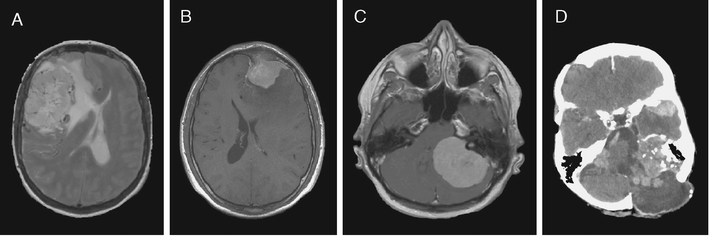
Radiological appearances of meningiomas. a Magnetic resonance imaging (MRI) showing a large convexity meningioma with edema and displacement of the midline to the anatomical left. b MRI section illustrating a WHO grade I convexity meningioma invading the frontal bone. c Large meningioma arising in the cerebello-pontine angle compressing the brain stem. d Computerized tomography showing multiple meningiomas and postoperative changes in a patient with neurofibromatosis type 2
The aim behind this article is to provide a review of the mRNA microarray studies that have been performed on meningiomas, and to reflect the advances in the biological understanding of the tumors gained by this technology. We also address some future prospects and challenges ahead in the field of gene expression profiling.
Material and methods
We performed a systematic search in the PubMed and EMBASE databases in May 2010 with the following medical subject headings (MeSH) terms: “meningioma”, “microarray analysis”, “oligonucleotide array sequence analysis”, “gene expression profiling” (Table 1). Upon literature review only original research articles in English that had used RNA hybridized to high-resolution microarray chips to generate gene expression profiles were included.
Table 1.
Search terms used to identify RNA microarray studies performed on human meningioma samples in the PubMed (http://www.ncbi.nlm.nih.gov/) and EMBASE (http://www.embase.com/) databases
| Search term no. | Search term | PubMed hits | EMBASE hits |
|---|---|---|---|
| 1 | “Meningioma and microarray analysis” | 44 | 14 |
| 2 | “Meningioma and oligonucleotide array sequence analysis” | 37 | 0 |
| 3 | “Meningioma and gene expression profiling” | 70 | 27 |
Results
Based on our search criteria we detected a total of 70 studies. There were no additional studies found in EMBASE to those located in PubMed. All microarray studies found by search term 1 were also found with search term 3. With search term 2, one additional study was identified; however, it was excluded since it was a review article. With search term 3, we found one additional RNA-based microarray study that was included. However, many of the studies were based on quantitative reverse transcriptase real time polymerase chain reaction (qRT-PCR) for selected genes with an a priori hypothesis. Other excluded studies were proteomic profiling projects, tissue arrays, review articles, and non-human material. We identified 13 original research articles that analyzed meningioma RNA with microarray chip technology (Table 2). All studies have been performed during the last decade.
Table 2.
mRNA microarray studies on human meningiomas identified with our search strategy
| Year | Author | Microarray platform | No. of transcripts | No. of meningiomas | Distribution WHO grades | Source of control | No. of controls | Main results | Selected interesting molecules or pathways |
|---|---|---|---|---|---|---|---|---|---|
| 2002 | Watson et al. [14] | Affymetrix HCG110 | 2,059 | 15 | 6 grade I | Meninges | 3 | Gene expression profiles associated with WHO grade subtypes | Growth hormone receptors, endothelin receptor A, IGF2, IGFBP-7 |
| 6 grade II | |||||||||
| 3 grade III | |||||||||
| 2003 | Sasaki et al. [15] | Affymetrix U95A | 12,000 | 3 | 1 grade I | Primary cultures of the samples | 3 | Differing transcriptomic profiles between original frozen meningioma samples and primary cultures | Wnt, Notch, PI3K, MAP kinase |
| 1 grade II | |||||||||
| 1 grade III | |||||||||
| 2003 | Fathallah-Shaykh et al. [16] | 19 K gene microarrays (Ontario Cancer Institute) | 19,200 | 10 | Not declared | Human brain | 4 | Differential gene expression between meningiomas and control human brain | PRKDC, PSIP2, NCOR2, TAF13 |
| 2005 | Wrobel et al. [17] | Custom made cDNA microarray | 4,211 | 30 | 3 grade I1 | Human RNA pool | Pool of 8 different tissues | Meningioma signature genes and differences in gene expression according to WHO grade | PTGDS, CLU, BAD, MGP, LIG1, ANXA2, MMP12, VIM, TIMP1, CCND1 |
| 12 grade II | |||||||||
| 5 grade III | |||||||||
| 2005 | Lusis et al. [18] | Affymetrix U133A and U133B | 39,000 | 10 | Not declared | – | – | Loss of transcripts in grade III meningiomas | NDRG2 |
| 2006 | Sayagues et al. [19] | Affymetrix U133A | 18,400 | 7 spinal and 11 intracranial | Only WHO grade I | – | – | Differential gene expression between intracranial and spinal meningiomas | NR4A3, DUSP5, HOXA5 |
