Abstract
The eukaryotic RNA splicing machinery is dedicated to the daunting task of excising intronic sequences on the many nascent RNA transcripts in a cell, and in doing so facilitates proper translation of its transcriptome. Notably, emerging evidence suggests that RNA splicing may also play direct roles in maintaining genome stability. Here we report the identification of the RNA/DNA-binding protein SON as a component of spliceosome that plays pleiotropic roles during mitotic progression. We found that SON is essential for cell proliferation, and that its inactivation triggers a MAD2-dependent mitotic delay. Moreover, SON deficiency is accompanied by defective chromosome congression, compromised chromosome segregation and cytokinesis, which in turn contributes to cellular aneuploidy and cell death. In summary, our study uncovers a specific link between SON and mitosis, and highlights the potential of RNA processing as additional regulatory mechanisms that govern cell proliferation and division.
Key words: SON, splicing factor, mitosis, MAD2
Introduction
Processing of precursor messenger RNAs (mRNAs) into mature mRNAs involves the concerted action of a complex repertoire of splicing factors and their interaction with RNA cis-elements. Through the active and selective removal of non-coding intervening sequences, mature RNAs are translated, and the sum of which contributes to the expressed proteome. Given the fundamental importance and critical link between RNA splicing and protein expression in higher eukaryotes, one can envision that failure to properly process pre-mRNAs will give rise to aberrant protein products and expression, and will compromise cellular functions that are instrumental for cell proliferation.1 Indeed, inactivation of various splicing factors results in developmental defects and lethality across different species, underscoring RNA splicing as a conserved and indispensible process essential for cell viability.2
Whereas the core splicing machinery is attributed to constitutive processing of pre-mRNAs, mounting evidence implicate specific splicing factors in cell cycle progression and apoptosis.3 Specifically, deficiency in the prototypic SR protein splicing factor ASF/SF2 results in G2 cell cycle arrest and apoptosis,4,5 presumably through the accumulation of DNA double-strand breaks. Similarly, inactivation of spliceosome factors SC-35 and PLRG1 coincided with elevated DNA damage and genome instability.6,7 Together, these observations lend credence to the idea that splicing factors may, directly or indirectly, modulate cell growth and survival. In line with this notion, patient mutations that render splice site recognition faulty give rise to hypomorphic tumor suppressor protein products, including cell cycle control and DNA repair proteins.8,9 With the vast number of splicing regulators that exhibit sequence-specific binding in a eukaryotic cell, it remains to be tested how each of them may have evolved to modulate distinct cellular processes in vivo.
Proper chromosome segregation involves the dynamic coordination of a host of factors that control and closely monitor each of the steps during mitosis. The demand for time-efficient separation of the genetic content renders mitosis most vulnerable to genome instability. In this regard, the Spindle Assembly Checkpoint (SAC) plays a pivotal role as a safeguard mechanism that halts sister chromatid separation and anaphase onset until all chromosomes are bi-oriented on the microtubule spindle.10 Weakened mitotic checkpoint signaling leads to cellular aneuploidy and tumorigenesis and even cell death.11 Although a plethora of factors, amongst which include numerous splicing factors, have been short-listed for roles in promoting orderly mitotic progression,12–15 it remains a challenging task to functionally dissect how each of them contributes to a successful cell division event.
Results
SON associates with spliceosomes.
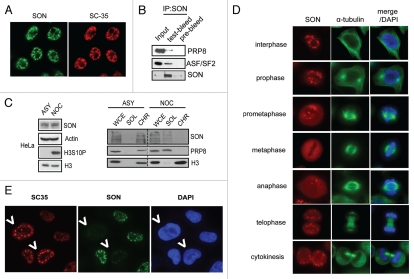
With the aim to uncover mechanistic links between RNA splicing and mitosis, we performed literature search and identified the RNA/DNA-binding domain containing protein SON as a candidate mitotic regulator. Consistent with previous reports,16,17 cytological analysis of SON proteins revealed distinct focal structures that overlap with the spliceosome marker SC-35 (Fig. 1A). Analysis of SON-associated proteins by affinity purification and mass spectrometry revealed the identity of a number of splicing factors (Suppl. Table 1), including PRP8 and ASF/SF2. Their in vivo interactions were confirmed by immunoprecipitating endogenous SON protein-complexes followed by western blot analysis (Fig. 1B). Since nuclear speckles enriched in splicing factors disassemble during mitosis,18 to strengthen the notion of a physical association of SON with spliceosome components, we performed biochemical fractionation experiments using lysates derived from asynchronised or nocodazole-treated HeLa cells (Fig. 1C). Immunoblotting results suggest that SON, similar to splicing factor PRP8, normally associates with chromatin in interphase cells but is released during mitosis (Fig. 1C). Similarly, indirect immunofluorescence studies indicate that SON is concentrated in nuclear speckles during interphase and becomes dispersed at metaphase (Fig. 1D). Finally, using SC-35 as a surrogate marker for spliceosome functions, we found that depletion of SON results in discernible alterations to SC-35 immunostaining (Fig. 1E), which resemble those observed upon drug-induced pre-mRNA splicing inhibition.19,20 Taken together, these results implicate SON as a potential spliceosome component which may be required for efficient processing of pre-mRNAs.
