Abstract
Loss of imprinting (LOI) of the insulin-like growth factor 2 gene (IGF2) is one of the most common epigenetic abnormalities seen in human neoplasms. LOI may be associated with the lack of Zinc-finger DNA binding protein CTCF-mediated enhancer insulation, presumably due to the gain of methylation on the maternal allele of the differentially methylated domain (DMD) of the imprinting control region. This results in an interaction between the IGF2 promoters and enhancers; and IGF2 is produced from both alleles. In this study we investigated the feasibility of a novel anti-cancer adenovirus (AdDC312-DT-A) driven by H19 enhancer DMD-H19 promoter complex. Cell lines with IGF2 LOI (HCT-8, HT-29 and H-522) that were infected with AdDC312-EGFP produced the EGFP protein. However, in cells in which imprinting was maintained (MOI) (MCF-7 and GES-1), no EGFP protein was produced. The AdDC312-DT-A significantly decreased cell viability and induced apoptosis only in LOI cells in vitro, and suppressed tumour development in HCT-8 xenografts in nude mice. In conclusion, the toxin gene therapy proves effective in inhibiting LOI cell growth in vitro and in vivo and provides a novel option for targeted gene therapy based on loss of IGF2 imprinting.
Key words: genomic imprinting, IGF2, CTCF, DT-A, tumor, gene therapy
Introduction
The mammalian genome contains a small number of genes that are subject to genomic imprinting.12 These genes are epigenetically marked with their parental origin so that a given parental allele is expressed while the other allele is repressed. The best-characterized gene is the insulin-like growth factor 2 gene (IGF2), which is a paternally expressed imprinted gene.3 In humans this gene resides at 11p15.5 and can be found conserved on distal chromosome 7 in mice. While the majority of studies on IGF2 have been performed on mice, many attributes of the gene, including the expression profile and regulatory mechanisms, are similar in humans. IGF2 encodes a protein that plays a major role in promoting embryonic and placental growth and development.4 IGF2 imprinting is regulated by an imprinting control region (ICR) containing differentially methylated domains (DMD).5,6 The ICR/DMD is located between the IGF2 and H19 and contains CTCF binding sites that are differentially methylated depending on their parental origin; normally, the maternal allele is unmethylated, while the paternal allele is methylated.8,9 In osteosarcomas, IGF2 LOI is associated with bialleleic methylation of the ICR.10 The ICR acts by regulating interactions between the H19 and IGF2 promoters and their shared enhancers lying downstream of H19. Deletion of the ICR/DMD results in the LOI of the IGF2 gene.7 Proper imprinting of IGF2 requires that the ICR/DMD is methylated on the paternal allele and is unmethylated on the maternal allele. The insulator protein CCCTC-binding factor (CTCF) binds to unmethylated CTCF-binding sites in the ICR.11 CTCF was initially shown to mediate insulator or enhancer blocking activity at similar sequences in the β-globin locus.12 CTCF is a ubiquitous, highly conserved, multivalent transcription factor which plays multiple roles in gene regulation such as activation, repression, silencing, chromatin insulation, and long range chromosome interactiosn. These roles are dependant on the combinatorial utilization of different zinc fingers to bind varying CTCF target sites. The human CTCF maps within one of the smallest regions of the overlap for common loss of heterozygosity at 16q22.1. Its deletion has been observed in many solid tumours.13
As an insulator of transcription CTCF may serve as one of the putative imprinting factors. When CTCF levels are diminished by RNA interference (RNAi) in mouse fibroblasts IGF2 imprinting is partially lost.14 Binding of CTCF to the ICR/DMD is vital to the establishment of IGF2 imprinting in mice. Deletion of the locus containing the ICR/DMD leads to biallelic expression of IGF2.15 Mutation of each of the CTCF binding sites in the ICR/DMD also alters IGF2 imprinting.16 When using a transgenic RNAi-based approach to generate oocytes with reduced CTCF protein, Bartolomei et al.17 discovered that CTCF protected the ICR/DMD from de novo methylation during oocyte growth and was required for normal pre-implantation development. However, numerous other factors have been implicated in the imprinting process, including the polycomb repressive complex genes,18–19 and the precise mechanisms and genes underlying the imprinting process remains unknown.
IGF2 LOI can be corrected by transferring nuclei from human tumour cells exhibiting loss of IGF2 imprinting (WTCL, H522, SKNEP and HRT18) into enucleated mouse and human fibroblasts (HBF1 and MBW2) that have maintained normal IGF2 imprinting. After nuclear transfer the abnormal biallelic expression of IGF2 in tumour nuclei transiently converted to normal monoallelic imprinted expression in the reconstructed diploid cells. However, in tetraploid hybrid cells, normal IGF2 imprinting was restored permanently in the tumour genome. Inhibition of the synthesis of putative transimprinting factors with cycloheximide leads to the loss of IGF2 imprinting in normal cultured fibroblasts. This suggests that normal cells produce proteins that act in trans to induce or maintain genomic imprinting.20 In this system, CTCF levels were not decreased in the LOI cells.