| 2007 | Carvalho et al. [20] | Affymetrix U133 Plus 2.0 | 47,400 | 23 | 8 grade I | – | – | Differential gene expression between grade I and III meningiomas | TGF-ß signaling pathway |
| 7 grade II | |||||||||
| 8 grade III | |||||||||
| 2008 | Aarhus et al. [21] | Applied Biosystems 1700 | 30,000 | 27 | 22 grade I | Arachnoid cyst membranes | 4 | Differential gene expression between arachnoidal cells and meningiomas, and between meningothelial and fibrous meningiomas. | WWOX, TYMS, BMPR1B, RAMP1, DMD |
| 5 grade II | |||||||||
| 2008 | Lillehei et al. [22] | Affymetrix U133 Plus 2.0 | 47,400 | 5 radiation induced and 6 sporadic | Not declared | Pediatric nervous system tumors | 62 | Differential gene expression between sporadic and radiation-induced meningiomas | RGS3, ZNF564, GYPA, CRHR1, MST1R, INPP5B |
| 2008 | Hankins et al. [23] | Affymetrix U95A | 12,000 | 6 | 6 grade I | Dura | 4 | Differential gene expression between meningiomas and dura | DLC1 |
| 2008 | Claus et al. [24] | Affymetrix U133 plus 2.0 | 47,400 | 31 | 31 grade I | – | – | Differential gene expression between progesterone receptor positive and negative meningiomas | SLC24A3, SERPINB1, PDGFRB, MAP4K4, FAT4 |
| 2009 | Fevre-Montange et al. [25] | CodeLink Uniset Human Whole Genome Bioarrays | 55,000 | 17 | 10 grade I | Human brain | 1 | Differential gene expression between fibrous and meningothelial meningiomas, and between WHO grades | CD44, LOX, CKS2, BIRC5, Notch1, MMP2, TIMP3, FAT3 |
| 6 grade II | |||||||||
| 7 grade III | |||||||||
| 2009 | Castells et al. [26] | The human CNIO oncochip 12 K cDNA microarray | 11,500 | 18 | 18 grade I | GBMa | 17 | Automated prediction of brain tumor biopsies | GFAP, DSP |
GBM glioblastoma multiforme
Microarray platforms
Six different microarray platforms had been used in the studies, with technology from Affymetrix being most frequently used (eight of 13 studies). Three of the studies used chips that were spotted with cDNA clones; the remaining used oligonucleotide microarrays. A high number of transcripts were represented on the chips; however, some of the early works included relatively few gene probes due to technical limitations at the time of study.
Meningioma subtypes
Altogether, 227 meningiomas were included in the studies; according to the WHO classification these were 151 grade I, 42 grade II, and 34 grade III. Some papers did not describe the histological subtypes present in their material; however, from the studies with histology data available we found the following subtypes present among the grade I tumors: fibrous (n = 31), meningothelial (48), transitional (n = 27), secretory (n = 1), and psammomatous (n = 1). Apart from two cases of clear cell meningiomas (WHO grade II), there was no further information on histological subtype for the grades II and III cases.
Source of control tissue
The aims of the various studies are different, and thus the source of control tissue is variable. For those studies aiming at detecting gene expression profiles separating meningiomas from tissue of origin the authors have extracted RNA from meninges, dura, arachnoid cyst membranes, normal brain, pooled normal tissue from various sources, and other brain tumors. Other studies have aimed at detecting gene expression changes due to irradiation, anatomical location (spinal vs. intracranial), and WHO grade; hence, samples from each of the categories have been compared with no external controls applied.
Statistics
Of the 13 microarray studies, ten provided a clear description of statistical procedure for the detection of differentially expressed genes between cases. Most frequently the Student’s t test was applied (seven studies), two studies used the Mann–Whitney test, one study used significance analysis of microarrays (SAM), and finally one study used receiver operating characteristic curve.
Summary of main results (Table 2)
In the pioneering oligonucleotide microarray study on meningiomas by Watson and colleagues [14], 15 meningiomas (WHO grade I, n = 6; WHO grade II, n = 6; WHO grade III, n = 3) and three post mortem leptomeninges were studied. The main result was the detection of gene expression profiles associated with WHO grade subtypes (growth hormone receptors, endothelin receptor A, IGF2, IGFBP-7). Selected genes were confirmed using qRT-PCR.