Figure 1.
SON is a component of spliceosomes. (A) SON colocalises with spliceosome marker SC-35. HeLa cells grown on coverslips were fixed, permeabilised and immunostained with indicated antibodes. (B) SON physically interacts with spliceosome components. Nuclease-treated HeLa cell extract was subjected to co-immunoprecipitation using Protein A agarose beads conjugated with anti-SON antibodies or pre-bleed control. Immunoprecipitates were washed with NETN buffer twice, eluted by boiling and separated by SDS-PAGE. Western blotting was performed using indicated antibodies. (C) SON associates with chromatin during interphase and is released during mitosis. HeLa cells were either untreated (ASY) or arrested in mitosis (NOC) by incubating with 100 ng/ml nocodazole for 16 hr. Cell lysates were biochemically fractionated according to Methods. Western blot analysis of SON expression in whole cell extract (WCE), soluble (SOL) and chromatin-enriched fractions (CHR). (D) SON concentrates in nuclear speckles and disperses during metaphase. Indirect immunofluorescence staining showing SON localization at different cell cycle phases. (E) SON deficiency compromises spliceosome function. HeLa cells treated with control or SON-specific siRNAs were mixed, seeded onto coverslip and processed 24 hr post-transfection. Indirect immunostaining was performed as described in Methods using indicated antibodies. SON-depleted cells are marked by arrows.
SON depletion activates the spindle assembly checkpoint.
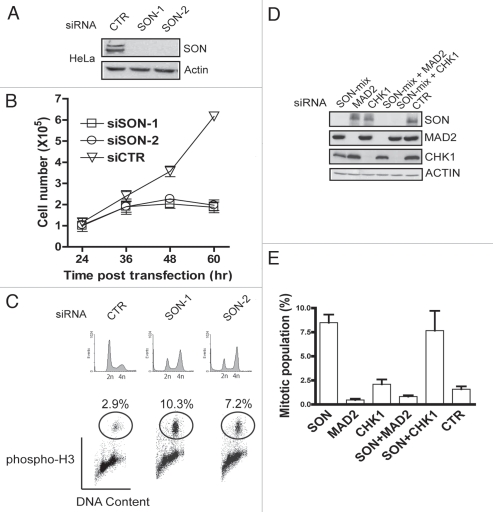
Recent evidence implicated splicing factors in maintaining genome stability,5,6,21 and that deficiencies in various splicing factors trigger cell cycle arrest and apoptosis.4,6,22 When we depleted HeLa cells of endogenous SON expression using two independent siRNAs (Fig. 2A), we observed prominent decrease in cell proliferation as compared to control (CTR) siRNA-treated cells (Fig. 2B; reviewed in ref. 23). Moreover, downregulation of SON was accompanied by dramatic morphological changes, including cell rounding (data not shown). To confirm that these rounded cells represent mitotic cells, we performed flow cytometry analysis using phospho-H3 as a mitotic indicator. Indeed, pre-treatment of both SON-specific siRNAs resulted in substantially elevated percentage of mitotic cell populations (Fig. 2C), suggesting that SON is required for mitotic progression. The spindle-assembly checkpoint (SAC) plays a pivotal role in maintaining genome stability, and represents an important safeguard via which cells ensure proper chromosome segregation.10 Failure to properly activate the spindle-assembly checkpoint is often associated with cellular aneuploidy and cell death.11 Given that SON deficiencies elevated the percentage of mitotic cell populations, we tested whether the SAC may be responsible for this phenotype by inactivating MAD2, an essential component of the mitotic checkpoint. In cells co-depleted of SON and MAD2 (Fig. 2D), the SON-associated increase in mitotic cell fraction was reversed (Fig. 2E), suggesting that SON deficiency activates the MAD2-dependent SAC. By contrast, co-depletion of SON and CHK1 did not alter the elevated percentage of mitotic cells as compared to SON depletion alone, indicating that the G2/M checkpoint kinase CHK1 is not involved in this process.