In this study we developed a recombinant adenoviral vector containing the transcriptional regulatory sequence of the enhancer DMD-H19 promoter complex to drive the expression of a toxin gene in several cancer cell lines. We hypothesized that in cells in which IGF2 imprinting was maintained, this construct would bind CTCF and the rest of the imprinting machinery to insulate the attached genes from the enhancer, so that those genes would not be expressed; we further hypothesized that in cells in which IGF2 imprinting was lost, the lack of imprinting factors would lead to a loss of enhancer blocking, and the attached genes would be expressed. Thus, only cells with LOI (i.e., cancer cells) would express the toxin and would be killed by the adenovirus. In contrast, the cells in which imprinting was maintained (normal cells) would not express the toxin and would survive. Thus, we would use the availability of the imprinting machinery to create a toxin-based therapy directed only to abnormal, LOI cells.
We selected the toxin gene diphtheria A (DT-A) as it has suitable properties for achieving the efficacious killing of cancer cells.21,22 DT-A is the component of diphtherias toxin that inhibits protein synthesis in susceptible cells. It binds directly to NAD+ and catalyses the transfer of ADP ribose from NAD+ to elongation factor 2 and irreversibly inhibits it.23 The toxic gene DT-A has previously been considered to be suitable for use in cancer gene therapy.24–26 Therefore, we predict that using a combination of therapeutic expression constructs driven by enhancer -DMD (four CTCF binding sites) H19 promoter complex will allow us to selectively kill cancer cells, which are the only cells to demonstrate LOI.
Results
EGFP protein expression in normal and cancer cell lines.
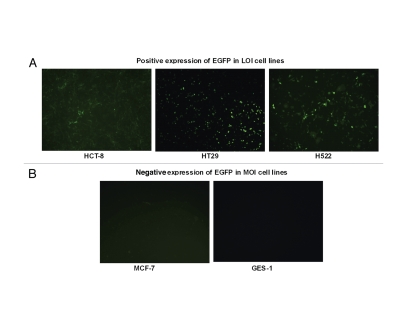
Initially, we utilized an EGFP reporter system to test the applicability of the expression system (Fig. 1). After infection with AdDC312-EGFP (10 PFU/cell) EFGP protein expression was observed in the LOI cells lines HCT-8, HT-29 and H-522. However, in the normal cell line (GES-1) and in mammary carcinoma cell line MCF-7 that maintains normal IGF2 imprinting, negative or only weakly positive expression of EGFP protein occurred (Fig. 2). Thus, the viral gene therapy system only expressed the reporter gene in cells in which IGF2 imprinting was abnormally maintained.
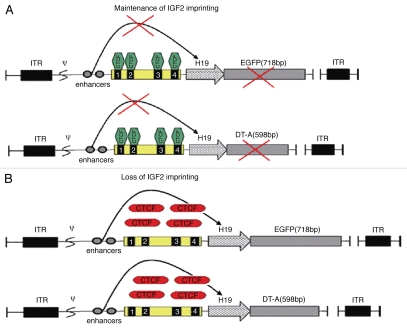
Figure 1.
Schematic diagrams of adenoviral construction. The enhancer, DMD (four CTCF binding sites) and H19 promoter were used to control the expression of the DT-A and EGFP genes, generating AdDC312-DT-A and AdDC312-EGFP, respectively. ITR, inverted terminal repeats; Ψ, adenovirus type 5 packaging signal.
Figure 2.
Cell-specific expression of the EGFP in five cell lines infected with recombinant adenoviral vectors. (A) Positive expression of EGFP in LOI cell lines. Infection with AdDC312-EGFP (10 PFU/cell) induced expression of EGFP in HCT-8, HT-29 and H-522 cells. (B) Negative expression of EGFP in MOI cell lines. Microscope images show that there is no EGFP expression in GES-1cells and only weak expression of EGFP in MCF-7 cells infected with AdDC312-EGFP (10 PFU/cell).
DT-A mRNA transcript and protein expression.
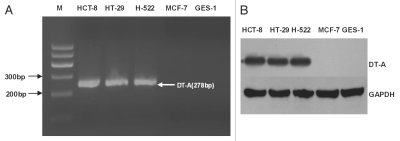
To investigate the expression of DT-A mRNA, normal and cancer cells were infected with AdDC312-DT-A (10 PFU/cell). The expression of DT-A mRNA was determined by RT-PCR 24 hours after infection, and DT-A protein expression was determined by Western blot 48 hours after infection. As shown in Figure 3, DT-A mRNA and DT-A protein were expressed in the LOI cell lines (HCT-8, HT-29 and H-522) but not in the MOI cell lines (GES-1 and MCF-7).