Sasaki and colleagues [15] aimed at comparing the transcriptomic profiles of original meningiomas (n = 3; one of each WHO grade) and primary cultures of the same samples. They found that 51 genes were up-regulated > fivefold, and 19 genes were down-regulated by twofold or more in the primary cultures. The results were validated with qRT-PCR.
Fathallah-Shaykh and colleagues [16] studied ten meningiomas and compared them with pooled post mortem brain RNA from the occipital lobes of four individuals. With their approach 364 genes were differentially expressed, and they found evidence of activation of different signaling pathways like Wnt, MAP kinase, PI3K, and notch. No validation of the findings with alternative approaches was performed.
In a study of 30 meningiomas (WHO grade I, n = 13; WHO grade II, n = 12; WHO grade III, n = 5) Wrobel and colleagues [17] investigated 2,600 genes using cDNA microarrays. The gene expression profiles of each category were compared with each other. The main finding was that 37 genes were decreased and 27 increases in grades II and II meningiomas compared with grade I. Compared with a pool of RNA from various human tissues (heart, spleen, placenta, kidney, skeletal muscle, liver, brain, and lung) a gene signature of the meningiomas was identified: PTGDS, CLU, BAD, MGP, LIG1, ANXA2, MMP12, VIM, TIMP1, and CCND1 were highly expressed in the meningiomas. Selected genes were validated with qRT-PCR, and for several candidates the results corroborated with those found by microarray. The authors concluded that the study showed that genes related to cell cycle regulation, cellular proliferation, as well as the IGF and WNT signaling pathways were up-regulated in grades II and III meningiomas. However, a main limitation of the study was the low number of cDNA probes, covering <10% of all human genes.
The aim of the study of Lusis et al. [18] was to identify genetic events responsible for malignant progression of meningiomas. Using the Affymetrix U133A/B GeneChip Microarrays the authors searched for transcripts that were lost in grade III meningiomas compared with grade I. They found that approximately 40% of down-regulated genes in grade III meningiomas were located at chromosomes 1p and 14q. One of the candidates, the NDRG2 genes, was consistently down-regulated in all grade III meningiomas both at the mRNA and protein level, and that this was correlated with hypermethylation of the corresponding promoter.
As a part of a larger study of the genomics of spinal meningiomas Sayagues et al. [19] performed gene expression profiling to compare spinal and intracranial meningiomas. They included seven spinal and 11 intracranial meningiomas in the study. The main result was differential expression of 1,555 genes, of which 35 genes showed the highest correlation (r 2 > 0.7 or r 2 < −0.7). Thirty of these had lower expression in the intracranial tumors, whereas the remaining five genes were up-regulated. Three genes were selected for qRT-PCR validation, and a significant correlation (p < 0.001) with microarray expression was found for all genes (NR4A3, DUSP5, and HOXA5).
In the study of Carvalho et al. [20] the purpose was to identify molecular signatures that characterize the different grades of meningiomas and molecular mechanisms driving meningioma tumorigenesis. They included 23 meningiomas (WHO grade I, n = 8; WHO grade II, n = 7; WHO grade III, n = 8). Using SAM, the authors found 28 genes differentially expressed between grades I and II meningiomas, and no differential expression between grades II and III. A total of 1,212 genes were differentially expressed between grades I and III meningiomas. In an unbiased unsupervised cluster analysis the 23 meningiomas grouped in two branches. All grades I and grade III meningiomas were located in separate branches, and the authors thus designated each branch as “low proliferative” and “high proliferative”, respectively. The grade II meningiomas were located in both branches, three of seven in the low-proliferative group, and the remaining four in the high-proliferative group. A selection of genes were validated using qRT-PCR.
In 2008, our group [21] published a microarray study on meningiomas of grades I (n = 22) and II (n = 5), where the aim was to study the gene expression profiles of meningiomas in comparison to progenitor meningioma tissue (arachnoid cells). Unsupervised cluster analysis of a filtered data set of 16,430 genes showed that five of seven fibrous meningiomas clustered together, while the remaining samples (meningothelial, transitional, and atypical) made no clear branching. As control tissue we used samples from the membranes of four arachnoid cysts (AC), and all these samples formed a separate cluster indicating a very homogeneous transcription profile. Using the t test, we detected 20 genes that differentiated between meningiomas and ACs (p < 4.3 × 10−7), in which the tumor suppressor gene WWOX was down-regulated and the oncogene TYMS was up-regulated. We also found 20 genes separating fibrous from meningothelial meningiomas (p < 1.1 × 10−5), where DMD and BMPR1B were up-regulated in the fibrous, and RAMP1 was down-regulated in the meningothelial meningiomas. qRT-PCR was performed on a selection of genes and showed similar expression profiles as those generated using microarray analysis.