Figure 2.
Activation of spindle assembly checkpoint in SON-depleted cells. (A) Western blot analysis of lysates derived from HeLa cells pre-treated with two independent SON-specific (SON-1 and SON-2) or control (CTR) siRNAs. (B) SON is required for cell proliferation. Immediately after siRNA transfection, cells were seeded onto separate 6 cm2 dishes. At indicated time points cells were harvested and counted using a hemocytometer. (C) SON-depletion results in accumulation of mitotic cells. 36 hr post-transfection, siRNA-treated cells were fixed in 70% ethanol, immunostained with anti-phospho-H3 antibodies and subjected to flow cytometry analysis to evaluate percentage of mitotic cells. (D and E) SON-depletion activates the MAD2-dependent spindle-assembly checkpoint. Western blot showing protein levels in lysates derived from cells pre-treated with indicated siRNAs (D). Percentage of mitotic cell population in siRNA-treated cells (E). Procedures were as described in Methods and in (C). Results represent mean ± S.D from three experiments.
SON inactivation leads to mitotic aberrations.
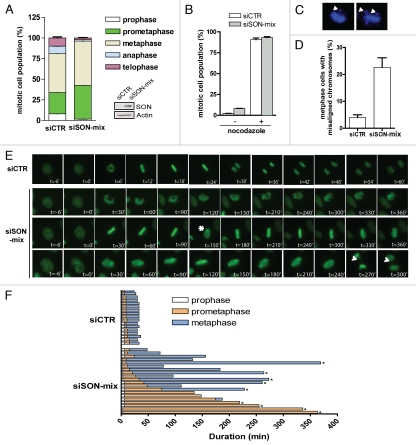
To determine specifically where SON-depleted cells accumulate in mitosis, we first compared the proportion of siRNA-treated cells in each phase of mitosis. Consistent with SON-depletion incurring a MAD2-dependent checkpoint delay, we observed significant increase of cell fractions in prometaphase and metaphase in SON siRNA-transfected cells (Fig. 3A). Since SON siRNA-treated cells were equally sensitive to the microtubule-depolymerising agent nocodazole as control cells (Fig. 3B), we asked whether SON deficiency may compromise chromosome congression, a common defect associated with SAC activation. Quantification of fixed cells revealed significantly higher percentage of SON siRNA-treated cells with mis-aligned chromosomes (Fig. 3C and D). In addition, by monitoring GFP-H2B-expressing HeLa cells using time-lapse microscopy, we found that SON deficiency is associated with frequent chromosome congression failure, which in turn leads to prolonged prometaphase and metaphase arrests (Fig. 3E and F).
Figure 3.
SON promotes mitotic fidelity. (A) Quantification of mitotic cells in different stages of mitosis: prophase, prometaphase, metaphase, anaphase and telophase. Note that the mitotic index is around five times higher in SON-depleted cells. (B) Microtubule depolymerisation arrested SON-depleted cells. siRNA-treated cells were incubated in 20 ng ml−1 nocodazole or DMSO for 16 hr. Thereafter cells were harvested, fixed and processed to evaluate the percentage of mitotic cells. (C and D) Misaligned chromosomes in SON-depleted cells. Representative figures showing misaligned chromosomes in SON-depleted cells (C). Quantification of cells with chromosome congression defects. HeLa cells transfected with indicated siRNAs were processed and metaphase cells were visualized by DAPI staining. Cells with misaligned chromosomes were scored, n = 100 (D). (E and F) SON depletion delays mitotic progression. Representative images of control (siCTR) or SON-specific (siSON-mix) siRNA-treated cells from nuclear envelope breakdown to anaphase onset (E). Live cells pretreated with indicated siRNAs were followed over a 8-hr period and the durations of prophase, prometaphase and metaphase are illustrated. SON-depleted cells often display cycles of chromosome condensation and decondensation (marked by asterisk) and internuclear bridges during anaphase can be observed (marked by arrow). (F). The asterisks are indicative of cells that did not go onto anaphase.
SON silencing results in defective cytokinesis and compromises cell survival.