Figure 3.
The expression of DT-A mRNA and protein from the adenoviral vectors carrying the DT-A gene. (A) RT-PCR analysis of DT-A mRNA expression in five cell lines 24h after infection with AdDC312-DT-A (10 PFU/cell). DT-A mRNA was expressed in HCT-8, HT-29 and H-522 cells, but not in GES-1 cells and MCF-7 cells. (B) Western blot analysis of DT-A protein expression in five cell lines 48h after infection with AdDC312-DT-A (10 PFU/cell). DT-A protein was observed in the LOI cells, abut not in the MOI cells.
Cytotoxic effect in cells by the activation of the diphtheria toxin gene.
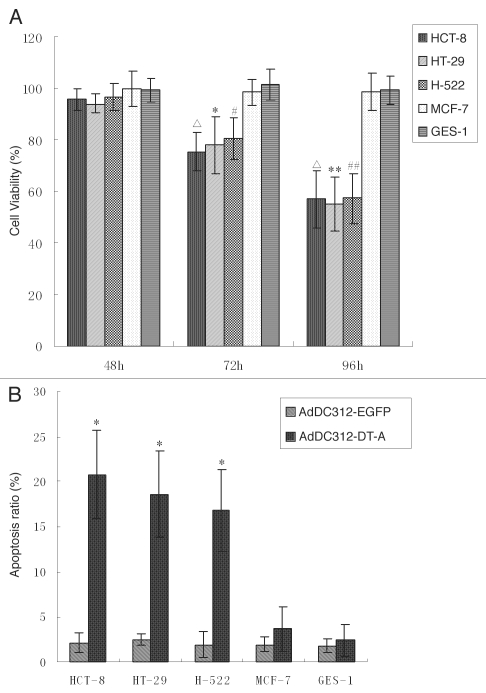
The cytotoxic effects of the adenoviral vectors expressing the DT-A gene was investigated in all cell lines. Cell viability was assessed by MTT assay at 48, 72 and 96 h after infection by recombinant adenoviral vectors (10 PFU/cell). Infection with AdDC312-EGFP provided a negative control to evaluate the cytopathic effect of the adenoviral infection itself. As shown in Figure 4a, the MOI cells lines GES-1 and MCF-7 that were infected with AdDC312-DT-A remained fully viable for at least 96 h after infection. However, the LOI cell lines (HCT-8, HT-29 and H-522) displayed decreased viability after 72 h (p < 0.05).
Figure 4.
The in vitro effect of adenoviral vectors carrying the DT-A gene. (A) Cell viability was determined by MTT assay 48, 72 and 96h after infection with recombinant adenoviral vectors (10 PFU/cell). The cell viability in MOI cell lines (GES-1 and MCF-7) infected with AdDC312-DT-A showed no significant decrease compared with the control group (p > 0.05), but the cell viability in LOI cell lines (HCT-8, HT-29 and H-522) infected with AdDC312-DT-A 72 and 96h showed a significant decrease compared with the control group (Δp < 0.01; *p < 0.05, **p < 0.01; #p < 0.05, ##p < 0.01). Graphs are representative of three separate experiments. (B) Apoptosis was investigated using flow cytometry analysis 72h after infection with AdDC312-DT-A or AdDC312-EGFP (10 PFU/cell). The apoptotic ratio in MOI cell lines (GES-1 and MCF-7) infected with AdDC312-DT-A showed no significant increase compared with the control group (p > 0.05), but the apoptotic ratio in LOI cell lines (HCT-8, HT-29 and H-522) infected with AdDC312-DT-A showed a significant increase compared with the control group (*p < 0.01). Graphs are representative of three separate experiments.
Cell apoptosis induced by the DT-A.
We analysed apoptosis in all cell lines using flow cytometry 72 h after infection. Infection with AdDC312-EFGP served as negative controls to enable us to evaluate the cytopathic effect of the adenoviral infection itself. As shown in Figure 4B, the percentages of cells undergoing apoptosis in the LOI cell lines HCT-8, HT-29 and H-522 were approximately 20, 18 and 16% respectively and all were higher than the apoptosis observed in the control group (p < 0.01) which was only 2%. However, the percentage of apoptosis in MOI cell lines MCF-7 and GES-1 cells was not increased as compared with the control group (p > 0.05).
In vivo antitumor effect of recombinant adenoviral vectors.