Since a recognized mechanism of meningioma initiation is irradiation, Lillehei et al. [22] performed a microarray study of five radiation-induced meningiomas (RIM) and six spontaneous meningiomas to find unique genes behind this phenomenon. Interestingly, based on a microarray of 54,675 genes unsupervised hierarchical cluster analysis did not show separate clustering of RIMs and spontaneous meningiomas. Using a t test to compare the gene expression profiles of RIMs and spontaneous meningiomas the authors found a small subset of 20 genes separating the two groups (p < 0.001).
Hankins et al. [23] studied the expression profiles of 12,000 genes in six meningiomas (WHO grade I) and four dural samples. By this approach, the authors found five up-regulated and 35 down-regulated genes in the meningiomas. The down-regulation of the DLC1 gene was confirmed with qRT-PCR and immunohistochemical staining. No evidence of CpG methylation of the corresponding promoter was found. The authors concluded that DLC1 may function as a tumor suppressor gene in meningiomas.
Claus and colleagues [24] studied 31 samples from sporadic meningiomas (WHO grade I, n = 25; WHO grade II, n = 6) with the aim of examining the gene expression profiles in relation to hormone receptor status. Estrogen receptor positivity was present in 33% and progesterone receptor positivity in 84%. In a comparison of PR+ and PR− meningiomas, the study showed up-regulation of ten genes, and down-regulation of 14 genes. No genes separated ER+ from ER− meningiomas. As the number of candidate genes was small, no single pathways or groups of genes were clearly identified.
In 2009, Fèvre-Montange and colleagues [25] published a transcriptomic study of 17 meningiomas (WHO grade I, n = 10; WHO grade II, n = 5; WHO grade III, n = 2). As control tissue RNA from a human whole brain (72 years of age) was used. The aim was to distinguish between the different WHO grades and histopathological subtypes, and to identify factors predicating recurrence. Unsupervised cluster analysis showed three groups of samples: group A consisted of seven of ten grade I cases, group B of the remaining three grade I samples and all five grade II meningiomas, and finally group C consisted of the two grade III tumors. Statistical analysis revealed that 346 and 2,995 genes showed more than twofold over-expression in groups B and C, respectively. Similarly, 184 and 1,380 genes were down-regulated, respectively. Furthermore, the study showed differential gene expression between fibrous and meningothelial meningiomas, with 12 up-regulated and 20 down-regulated genes in the fibrous subset. Selected genes were validated with qRT-PCR.
The last published microarray study on meningiomas so far was published in 2009 and performed by Castells et al. [26]. The aim was to assess whether automated categorization of brain tumors can be made by the use of microarray. Biopsies from 35 patients (17 glioblastomas and 18 meningothelial meningiomas) were subjected to cDNA-based microarray analysis. The study showed up to 100% prediction accuracy by using microarrays, thus providing evidence of possible clinical diagnostic use of this technology.
Discussion
During the last decade, a total of 13 microarray-based studies have been performed on RNA from altogether 227 meningiomas. The aims, sources of control tissue, microarray platforms, and statistical approaches vary between the studies. The main results of the studies can be grouped in three categories: (1) several groups have identified meningioma-specific genes, and genes associated with the three WHO grades and the main histological subtypes of grade I meningiomas. (2) One publication has shown that the general transcription profile of samples of all WHO grades differs in vivo and in vitro. (3) One report provides evidence that microarray technology can be used in an automated fashion to classify tumors.
We found no clear overlap between the studies regarding individual candidate genes. Other than the gross signature differences identified in the studies, we were not able to detect any shared deregulated genes. This illustrates a big challenge in gene expression profiling studies: the vast number of transcripts present on the chips makes it very difficult to compare the studies and isolate key candidate genes with a possible role in the biology. Critiques may regard this as a weakness of the technology; however, the lack of external validity may in fact be explained by the variation in microarray platforms, study populations, chip manufacturer, statistical approach, and quality of RNA in the tissue at study. In fact, the Microarray Quality Control (MAQC) project (http://www.fda.gov/ScienceResearch/BioinformaticsTools/MicroarrayQualityControlProject/default.htm) has shown that the microarray quality at present may be higher than that of, e.g., immunohistochemical analysis [27, 28]. So the question is how should we perform microarray studies? Would it be appropriate to standardize some factors, like the source of control tissue and statistical procedure? And should it be mandatory to post microarray data in publicly available databases so that independent researchers could perform meta-analyses with the aim of identifying biological markers that could be used clinically? If so, would it be reasonable to demand posting of certain data, e.g., the overall best candidate genes for comparison purposes? Some journals have implemented this policy, however it is our opinion that it should be adopted globally.
