Abstract
MicroRNAs (miRNAs) are single-stranded, non-coding RNA molecules that regulate gene expression at the post-transcriptional level. Genes encoding miRNAs are located in regions of the genome that are commonly amplified, deleted or rearranged. They are commonly dysregulated in human cancers and known to act as oncogenes or tumor suppressors. Members of the miR-200 miRNA family are downregulated in human cancer cells and tumors due to aberrant epigenetic gene silencing and play a critical role in the suppression of epithelial-to-mesenchymal transition (EMT), tumor cell adhesion, migration, invasion and metastasis, by targeting and repressing the expression of key mRNAs that are involved in EMT (ZEB1 and ZEB2), β-catenin/Wnt signaling (β-catenin), EGFR inhibitor resistance (ERRFI-1) and chemoresistance to therapeutic agents (TUBB3). Since the miR-200 family functions as putative tumor suppressors and represent biomarkers for poorly differentiated and aggressive cancers, restoration of miR-200 expression may have therapeutic implications for the treatment of metastatic and drug-resistant tumors.
Key words: miRNAs, mir-200, epithelial-mesenchymal transition, β-catenin/Wnt signaling, microenvironment, metastasis, RNA
MicroRNAs (miRNAs) are non-coding RNA molecules approximately 21–23 nucleotides long that regulate gene expression at the post-transcriptional level.1–6 They are differentially and temporally expressed in a tissue and developmental-specific manner. Approximately 30% of messenger RNA transcripts are estimated to be regulated by miRNAs.6 Genes encoding miRNAs are located within the introns of transcriptional units and are also frequently clustered, such that a single primary miRNA transcript may contain multiple miRNA genes. miRNA genes are transcribed in a RNA polymerase II-dependent manner producing the primary miRNA (pri-miRNA), which contains a 5′ cap and poly-A tail, and can be several kilobases in length. The pri-miRNA is further processed by the RNAse III endonuclease enzyme, Drosha, in the nucleus and by Dicer in the cytoplasm, producing the pre-miRNA and mature miRNA, respectively.1,7–9 The mature miRNA will then bind to the “seed” sequence spanning nucleotides 2–8 of the miRNA and its complementary sequence within the 3′ untranslated regions (UTRs) of its target mRNAs. Perfect complementarity between the miRNA and its target mRNA leads to endonucleotic cleavage of the mRNA transcript whereas imperfect complementarity results in either transcript degradation or translational inhibition. miRNAs regulate various physiological and pathological processes by modulating the expression of their target mRNAs that play important roles in diverse cellular processes including differentiation, proliferation, growth, migration and survival.
miRNA expression profiling analysis reveals a global downregulation of mature miRNAs levels in primary human tumors relative to normal tissue.10,11 Thus, miRNAs may function as tumor suppressors or oncogenes and dysregulation in their expression may contribute to tumor cell metastasis. We will focus on the miR-200 family, members of which are downregulated in aggressive human tumors and are potent inhibitors of epithelial-mesenchymal transition (EMT), tumor cell adhesion, migration, invasion, metastasis and β-catenin/Wnt signaling.
Epithelial-Mesenchymal Transition (EMT)
EMT is a critical step in tumor cell invasion and metastasis, and correlates positively with poor patient prognosis.12,13 The loss of epithelial markers and the acquisition of mesenchymal morphological features are characterized by the disassembly of tight junctions and loss of apical-basal polarity due to a repression of the transmembrane adhesion receptor E-cadherin and a gain in expression of the mesenchymal markers, including vimentin, collagen, fibronectin and the E-cadherin transcriptional repressors, ZEB1 and Smad-interacting protein SIP1, also known as ZEB2, thereby leading to extracellular matrix (ECM)-induced stimulation of integrin signaling and focal adhesion formation, with the facilitation of cell motility and invasion.14,15 The EMT-inducing transcriptional factors ZEB1 and ZEB2 repress E-cadherin expression and promote cancer cell migration and invasion.16–19 Transforming growth factor beta (TGFβ) controls cell proliferation and differentiation and is a major inducer of EMT in epithelial cells during embryonic development and cancer progression.