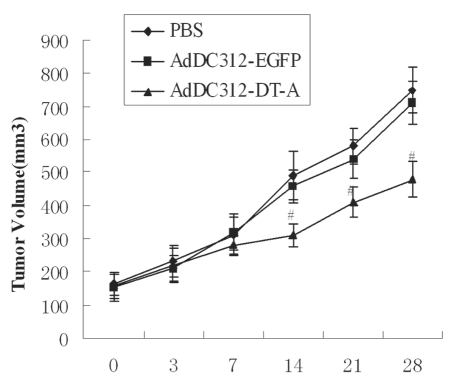
We examined the antitumor effect of this adenoviral construct by injecting the indicated adenoviruses (a total dosage of 109 PFU/mouse) into nude mice which were carrying HCT-8 tumours. We measured tumour volume after injecting AdDC312-DT-A or AdDC312-EGFP or PBS (n = 5 per group). Tumour volume was measured twice weekly for 28 d. The mice displayed no serious adverse side effects during the 28 d as assessed by weight loss, ruffling of fur, life span, behaviour and feeding time. Tumour growth was observed to be similar and consistent in all 3 groups initially. However, after 14 d of observation, tumour growth was suppressed in the AdDC312-DT-A group. This group displayed tumour inhibition rates of approximately 40% compared with only 5% in the AdDC312-EGFP treated group (p <0.01). No significant antitumor effect was observed in the AdDC312-EGFP treated group compared to the PBS-control group (p > 0.05) (Fig. 5).
Figure 5.
In vivo effects of adenoviral vectors carrying the DT-A gene. Nude mice were given subcutaneous injections of HCT-8 cells to establish tumor xenografts. After intratumoral injections with recombinant adenoviral vectors (a total dosage of 109 PFU/mouse), the tumor volume was observed for 28 d. The antitumor effect was marked in the AdDC312-DT-A groups (#p < 0.01 for AdDC312-DT-A versus control group after 14 d).
Immunohistology by TUNEL assay.
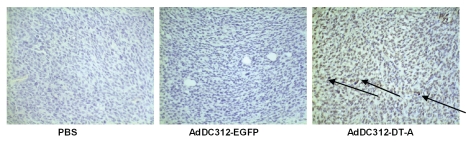
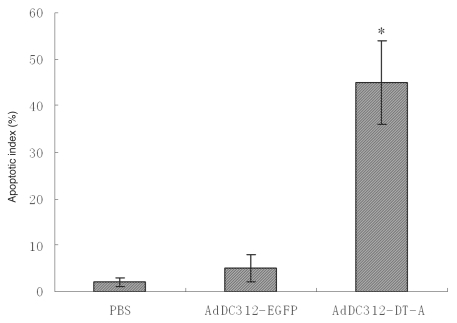
Apoptosis in the nude mouse model was confirmed after treatment with AdDC312-DT-A, AdDC312-EGFP or PBS by TUNEL assay. As shown in Figure 6, there was a significant increase in apoptotic bodies in the AdDC312-DT-A treated mice tumors compared to the AdDC312-EGFP and PBS groups. The apoptosis index was measured as the percentage of TUNEL-positive cells. The apoptotic indexes of PBS, AdDC312-EGFP and AdDC312-DT-A were 2±1, 5±3 and 45±9 respectively p < 0.01) (Fig. 7).
Figure 6.
Immunohistochemical staining for tumor apoptosis (TUNEL) after treatment with recombinant adenoviral vectors. Arrows indicate TUNEL-positive cells. Representative pictures are at ×400 magnification.
Figure 7.
Quantification of apoptotic index after therapy with recombinant adenoviral vectors. The number of TUNEL-positive cells counted from the randomly selected fields of each tumor section (*p < 0.01 compared to the PBS control).
Discussion
IGF2 imprinting is lost in a variety of human neoplasms and this results in a biallelic IGF2 expression.27,28 and, theoretically, an abnormally high level of mitogenic IGF-II protein. IGF2 LOI has been seen in more than 20 different malignancies, including Wilms' tumour, colorectal, lung, breast and prostrate cancers.29
The current model of the regulation of IGF2 imprinting involves the binding of CTCF to the unmethylated maternal ICR/DMD. This protects it from de novo methylation and prevents downstream enhancers from activating IGF2. However, CTCF is unable to bind the methylated paternal ICR/DMD and this results in the expression of IGF2. In this way, the ICR/DMD acts as a CTCF-dependant insulator/enhancer blocker yet the mechanism of insulation remains incomplete. Chromosome conformation capture (3C) experiments in mice in humans, which assay for physical interactions between chromosomal regions, have suggested that the chromosomal looping is involved in the imprinting mechanism.19 However, the precise nature and function of the looping remains a subject for debate.30–32 While there is some consensus that the enhancers physically interact with the IGF2 promoters on the paternal chromosome, interactions on the maternal chromosome remain unclear.33 Kurukuti et al report that maternal-specific silencing of IGF2 occurs when the ICR/DMD interacts with a matrix attachment region and a differently methylated region at the IGF2 locus. This interaction generates a tight loop around the IGF2 gene which then physically impedes IGF2 expression.34 In contrast Yoon et al demonstrated that the ICR/DMD forms a transcriptionally unproductive association with the enhancers and the inactive IGF2 promoters on the maternal chromosome which leads to silencing of the IGF2.35
Previous studies in our lab20 have shown that IGF2 aberrant epigenotype can be corrected by transferring nuclei from human tumour cells (LOI of IGF2) into enucleated mouse and human fibroblasts (MOI of IGF2). These results demonstrated that an abnormal tumour epigenotype could be corrected using in vitro reprogramming. While we do not completely understand the mechanisms for initiating and maintaining imprinting or LOI, we hypothesized that if we place a construct similar to the IGF2/H19 imprinting control region into cells with intact imprinting machinery, that the construct would not be expressed (it would be “imprinted”). On the other hand, we predicted that if the construct were placed in LOI cells, the construct would be expressed. Our prediction was borne out in these experiments, although we do not know the mechanism. We might speculate that the CTCF binding sites were methylated in the LOI cells, preventing CTCF from binding and insulating the toxin gene from the enhancer.