To draw firmer conclusions of the studies, consensus on source of control tissue and statistical approach, as well as replication of the most biological relevant findings so far is required. The time is therefore now due for a large replication study that includes the most significant and biologically relevant candidate genes generated from the various transcription studies that have been performed. This means that a custom-made microarray chip that includes these consensus genes should be designed, and quality controlled samples from several laboratories included. The data should also be compared with clinical outcome in order to identify clinically relevant genes. Recently, The Cancer Genome Atlas Research Network successfully applied this method to a large panel of glioblastomas [29]. Here, 206 glioblastomas were subjected to gene expression profiling, DNA copy number variation analysis, and CpG methylation status assessment of 601 selected genes. It follows that such a project demands considerable dedication, coordination, and financial support.
An interesting result was the difference in transcriptional profiles between original frozen specimens and primary culture of all three WHO grades. The study provides evidence that as tumors are removed from their native environment, the gene expression changes accordingly. Thus, results gained from cell culture experiments have to be interpreted with caution. Validation in vivo seems to be mandatory before firm conclusion can be made. It is however no surprise that the gene expression profile is altered when a tumor is removed from its site of origin. The dynamic transcriptome will rapidly adjust to changes in local environment to provide the substrates necessary to maintain the tumor.
Medicine has been transformed by the genomic revolution, and classical population risk assessment and empirical treatment has been challenged by molecular classification and prospects of individualized therapy [1]. With the help of microarray technology novel candidate genes, pathways, and networks may be linked with clinical scenarios, such as treatment response or environmental exposure. Thus, microarrays may be helpful in identifying biomarkers, developing new treatment strategies, and in tumor classification [2] (Fig. 3). Hence, a reasonable application of global gene expression profiling in oncology is to subclassify brain tumors of WHO grade II. Such tumors are generally infiltrative, have low-proliferative activity, tend to recur, and can undergo malignant transformation. Often there is marked clinical and morphological heterogeneity within tumors of this grade. Thus, the categorization of grade II tumors may be more challenging than that of purely benign or malignant entities. With microarray analysis, high- or low-proliferative gene expression signatures, or hyper- or hypo-mutator phenotypes may be revealed. Consequently, signatures indicating high or low risk of recurrence or transformation may be found. Such information may be useful when the addition of adjuvant therapy or follow-up schedule is discussed.
Fig. 3.
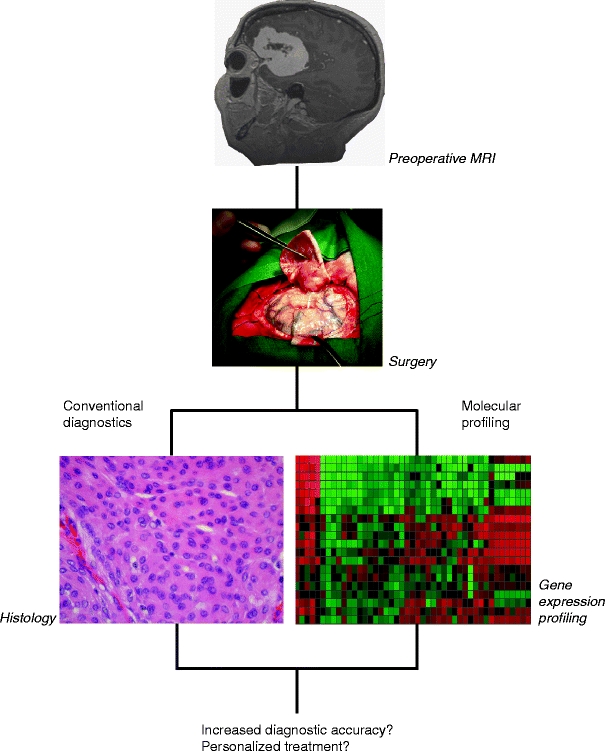
Outline of possible application of microarray technology in the diagnostic armamentarium
However, some technical issues make the use of this powerful technology difficult: (1) Since only minute amounts of RNA are required and probes corresponding to all known genes are analyzed, one must ensure that only representative tissue is subjected to hybridization to the microarray chips. Contamination with leucocytes, neighboring blood vessels or other normal tissue will contribute to the overall transcriptomic profile. (2) If the goal of the analysis is to measure therapeutic response to e.g. irradiation or cytotoxic drugs one must sample tissue at first-time surgery as well as after completed adjuvant therapy. Thus, the patient must undergo repeated surgery with the inherent risk factors. (3) RNA is very susceptible to time dependent degradation due to abundant RNase in the environment. Low quality RNA cannot be subjected to microarray analysis. Hence, the personnel involved in the sampling and handling of tissue must be dedicated and have a proper logistic procedure in order to preserve RNA. This means that tissue must be snap-frozen in liquid nitrogen in the operating theater and then transferred to permanent storage at ultra cold temperatures or in media containing RNase inhibitors. 4) Since transcriptome analysis can reveal genes that differ between tissue types, appropriate controls and groups of e.g. tumors have to be selected long before time of study. As meningiomas develop from arachnoid cells [4], it follows that such cells should be selected if the purpose was to find meningioma-specific genes. If dura or whole brain were used as controls the transcriptional profile would presumably be different due to the comparison of different tissue types. It would thus be impossible to call these genes different because of tumorigenesis per se. Only one of the 13 studies states that arachnoid tissue has been used as controls [21]. However, if the purpose is to identify genes associated with the different WHO grades it follows that no additional controls would be needed.