miR-200 Family of miRNAs
The microRNA (miR)-200 family comprises five members (miR-200a, -200b, -200c, -141 and -429) that are clustered and expressed as two separate polycistronic pri-miRNA transcripts, miR-200b-200a-429 and miR-200c-141, located on human chromosomes 1 and 12, respectively. In a screen to identify novel miRNAs that are involved in the regulation of EMT, the expression level of all five members was significantly reduced in cells following TGFβ-mediated EMT.20 Enforced constitutive expression of the miR-200 miRNAs in mesenchymal cells promoted mesenchymal epithelial transition (MET).20 Conversely, inhibition of miR-200 induces a mesenchymal-like spindle cell morphology, accompanied by an increase in ZEB1 expression and cell migration.20 Furthermore, loss of miR-200 correlates with a lack of E-cadherin expression in invasive breast cancer cell lines and in breast tumor specimens, supporting an in vivo role for the miR-200 family in EMT repression.20 In addition, both miR-200 miRNA clusters are markedly downregulated in a TGFβ inducible mouse model of mammary tumor with EMT, suggesting a suppressor function for miR-200 in EMT. Indeed, overexpression of the individual miR-200 members or separate clusters represses EMT by directly targeting and downregulating ZEB1 and ZEB2 via miR-200-binding sites located within their 3′ UTRs, resulting in enhanced E-cadherin expression and inhibition of murine mammary tumor cell migration and cancer cell motility.21,22
Importantly, miRNA profiling analysis of NCI60 human cancer cell lines identified the miR-200 family as an epithelial marker that is selectively expressed in only E-cadherin-positive and vimentin-negative cancer cell lines and primary ovarian cancer patient tumor tissues that lack ZEB1 and ZEB2, demonstrating a strong correlation between loss of miR-200 expression and tumor cell aggressiveness.22 Consistent with this notion, exogenous expression of miR-200 induced E-cadherin expression and MET.22 By contrast, inhibition of miR-200 reduced E-cadherin and increased vimentin, ZEB1 and ZEB2 levels are associated with a mesenchymal-like cell morphology.22 miRNA profiling analysis of lung adenocarcinoma cells derived from mutant K-ras and p53 mice that form polarized epithelial spheres in three-dimensional Matrigel culture and undergo EMT following TGFβ treatment or when injected into syngeneic mice, revealed a significant reduction in miR-200 levels and concomitant increase in mesenchymal markers, vimentin and ZEB1.23 By contrast, constitutive miR-200 expression blocked TGFβ-induced EMT, reverting cells to a non-metastatic mRNA expression profile, inhibited cell migration and invasion and abrogated tumor growth and metastasis in vivo.23 Strikingly, in a panel of 40 human non-small cell lung cancer cell lines, miR-200 correlated with EMT markers, distinguishing between cell lines derived from primary lung tumors from those derived from metastatic lesions.23
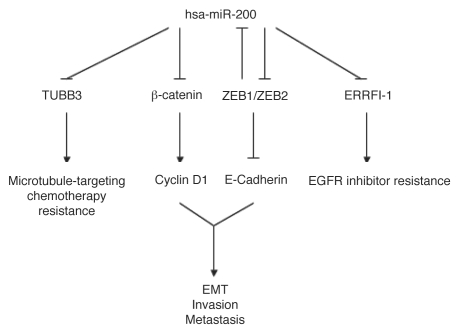
A recent study identified ErbB receptor inhibitor-1 (ERRFI-1) as a direct target that is downregulated by miR-200 and plays a key role in reversing EMT-induced EGFR inhibitor resistance in mesenchymal bladder carcinoma cells, improving sensitivity to growth inhibition by anti-EGFR-targeted therapy.24 Interestingly, in a genome-wide miRNA array screen, miR-200a was identified as the most downregulated miRNA in human meningiomas.25 Restoration of miR-200a decreased ZEB1 and SIP1, thereby leading to increased E-cadherin expression, and was associated with a suppression of meningioma cell growth in culture and tumor formation in vivo due to an induction of caspase-mediated apoptosis.25 Importantly, miR-200a directly targeted β-catenin mRNA, which contains a functionally conserved miR-200a-binding site in its 3′ UTR and suppresses β-catenin/Wnt signaling, which is implicated in tumorigenesis in human colon cancer, hepatocellular carcinoma, melanoma, ovarian cancer and prostate cancer.25–29 Consistent with this finding, miR-200a expression correlates inversely with β-catenin and one of its downstream targets, cyclin D1, in human meningioma tumor tissues.25 Interestingly, reintroduction of miR-200c reduces aggressive breast, ovarian and endometrial cancer cell migration, invasion and adhesion to extracellular matrix (ECM) independent of E-cadherin restoration, and enhances their sensitivity to microtubule-targeting chemotherapeutic agent-induced cell death mediated by direct targeting and downregulation of class III β-tubulin (TUBB3).30,31 Conversely, ectopic expression of TUBB3 lacking the miR-200c target site in its 3′UTR reversed chemosensitivity to microtubule-directed agents in long-term clonogenic assays.30 Furthermore, in addition to ZEB1 and ZEB2, microarray profiling identified several mesenchymal markers including fibronectin-1 (FN), neurotrophic tyrosine kinase (NTRK2) and QKI, that are downregulated by miR-200c overexpression.31 Together, these findings suggest a novel role for miR-200 in controlling EMT reversion by down-modulation of β-catenin/Wnt signaling, and improved sensitivity to EGFR inhibition and microtubule-targeting chemotherapeutic agents in aggressive drug-resistant cancers.