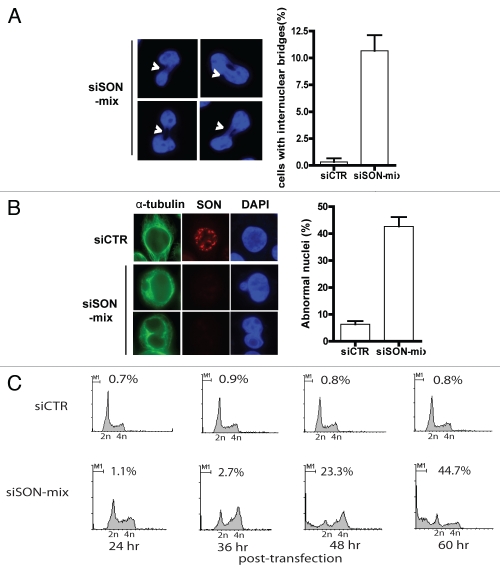
Significantly, although a sub-population of SON-depleted cells eventually progressed into anaphase and telophase, they often do so at the expense of aberrant chromosome segregation. Indeed, internuclear bridges as well as abnormal nuclei can be readily detected in SON-depleted cells (Fig. 4A and B). Furthermore, we found that SON depletion resulted in accumulation of cells with sub-G1 DNA content over time (Fig. 4C), which is a hallmark of apoptotic cell death. Taken together, these observations indicate that SON is essential for proper chromatin dynamics in preparation for mitotic division, and that perturbing SON functions compromises genome integrity, contributes to cellular aneuploidy and reduces cell survival.
Figure 4.
SON deficiency results in defective cytokinesis and induces cell death. (A) SON depletion is accompanied by aberrant chromosome segregation. Representative images showing SON-depleted cells with defective cytokinesis and quantification of the number of internuclear bridges indicative of chromosome missegregation. (B) SON deficiency is associated with abnormal nuclear morphology. Quantification and representative images showing cells with abnormal nuclei in the absence of SON protein. siRNA-treated cells were fixed, permeabilised and immunostained with indicated antibodies. (C) SON depletion results in cell death. Following siRNA transfection, cells were harvested at different time points (24–60 hr post-transfection). Representative cell cycle profiles of control (siCTR) or SON-specific siRNA (siSON-mix)-treated cells. Cells were fixed in 70% ethanol, stained with propidium iodide and subjected to flow cytometry analysis. Percentages of sub-G1 cell populations (M1) are shown.
Discussion
Full-length human SON is a polypeptide of 2, 426 amino acids and harbors domains typical of RNA processing factors.17 Here we propose that SON is a spliceosome component by virtue of its physical interactions with spliceosomal factors (Fig. 1B) and its distinct subcellular localization (Fig. 1A and D). The fact that SON depletion phenocopies spliceosome inhibition,19,20 rendering discernible changes in SC-35 focal accumulation (Fig. 1E), suggests that SON deficiency may hinder processing of pre-mRNAs important for a number of cellular processes. Whether SON modulates or is endowed with RNA splicing activity will require further work. However, since SON deficiency is associated with pleiotropic defects during mitosis, it is tempting to speculate that SON may have evolved to target and regulate a sub-set of mitotic regulators.
Dysregulation of chromosome dynamics during mitosis has profound effects on cell proliferation and survival. Indeed, our data indicated that SON promotes mitotic fidelity, failure of which compromises cell survival (Fig. 4C). In support of a prominent and specific role of SON in mitosis, SON silencing activated the MAD2-dependent SAC and resulted in defective chromosome dynamics. Although SON is clearly essential for orderly mitotic progression, we do not currently know if these mitotic defects are associated with its putative role in regulating spliceosome function, largely because the large size of SON cDNA has precluded us from testing whether its putative splicing activity is required. Future experiments will be needed to address this possibility.
SON protein concentrates in nuclear speckles that perfectly overlap with those of the spliceosome marker SC-35 during interphase (Fig. 1A). Importantly, by following SON proteins at different phases of mitosis, we observed dynamic changes of its sub-cellular localisation (Fig. 1D). Resembling those of SC-35,18 our results clearly showed that SON becomes diffusely localized at metaphase. Thus, given that SON deficiency first arrests cells at prometaphase (Fig. 3A, E and F), we favor the possibility that SON ensures proper chromosome dynamics prior to mitotic entry.
In conclusion, our study identifies SON as an important cellular component required for mitotic progression and cell survival. Interestingly, SON was recently implicated a role in influenza virus replication,24 thus it remains to be tested whether SON, via its spliceosome-associated functions, may regulate diverse cellular processes through specific recognition of a subset of pre-mRNAs. In an era where RNA processing is emerging as an important gene regulatory mechanism, much of the inner-workings of RNA splicing remains obscure, and has hampered advances in understanding the functional specificities of splicing regulators. Thus, the intimate link between RNA splicing, its regulation of cellular functions, and its contributions to human diseases deserve to be systematically investigated.25
Methods and Materials
Antibodies.