The main limitations of the studies were small sample size and unequal group sizes when performing statistical comparison. This illustrates how difficult it is to recruit patients and controls to such studies. It may also be due to the relatively high cost of performing microarray analysis. Furthermore, the results of the studies have to be functionally tested in order to safely identify biological markers and key players in meningioma pathogenesis.
As in most microarray studies of the transcriptome, new genes and pathways have been revealed in all the meningioma studies. The challenge is to make use of the results clinically. At present, microarray chip technology is mature and with low technical error rate. Thus, the main issues that have to be handled in transcriptomic projects are study design, representative tissue source, logistics, and data interpretation. Since transcriptome analysis only provides snapshots of the gene expression state in cells constituting the tissue at study, clear improvements in diagnostic accuracy or clinical endpoints (e.g., improved recurrence free survival) must be objectively shown before the results of such studies can be safely implemented in the clinical armamentarium. This notion is by no means reserved for microarray studies, as all new methods have to be quality checked accordingly before clinical approval. Microarray studies have been taken into account for a trend towards personalized medicine, e.g. the genetic profile of a patient’s tumor could tell which pathways that could be pharmacologically inhibited. However, as long as there are no individually designed drugs, such treatment is reserved for the future. How eager the pharmacological industry is to make such designed drugs, which presumably are more expensive than mass production, remains to be seen. Nevertheless, the molecular technology is continuously developing new and faster methods for the analysis of the nucleic acids. Next-generation sequencing is already “this generation”; the first people and cancers have been deciphered at base-pair resolution by the so-called deep sequencing technology. The complete sequencing of the human genome that took 10 years in the last decade of the twentieth century can now be performed in a few days. Therefore, awaiting individualized therapy, the era of the personalized genome is inevitably about to begin.
Acknowledgments
Conflicts of interest
None.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Comment
As a frequent reviewer for Acta, I have done my best to shoot down reports of immunostaining of this or that protein in this or that tumor as useless philately from by-gone decades.
At last—transcriptomic data on meningioma reviewed by the Norwegian colleagues in the era of genomics/transcriptomics/signalomics/metabolomics/glycomics/et al. mics (1–4). RNA sequencing of transcriptome of target tissue—analysis of differential signaling pathways—probing of synthetic molecules from drug libraries into key receptors—new ideas for phase I–II clinical drug trials. Furthermore, with the techniques cheaper, honed and widespread, each tumor in each carrier could be individually profiled for optimal therapies and outcome—we hope.