Under the control of a cell-type or tumour-specific promoter, the DT-A gene has previously been tested for use in cancer treatment.24,36,37 This bacterial protein is highly toxic when introduced into the cytoplasm of eukaryotic cells. It inhibits protein synthesis by catalysing ADP ribosylation of the diphthamide group of cellular elongation Factor 2 and destroys cells through an apoptotic pathway.38 The toxicity of DT-A in mammalian cells necessitates the use of an expression system with tumour selectivity in order to control its expression and avoids unintended deleterious effects on non-target cells. In this study we examined the effectiveness of the enhancer DMD-H19 promoter driven DT-A expression for destroying tumour cells based on loss of IGF2 imprinting. We constructed recombinant adenoviral vectors to express DT-A protein only in the cells which demonstrated LOI.
Initially the usefulness of our expression system was identified using EGFP reporter assays. The results showed that green fluorescence was observed only in the LOI cells. The cytotoxic effects of the adenoviral vectors were also examined in normal and MOI cancer cells to assess whether the diphtheria toxin expression driven by the enhancer DMD-H19 promoter is cell-type specific. There was no detectable cytotoxicity in GES-1 and MCF-7 cells which were co-infected with 10 PFU/cell of AdDC312-DT-A but there was substantial cytotoxicity in the LOI cell lines. Therefore, we observed imprinting-dependant induction of apoptosis in tumour cells infected with the DT-A expressed adenovirus.
Our study also utilised the HCT-8 cell line to establish a xenograft animal model. As DT-A was unable to penetrate the cell membrane without the assistance of the B chain of DT we injected the virus in several areas within the tumour. The results showed a significant tumour growth inhibition in AdDC312-DT-A treated groups in comparison with the control group. The apoptosis in the HCT-8 animal model were also confirmed using an immunohistochemicalstaining method. The results showed that AdDC312-DT-A increased the apoptotic index and suggests that AdDC312-DT-A resulted in an increased apoptosis of tumour cells. This led to tumour growth inhibition, indicating that the DT-A expression vector may serve as promising candidate for cancer therapy in humans.
In conclusion, an IGF2 imprinting-based toxin gene therapy developed in this study is effective in inhibiting the growth of human cancer cells in vitro and in vivo. This study is, to the best of our knowledge, the first demonstration of the use of this system for cancer gene therapy both in vitro and in vivo. Although our study only involved a small number of types of cancer, we believe that the unique attributes of the vectors are equally relevant for the treatment of other tumours which have loss of IGF2 imprinting.
Material and Methods
Cell lines and cell culture.
Human colon cancer cells (HCT-8 and HT-29), human lung cancer cells (H-522), human breast cancer cells (MCF-7) and human gastric epithelial cells (GES-1) were obtained from the American Type Culture Collection (Manassas, VA). All of the cell lines except H-522 and HT-29 were maintained in DMEM (Hyclone, UT), and supplemented with 10% fetal bovine serum (Hyclone, UT). H-522 cells were maintained in RPMI 1640 (Invitrogen, CA) and 10% FBS.39 Cells were incubated in a 100% humidified incubator at 37°C with 5% CO2.
Plasmid construction and its incorporation into adenoviral vectors.