Regulation of miR-200 Expression in Normal and Cancer Cells
The mechanisms controlling miR-200 transcriptional regulation during the phenotypic conversion of normal epithelial cells in EMT and tumor cell invasion and metastasis are poorly understood. Interestingly, ZEB1 and SIP1 have been found to bind directly to an E-box proximal minimal promoter element and repress primary transcript and mature miR-200 miRNA expression in mesenchymal human breast cancer cells, demonstrating a potential double-negative feedback loop between ZEB1/SIP1 and the miR-200 family during EMT and tumorigenesis.32 In a recent study, DNA methylation was shown to play an important role in regulating normal cell and tissue-specific expression of the miR-200c-141 cluster.33 However, loss of the miR-200 cluster correlated also with aberrant DNA CpG island methylation and histone modifications in breast and prostate cancer lines relative to normal human epithelial cells and tissues, indicating that repression of miR-200 expression due to inappropriate epigenetic silencing may contribute to EMT and tumor progression in certain human cancers.33 Together, these findings suggest a potential tumor suppressor role for miR-200 that is downregulated in human cancers leading to EMT and facilitating tumor cell invasion and metastasis, and loss of miR-200 family members is associated with an aggressive cancer cell phenotype.
The miR-200 family of miRNAs functions as a potential tumor suppressor in EMT (Fig. 1), a hallmark feature of malignant tumor progression.20,22,34–37 Future work will undoubtedly focus upon the identification and functional characterization of additional direct miR-200 downstream targets such as WAVE3, an actin cytoskeleton remodeling and metastasis promoter protein,38 as well as regulation of mir-200 by AKT,39 and other yet to be determined factors. This will improve our understanding of the events contributing to EMT and metastasis, leading to the development of novel therapeutic strategies.
Figure 1.
miR-200 mediated functions in the maintenance of the epithelial cell phenotype.
Acknowledgements
This work was supported by NIH DK056645, the National Colon Cancer Research Alliance and the Hansen Foundation.
Abbreviations
- miRNAs
microRNAs
- EMT
epithelial-to-mesenchymal transition
- ERRFI-1
ErbB receptor inhibitor-1
- TUBB3
class III β-tubulin
- TGFβ
transforming growth factor beta
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/12548
References
- 1.Rana T. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 2.Valencia-Sanchez M, Liu J, Hannon G, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 3.Pillai R, Bhattacharyya S, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Standart N, Jackson R. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen T. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya S, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Sontheimer E. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 8.Kim V, Nam J. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Filipowicz W, Jaskiewicz L, Kolb F, Pillai R. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, et al. Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 13.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 14.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imamichi Y, Menke A. Signaling pathways involved in collagen-induced disruption of the E-cadherin complex during epithelial-mesenchymal transition. Cells Tissues Organs. 2007;185:180–190. doi: 10.1159/000101319. [DOI] [PubMed] [Google Scholar]
- 16.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 17.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell. 2007;18:3533–3544. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 19.Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 21.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saydam O, Shen Y, Wurdinger T, Senol O, Boke E, James MF, et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol. 2009;29:5923–5940. doi: 10.1128/MCB.00332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, He X. Wnt/beta-catenin signaling: new (and old) players and new insights. Curr Opin Cell Biol. 2008;20:119–125. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 28.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Cochrane DR, Howe EN, Spoelstra NS, Richer JK. Loss of miR-200c: A Marker of Aggressiveness and Chemoresistance in Female Reproductive Cancers. J Oncol. 2010:821717. doi: 10.1155/2010/821717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009 doi: 10.1158/1535-7163.MCT-08-1046. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 33.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 5:8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, et al. WNT7a induces E-cadherin in lung cancer cells. Proc Natl Acad Sci USA. 2003;100:10429–10434. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spoelstra NS, Manning NG, Higashi Y, Darling D, Singh M, Shroyer KR, et al. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006;66:3893–3902. doi: 10.1158/0008-5472.CAN-05-2881. [DOI] [PubMed] [Google Scholar]
- 36.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 37.Olson P, Lu J, Zhang H, Shai A, Chun M, Wang Y, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sossey-Alaoui K, Bialkowska K, Plow E. The miR200 family of microRNAs regulates WAVE3-dependent cancer cell invasion. J Biol Chem. 2009;284:33019–33029. doi: 10.1074/jbc.M109.034553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iliopoulos D, Polytarchou C, Hatziapostolou M, Kottakis F, Maroulakou I, Struhl K, et al. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal. 2009;2:62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]