Juha E Jääskeläinen
Kuopio, Finland
1. Aarhus M, Bruland O, Sætran HA, Mork SJ, Lund-Johansen M, Knappskog PM. Global gene expression profiling and tissue microarray reveal novel candidate genes and down-regulation of the tumor suppressor gene CAV1 in sporadic vestibular schwannomas. Neurosurgery 2010;67:998–1019
2. Wibom C, Mörén L, Aarhus M, Knappskog PM, Lund-Johansen M, Antti H, Bergenheim AT. Proteomic profiles differ between bone invasive and noninvasive benign meningiomas of fibrous and meningothelial subtype. J Neurooncol 2009;94:321–31
3. Aarhus M, Bruland O, Bredholt G, Lybaek H, Husebye ES, Krossnes BK, Vedeler C, Wester K, Lund-Johansen M, Knappskog PM. Microarray analysis reveals down-regulation of the tumor suppressor gene WWOX and up-regulation of the oncogene TYMS in intracranial sporadic meningiomas. J Neurooncol 2008;88:251–9
4. Kurki M, Häkkinen S-K, Frösen J, Tulamo R, von und zu Fraunberg M, Wong G, Tromp G, Niemelä M, Hernesniemi J, Jääskeläinen JE, Ylä-Herttuala S. Rupture of saccular intracranial aneurysm wall associates to inflammation, leukocyte infiltration, oxidative stress, extracellular matrix degradation, and apoptosis in gene expression profiling. Neurosurgery (in press)
References
- 1.Ramaswamy S. Translating cancer genomics into clinical oncology. N Engl J Med. 2004;350:1814–1816. doi: 10.1056/NEJMp048059. [DOI] [PubMed] [Google Scholar]
- 2.Mischel PS, Cloughesy TF, Nelson SF. DNA-microarray analysis of brain cancer: molecular classification for therapy. Nat Rev Neurosci. 2004;5:782–792. doi: 10.1038/nrn1518. [DOI] [PubMed] [Google Scholar]
- 3.Stekel D. Microarray bioinformatics. Cambridge: Cambridge University Press; 2003. [Google Scholar]
- 4.OH LDN, Wiestler OD, Cavenee WK. WHO classification of tumours of the central nervous system. Lyon: World Health Organization IARC; 2007. [Google Scholar]
- 5.Arai E, Ikeuchi T, Karasawa S, Tamura A, Yamamoto K, Kida M, Ichimura K, Yuasa Y, Tonomura A. Constitutional translocation t(4, 22) (q12;q12.2) associated with neurofibromatosis type 2. Am J Med Genet. 1992;44:163–167. doi: 10.1002/ajmg.1320440209. [DOI] [PubMed] [Google Scholar]
- 6.Rouleau GA, Seizinger BR, Wertelecki W, Haines JL, Superneau DW, Martuza RL, Gusella JF. Flanking markers bracket the neurofibromatosis type 2 (NF2) gene on chromosome 22. Am J Hum Genet. 1990;46:323–328. [PMC free article] [PubMed] [Google Scholar]
- 7.Wertelecki W, Rouleau GA, Superneau DW, Forehand LW, Williams JP, Haines JL, Gusella JF. Neurofibromatosis 2: clinical and DNA linkage studies of a large kindred. N Engl J Med. 1988;319:278–283. doi: 10.1056/NEJM198808043190505. [DOI] [PubMed] [Google Scholar]
- 8.Wolff RK, Frazer KA, Jackler RK, Lanser MJ, Pitts LH, Cox DR. Analysis of chromosome 22 deletions in neurofibromatosis type 2-related tumors. Am J Hum Genet. 1992;51:478–485. [PMC free article] [PubMed] [Google Scholar]
- 9.Bello MJ, de Campos JM, Kusak ME, Vaquero J, Sarasa JL, Pestana A, Rey JA. Allelic loss at 1p is associated with tumor progression of meningiomas. Genes Chromosom Cancer. 1994;9:296–298. doi: 10.1002/gcc.2870090411. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda K, Saeki Y, Gonzalez-Agosti C, Ramesh V, Chiocca EA. Inhibition of NF2-negative and NF2-positive primary human meningioma cell proliferation by overexpression of merlin due to vector-mediated gene transfer. J Neurosurg. 1999;91:85–92. doi: 10.3171/jns.1999.91.1.0085. [DOI] [PubMed] [Google Scholar]
- 11.Peyrard M, Pan HQ, Kedra D, Fransson I, Swahn S, Hartman K, Clifton SW, Roe BA, Dumanski JP. Structure of the promoter and genomic organization of the human beta’-adaptin gene (BAM22) from chromosome 22q12. Genomics. 1996;36:112–117. doi: 10.1006/geno.1996.0431. [DOI] [PubMed] [Google Scholar]
- 12.Zucman-Rossi J, Legoix P, Thomas G. Identification of new members of the Gas2 and Ras families in the 22q12 chromosome region. Genomics. 1996;38:247–254. doi: 10.1006/geno.1996.0625. [DOI] [PubMed] [Google Scholar]
- 13.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson MA, Gutmann DH, Peterson K, Chicoine MR, Kleinschmidt-DeMasters BK, Brown HG, Perry A. Molecular characterization of human meningiomas by gene expression profiling using high-density oligonucleotide microarrays. Am J Pathol. 2002;161:665–672. doi: 10.1016/S0002-9440(10)64222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki T, Hankins GR, Helm GA. Comparison of gene expression profiles between frozen original meningiomas and primary cultures of the meningiomas by GeneChip. Neurosurgery. 2003;52:892–898. doi: 10.1227/01.NEU.0000053368.73552.BB. [DOI] [PubMed] [Google Scholar]