The original adenoviral shuttle plasmid that we used in this study was pDC312. The mouse H19 enhancer exon 1 (258bp) and the mouse H19 enhancer exon 2 (360bp) were amplified by PCR from mouse genomic DNA and then linked to a single fragment by PCR. The enhancer was cloned into the plasmid pDC312 using restriction endonuclease Xbal and Sal I. Subsequently, the mouse DMD exon 1–2 (429bp), DMD exon 3 (207bp) and DMD exon 4 (156bp) were also amplified by PCR from genomic mouse DNA and then linked to a single fragment using PCR. The DMD was cloned into the downstream of the enhancer using restriction endonuclease Sal I and Hind III. The mouse H19 promoter (302bp) was then amplified by PCR from mouse genomic DAN using the upper primer 5′ GCAAGCTTCCACCGTTCTATGAAGGGCTTC 3′(5′ primer for mouse H19 with Hind III) and lower primer 5′ AAGAATTCTCATCAGCGCCCATCTCTAGCC 3′ (5′ primer for mouse H19 with EcoR I) cloned into the downstream of the DMD using restriction endonuclease Hind III and EcoR I. The diphtheria toxin A (DT-A) sequence (598bp) was then amplified by PCR from diphtheria bacillus genomic DNA using the upper primer 5′ TAGGAATTCCTAGAGATGGGCGCTGATGAT 3′ (5′ primer for DT-A with EcoR I) and lower primer 5′ CGGGATCCTCATCGCCTGACACGATTTCCTGCACAG 3′ (5′ primer for DT-A with BamH I).
The enhanced green fluorescent protein (EGFP) reporter gene from pEGFP-C1 vector (Clonetech, Mountain View, CA) and the DT-A gene were then inserted into the downstream of the H19 promoter using restriction endonuclease EcoR I and BamH I to construct pDC312-enhancer-DMD-H19-EGFP and pDC312-enhancer-DMD-H19-DT-A. The product that resulted from this insertion was confirmed by DNA sequencing.
The plasmids pDC312-enhancer-DMD-H19-EGFP and pDC312-enhancer-DMD-H19-DT-A were then transfected into the HEK293 Cells using the PolyFect transfection reagent (QIAGEN Inc., Valencia, CA) together with adenoviral vector Ad5. After a homologous recombination in HEK293 cells we obtained two sets of the packaged adenoviruses: AdDC312-EGFP and AdDC312-DT-A (Fig. 1).
Virus infection.
In this study the cells were seeded in a total of 96 well plates at a density of 10,000 per well for MTT assay and in 6 well plates with a density of 1000,000 per well for RT-PCR and flow cytometry analysis. The cells were then incubated with a series of amounts of AdDC312-EGFP and AdDC312-DT-A in serum-free DMEM at 37°C for 1 h. After the incubation period, serum-free DMEM containing the viruses was replaced by a normal growth medium. The infected cells were further cultured at 37°C until they were used for further assays.
EGFP and DT-A expression analysis in the constructed plasmids.
EGFP expression was examined 48 h after infection with adenoviral vectors (10 plaque forming units/cell) using an Olympus™ microscope (Axioscope, Carl Zeiss Inc., Oberkochen, Germany) with a fluorescent filter set (excitation 450–490 nm). The DT-A mRNA expression was determined by PT-PCR. Complete RNA was extracted from each sample using Trizol (Invitrogen Life Technologies, CA) according to the manufacturer's instructions. First strand cDNA synthesis was performed in a total volume of 25 µ using 2 µg of each RNA sample which was primed with 0.5 µg of primer and 200 units M-MLV reverse transcriptase added. The cDNA was subsequently amplified in 50µl of reaction volume containing 0.4 µmol/L of sense primer (5′ CGTACCACGGGACTAAACCTG 3′) and anti-sense primer (5′ TAGTTTCGGCATTATCCACTT 3′) as well as 1.25 U Taq DNA polymerase (TaKaRa, Tianjin, China). The amplification conditions were pre-denaturation at 94°C for 5 min followed by 35 cycles of 94°C for 40 s, 60°C for 40 s and 72°C for 60 s with a final extension of 72C for 7 min. The PCR products were 278 bp and were then electrophoresed on a 2% ethidium stained agarose gel before being visualised using UV.
The DT-A protein expression was evaluated using a western blot method. Cells were harvested and lysed during three cycles of freeze/thaw at −80°C. Ttotal protein was separated by SDS-PAGE in 12% gels and then transferred onto a polyvinyllidene difluoride membrane. The immunoblotting was performed as described by Adler et al.40 The horseradish peroxidase (HPR)-conjugated goat anti-rabbit IgG antibodies were used against the primary antibody (mouse anti-diphtheria toxin A subunit monoclonal antibody) (American Research Products, MA). These proteins were visualised using Lumi-Light Western Blotting Substrate (Roche Molecular Biochemicals).
Cytotoxic effect of the DT-A on normal and cancer cell lines.
Cytotoxicity was assessed using MTT assay. This assay was based on the ability of the viable cells to reduce MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) (Sigma Aldrich, St Louis, MO) to insoluble coloured formazan crystals. The cells were seeded in 96 well plates at a density of 10,000 1 d before virus infection. The cells were then infected with recombinant adenoviral vectors (10 PFU/cell). Fresh medium was added 5 h after infection and then every 2 d afterwards. At the indicated points in time 20 µl of MTT (5 mg/ml in PBS sterilized by filtration) were added to each cell. After 4 hours of incubation at 37°C the medium was removed and 200 µl of DMSO was added into each well to dissolve the crystals. The absorbance was measured using a micro plate reader at 550 nm (Bio-Rad, Richmond, CA).