Anti-SON polyclonal antibody was raised against GST-SON fusion protein (residues 790–1409) and affinity purified using column coated with MBP-SON fusion protein. Antibodies specifically recognizing PRP8 and ASF/SF2 were obtained from Santa Cruz. Anti-MAD2 and anti-SC-35 antibodies were obtained from BD Biosciences. Anti-actin and anti-alpha-tubulin antibodies were obtained from Sigma. Anti-histone H3, anti-phospho-H3 and anti-CHK1 antibodies were previously described.26
Interaction studies and biochemical fractionation.
Tandem affinity purification using strepatavidin binding peptide-S peptide-tagged SON cDNA fragment (Image clone: 3161999) was performed essentially as described.27 In vivo interactions between SON and various splicing factors were done by co-immunoprecipitating SON complex from benzoase-treated cell lysate using protein-A beads conjugated with anti-SON antibodies. Biochemical fractionation experiments were done as described previously.26
Cell culture, siRNA-mediated gene silencing, cell cycle analysis and immunofluorescent staining.
HeLa cells were cultured in DMEM media supplemented with 10% fetal bovine serum (FBS) and maintained in 5% CO2 at 37°C. SON-targeting siRNAs were purchased from Dharmacon and were transfected using Oligofectamine (Invitrogen) following manufacturer's protocol. Percentage of mitotic populations (phospho-H3 positive cells) were analysed by flow cytometry. Immunofluorescence studies were performed as described.26
Time-lapse microscopy.
H2B-GFP expressing HeLa cells were seeded, transfected with control or SON siRNAs, and were monitored for 8 hours. Cells were placed in a live cell stage-mounted environment chamber and images were captured at 3-minute intervals using an automated microscope (LSM510 Meta Zeiss) equipped with Axiovision (V4.6).
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (CA113381 to J.C.) and Seed Funding for Basic Research (Project Code: 200908159008; HKU to M.S.Y.H.). M.S.Y.H. would like to thank Drs. Chris Klug and Jim Mulloy for valuable reagents, Drs. Zhiwei Chen and Henggui Liu for help with flow cytometry analysis, Mr. Tony Chan, Prof. George Tsao and the Faculty Core Imaging Facility for technical support with time-lapse microscopy, and is grateful to J.C. for his continuous support. J.C. is a recipient of an Era of Hope Scholar award from the Department of Defence and a member of the Mayo Clinic Breast SPORE program (P50 CA116201).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12151
Supplementary Material
References
- 1.Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Manley JL. Regulation of pre-mRNA splicing in metazoa. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 3.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, et al. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleinridders A, Pogoda HM, Irlenbusch S, Smyth N, Koncz C, Hammerschmidt M, et al. PLRG1 is an essential regulator of cell proliferation and apoptosis during vertebrate development and tissue homeostasis. Mol Cell Biol. 2009;29:3173–3185. doi: 10.1128/MCB.01807-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 9.Bartel F, Taubert H, Harris LC. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell. 2002;2:9–15. doi: 10.1016/s1535-6108(02)00091-0. [DOI] [PubMed] [Google Scholar]
- 10.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 11.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draviam VM, Stegmeier F, Nalepa G, Sowa ME, Chen J, Liang A, et al. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat Cell Biol. 2007;9:556–564. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- 13.Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 14.Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- 15.Smith E, Dejsuphong D, Balestrini A, Hampel M, Lenz C, Takeda S, et al. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat Cell Biol. 2009;11:278–285. doi: 10.1038/ncb1835. [DOI] [PubMed] [Google Scholar]
- 16.Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun CT, Lo WY, Wang IH, Lo YH, Shiou SR, Lai CK, et al. Transcription repression of human hepatitis B virus genes by negative regulatory element-binding protein/SON. J Biol Chem. 2001;276:24059–24067. doi: 10.1074/jbc.M101330200. [DOI] [PubMed] [Google Scholar]
- 18.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 19.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of premRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 20.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 21.Montembault E, Dutertre S, Prigent C, Giet R. PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1 and MAD2 localization to the kinetochores. J Cell Biol. 2007;179:601–609. doi: 10.1083/jcb.200703133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacheco TR, Moita LF, Gomes AQ, Hacohen N, Carmo-Fonseca M. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol Biol Cell. 2006;17:4187–4199. doi: 10.1091/mbc.E06-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn EY, Yan M, Malakhova OA, Lo MC, Boyapati A, Ommen HB, et al. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc Natl Acad Sci USA. 2008;105:17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 25.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.