- 16.Fathallah-Shaykh HM, He B, Zhao LJ, Engelhard HH, Cerullo L, Lichtor T, Byrne R, Munoz L, Von Roenn K, Rosseau GL, Glick R, Sherman C, Farooq K. Genomic expression discovery predicts pathways and opposing functions behind phenotypes. J Biol Chem. 2003;278:23830–23833. doi: 10.1074/jbc.M302800200. [DOI] [PubMed] [Google Scholar]
- 17.Wrobel G, Roerig P, Kokocinski F, Neben K, Hahn M, Reifenberger G, Lichter P. Microarray-based gene expression profiling of benign, atypical and anaplastic meningiomas identifies novel genes associated with meningioma progression. Int J Cancer. 2005;114:249–256. doi: 10.1002/ijc.20733. [DOI] [PubMed] [Google Scholar]
- 18.Lusis EA, Watson MA, Chicoine MR, Lyman M, Roerig P, Reifenberger G, Gutmann DH, Perry A. Integrative genomic analysis identifies NDRG2 as a candidate tumor suppressor gene frequently inactivated in clinically aggressive meningioma. Cancer Res. 2005;65:7121–7126. doi: 10.1158/0008-5472.CAN-05-0043. [DOI] [PubMed] [Google Scholar]
- 19.Sayagues JM, Tabernero MD, Maillo A, Trelles O, Espinosa AB, Sarasquete ME, Merino M, Rasillo A, Vera JF, Santos-Briz A, de AE G-M, MC OA. Microarray-based analysis of spinal versus intracranial meningiomas: different clinical, biological, and genetic characteristics associated with distinct patterns of gene expression. J Neuropathol Exp Neurol. 2006;65:445–454. doi: 10.1097/01.jnen.0000229234.13372.d8. [DOI] [PubMed] [Google Scholar]
- 20.Carvalho LH, Smirnov I, Baia GS, Modrusan Z, Smith JS, Jun P, Costello JF, McDermott MW, Vandenberg SR, Lal A. Molecular signatures define two main classes of meningiomas. Mol Cancer. 2007;6:64. doi: 10.1186/1476-4598-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aarhus M, Bruland O, Bredholt G, Lybaek H, Husebye ES, Krossnes BK, Vedeler C, Wester K, Lund-Johansen M, Knappskog PM. Microarray analysis reveals down-regulation of the tumour suppressor gene WWOX and up-regulation of the oncogene TYMS in intracranial sporadic meningiomas. J Neurooncol. 2008;88(3):251–259. doi: 10.1007/s11060-008-9569-6. [DOI] [PubMed] [Google Scholar]
- 22.Lillehei KO, Donson AM, Kleinschmidt-DeMasters BK. Radiation-induced meningiomas: clinical, cytogenetic, and microarray features. Acta Neuropathol. 2008;116:289–301. doi: 10.1007/s00401-008-0401-3. [DOI] [PubMed] [Google Scholar]
- 23.Hankins GR, Sasaki T, Lieu AS, Saulle D, Karimi K, Li JZ, Helm GA. Identification of the deleted in liver cancer 1 gene, DLC1, as a candidate meningioma tumor suppressor. Neurosurgery. 2008;63:771–780. doi: 10.1227/01.NEU.0000325488.72518.9E. [DOI] [PubMed] [Google Scholar]
- 24.Claus EB, Park PJ, Carroll R, Chan J, Black PM. Specific genes expressed in association with progesterone receptors in meningioma. Cancer Res. 2008;68:314–322. doi: 10.1158/0008-5472.CAN-07-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fevre-Montange M, Champier J, Durand A, Wierinckx A, Honnorat J, Guyotat J, Jouvet A. Microarray gene expression profiling in meningiomas: differential expression according to grade or histopathological subtype. Int J Oncol. 2009;35:1395–1407. doi: 10.3892/ijo_00000457. [DOI] [PubMed] [Google Scholar]
- 26.Castells X, Garcia-Gomez JM, Navarro A, Acebes JJ, Godino O, Boluda S, Barcelo A, Robles M, Arino J, Arus C. Automated brain tumor biopsy prediction using single-labeling cDNA microarrays-based gene expression profiling. Diagn Mol Pathol. 2009;18:206–218. doi: 10.1097/PDM.0b013e31818f071b. [DOI] [PubMed] [Google Scholar]
- 27.Shi L, Tong W, Fang H, Scherf U, Han J, Puri RK, Frueh FW, Goodsaid FM, Guo L, Su Z, Han T, Fuscoe JC, Xu ZA, Patterson TA, Hong H, Xie Q, Perkins RG, Chen JJ, Casciano DA. Cross-platform comparability of microarray technology: intra-platform consistency and appropriate data analysis procedures are essential. BMC Bioinform. 2005;6(Suppl 2):S12. doi: 10.1186/1471-2105-6-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 29.Comprehensive genomic characterization defines human glioblastoma genes and core pathways (2008). Nature 455(7216):1061–1068 [DOI] [PMC free article] [PubMed]