Quantative evaluation of apoptosis by flow cytometry.
Quantative evaluation of apoptosis was performed using flow cytometry after double staining with an annexin V fluorescein isothyocyanate (FITC) apoptosis detection kit. This allowed discrimination amongst early apoptotic (single annexin V positive) and necrotic cells (double annexin V propidium iodide (PI) positive). This enabled us to differentiate the cells that had lost their membrane integrity i.e. necrotic cells from living cells by means of red staining of their nuclei with PI. Cells (100,000 cells per well) were cultured in six well dishes and then AdDC312-DT-A was infected at 10PFU/cell. Cell apoptosis was analysed 72 h after infection with AdDC312-EFGP (10 PFU/cell) serving as a negative control.
Treatment of tumor-bearing nude mice with the adenoviral vectors.
HCT-8 cells were trypsinized to a single cell suspension and then resuspended in 109 cells/100 µl PBS. They were then subcutaneously injected into the flank areas of adult (8 w old) athymic male nude mice (approved by The Experimental Animal Center of University of Yangzhou, Yangzhou, China). When the tumours reached approximately 100 mm, the mice were randomly divided into 3 treatment groups of 5 mice each. A dosage of 109 PFU/mouse of AdDC312-DT-A was then injected into the growing tumours from 3 different directions on 3 successive days. The tumours which were injected with AdDC312-EGFP at the same dosage served as a viral vector control and those injected with PBS served as a negative control. The volume of the tumors were then observed for 28 d and their dimensions measured and volume calculated using the formula (width) 2 × length × 0.5 length. The mice were then euthanized by cervical dislocation at a predetermined interval of observation. The tumours were dissected and collected for histological analysis. The tumours were stored in 30% formalin, embedded in OCT compound and then cut in 5–7 mm using cryocut microtone (Leica, Germany).
TUNEL assay.
Apoptosis in the tumour cells was detected using terminal deoxynucleotide transferase (TdT)-mediated dUTP nick-end labelling (TUNEL) assay. It was performed using the In Situ Cell Death Detection Kit (Roche, Mannheim, Germany) following the manufacturer's guidance. Sections were fixed in 30% formalin before being rinsed. Next, the sections were incubated in 3% H2O2 and then permeabilized with 0.5% Triton X-100. Finally, they were rinsed again and incubated in the Tunel reaction mixture. The sections were rinsed and visualized using Converter-POD with diamonobenzidine (DAB). Harris' hematoxylin was used for counter-staining. The slides were air-dried at room temperature and Permount cover slips were used. The number of TUNEL positive cells was counted in six fields at 400 magnification of a light microscope, selected at random, before the apoptosis index for each field was calculated and expressed as a percentage of TUNEL-positive cells relative to the total.
Statistical analysis.
Experimental data were presented as the mean ± standard deviation and assessed using a Student's t-test and one-way ANOVA at a significance level of p < 0.05.
Acknowledgements
This project was supported by a grant from The National Nature Science Foundation of China (30223087), a grant from The Personnel Ministry of China (2008-86) to S.K.W, a NIH grant (1R43 CA103553-01) and The Department of Defense Grant (W81XWH-04-1-0597) to J.F.H, and the research sources of the Department of Veterans Affairs.
Abbreviations
- IGF2
insulin-like growth factor 2 gene
- DMD
differentially methylated domains
- LOI
loss of imprinting
- MOI
maintenance of imprinting
- DT-A
diphtheria toxin A
- PFU
plaque forming units
- MTT
3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide
- ICR
imprinting control region
- Ad
adenovirus
- EGFP
enhanced green fluorescent protein
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/12442
References
- 1.Verona RI, Mann MR, Bartolomei MS. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- 2.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 3.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 4.DeChiara TM, Efstratiadis A, Robertson EJ. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature. 1990;345:78–80. doi: 10.1038/345078a0. [DOI] [PubMed] [Google Scholar]
- 5.Reik W, Constancia M, Dean W, Davies K, Bowden L, Murrell A, et al. Igf2 imprinting in development and disease. Int J Dev Biol. 2000;44:145–150. [PubMed] [Google Scholar]
- 6.Arney KL. H19 and Igf2—enhancing the confusion? Trends Genet. 2003;19:17–23. doi: 10.1016/s0168-9525(02)00004-5. [DOI] [PubMed] [Google Scholar]
- 7.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 9.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 10.Ulaner GA, Vu TH, Li T, Hu JF, Yao XM, Yang Y, et al. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal changes in methylation of a CTCF-binding site. Hum Mol Genet. 2003;12:535–549. doi: 10.1093/hmg/ddg034. [DOI] [PubMed] [Google Scholar]
- 11.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 12.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 13.Filippova GN, Lindblom A, Meincke LJ, Klenova EM, Neiman PE, Collins SJ, et al. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- 14.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 15.Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 16.Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 17.Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 18.Qiu XW, Vu TH, Lu QC, Ling JQ, Li T, Hou A, et al. A complex deoxyribonucleic acid looping configuration associated with the silencing of the maternal Igf2 allele. Mol Endocrinol. 2008;22:1476–1488. doi: 10.1210/me.2007-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Hu JF, Qiu XW, Ling JQ, Chen H, Wang S, et al. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex-2 intra-chromosomal loop. Mol Cell Biol. 2008;22:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen HL, Li T, Qiu XW, Wu J, Ling JQ, Sun ZH, et al. Correction of aberrant imprinting of IGF2 in human tumors by nuclear transfer-induced epigenetic reprogramming. EMBO J. 2006;25:5329–5338. doi: 10.1038/sj.emboj.7601399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pastan I, Chaudhary V, FitzGerald DJ. Recombinant toxins as novel therapeutic agents. Annu Rev Biochem. 1992;61:331–354. doi: 10.1146/annurev.bi.61.070192.001555. [DOI] [PubMed] [Google Scholar]
- 22.Breitman ML, Clapoff S, Rossant J, Tsui LC, Glode LM, Maxwell IH, et al. Genetic ablation: targeted expression of a toxin gene causes microphthalmia in transgenic mice. Science. 1987;238:1563–1565. doi: 10.1126/science.3685993. [DOI] [PubMed] [Google Scholar]
- 23.Collier RJ. Diphtheria toxin: mode of action and structure. Bacteriol Rev. 1975;39:54–85. doi: 10.1128/br.39.1.54-85.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, McCadden J, Ferrer F, Kruszewski M, Carducci M, Simons J, et al. Prostate-specific expression of the diphtheria toxin A chain (DT-A): studies of inducibility and specificity of expression of prostate-specific antigen promoter-driven DT-A adenoviral-mediated gene transfer. Cancer Res. 2002;62:2576–2582. [PubMed] [Google Scholar]
- 25.Kunitomi M, Takayama E, Suzuki S, Yasuda T, Tsutsui K, Nagaike K, et al. Selective inhibition of hepatoma cells using diphtheria toxin A under the control of the promoter/enhancer region of the human alpha-fetoprotein gene. Jpn J Cancer Res. 2000;91:343–350. doi: 10.1111/j.1349-7006.2000.tb00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall PD, Willingham MC, Kreitman RJ, Frankel AE. DT388-GM-CSF, a novel fusion toxin consisting of a truncated diphtheria toxin fused to human granulocyte-macrophage colony-stimulating factor, prolongs host survival in a SCID mouse model of acute myeloid leukemia. Leukemia. 1999;13:629–633. doi: 10.1038/sj.leu.2401357. [DOI] [PubMed] [Google Scholar]
- 27.Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 28.Feinberg AP. Genomic imprinting and gene activation in cancer. Nat Genet. 1993;4:110–113. doi: 10.1038/ng0693-110. [DOI] [PubMed] [Google Scholar]
- 29.Kaneda A, Feinberg AP. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65:11236–11240. doi: 10.1158/0008-5472.CAN-05-2959. [DOI] [PubMed] [Google Scholar]
- 30.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 31.Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forné T, et al. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum Mol Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- 32.Engel N, Raval AK, Thorvaldsen JL, Bartolomei SM. Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking. Hum Mol Genet. 2008;17:3021–3029. doi: 10.1093/hmg/ddn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng W, Verbitsky A, Bao Y, Sawicki J. Regulated expression of diphtheria toxin in prostate cancer cells. Mol Ther. 2002;6:537–545. doi: 10.1006/mthe.2002.0694. [DOI] [PubMed] [Google Scholar]
- 37.Massuda ES, Dunphy EJ, Redman RA, Schreiber JJ, Nauta LE, Barr FG, et al. Regulated expression of the diphtheria toxin A chain by a tumor-specific chimeric transcription factor results in selective toxicity for alveolar rhabdomyosarcoma cells. Proc Natl Acad Sci USA. 1997;94:14701–14706. doi: 10.1073/pnas.94.26.14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michl P, Gress TM. Bacteria and bacterial toxins as therapeutic agents for solid tumors. Curr Cancer Drug Targets. 2004;4:689–702. doi: 10.2174/1568009043332727. [DOI] [PubMed] [Google Scholar]
- 39.Akagi Y, Liu W, Zebrowski B, Xie K, Ellis LM. Regulation of vascular endothelial growth factor expression in human colon cancer by insulin-like growth factor-I. Cancer Res. 1998;58:4008–4014. [PubMed] [Google Scholar]
- 40.Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD, et al. Regulation of JNK signaling by GSTp. EMBO J. 1999;18:1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]