Abstract
The present studies have examined approaches to suppress MCL-1 function in breast cancer cells, as a means to promote tumor cell death. Treatment of breast cancer cells with CDK inhibitors (flavopiridol; roscovitine) enhanced the lethality of the ERBB1 inhibitor lapatinib in a synergistic fashion. CDK inhibitors interacted with lapatinib to reduce MCL-1 expression and overexpression of MCL-1 or knock down of BAX and BAK suppressed drug combination lethality. Lapatinib-mediated inhibition of ERK1/2 and to a lesser extent AKT facilitated CDK inhibitor-induced suppression of MCL-1 levels. Treatment of cells with the BH3 domain/MCL-1 inhibitor obatoclax enhanced the lethality of lapatinib in a synergistic fashion. Knock out of MCL-1 and BCL-XL enhanced lapatinib toxicity to a similar extent as obatoclax and suppressed the ability of obatoclax to promote lapatinib lethality. Pre-treatment of cells with lapatinib or with obatoclax enhanced basal levels of BAX and BAK activity and further enhanced drug combination toxicity. In vivo tumor growth data in xenograft and syngeneic model systems confirmed our in vitro findings. Treatment of cells with CDK inhibitors enhanced the lethality of obatoclax in a synergistic fashion. Overexpression of MCL-1 or knock down of BAX and BAK suppressed the toxic interaction between CDK inhibitors and obatoclax. Obatoclax and lapatinib treatment or obatoclax and CDK inhibitor treatment or lapatinib and CDK inhibitor treatment radiosensitized breast cancer cells. Lapatinib and obatoclax interacted to suppress mammary tumor growth in vivo. Collectively our data demonstrate that manipulation of MCL-1 protein expression by CDK inhibition or inhibition of MCL-1 sequestering function by Obatoclax renders breast cancer cells more susceptible to BAX/BAK-dependent mitochondrial dysfunction and tumor cell death.
Key words: MCL-1, Lapatinib, Obatoclax, Flavopiridol, Roscovitine, CDK inhibitor, RTK inhibitor, BCL-2 inhibitor, BAK
Introduction
Flavopiridol (NSC 649890), is a semi-synthetic alkaloid that inhibits to varying degrees all known cyclin-dependent kinases (CDKs), including the cyclin T/CDK9 transcriptional regulatory complex (positive transcription elongation factor-b; PTEF-b).1,2 Other CDK9 inhibitors, such as roscovitine and its derivatives, are also being actively explored in the clinic.3 Inhibition of CDK9 results in the dephosphorylation of the carboxyl-terminal domain of RNA Pol II and reduced levels of transcription.4 Flavopiridol was the first CDK inhibitor to enter clinical trials.5 In vitro, clinically relevant low concentrations (<200 nM) of flavopiridol induce G1 arrest in tumor cells and variably trigger tumor cell apoptosis.6,7 Flavopiridol toxicity correlates with the transcription repression of various genes that promote cell survival, including those encoding short-lived proteins such as MCL-1.8,9 Studies from several laboratories have linked some of the lethal actions of flavopiridol in leukemia cells to inhibition of IκB kinases and to inactivation of the transcription factor NFκB, a transcription factor involved in diverse cellular processes, including cell survival, proliferation and differentiation.10 Treatment of cells with flavopiridol has also been shown to inhibit the activities of many signal transduction pathways that are frequently associated with cell survival and the regulation of cell survival protein expression e.g., AKT.11,12
Inhibitors of receptor tyrosine kinases, particularly of ERBB1 and ERBB2, have been under pre-clinical and clinical development for over 10 years.13,14 In vitro, numerous tumor cell types have been shown to exhibit growth reduction following inhibition of growth factor receptors, e.g., ERBB1 or inhibition of signaling pathways, e.g., MEK1/2.15 However, in many such studies the primary effect of a single kinase inhibitory agent at low “target specific” doses on tumor cells was cyto-static, rather than cyto-toxic.16 And, in contrast to the relatively encouraging findings from pre-clinical in vitro work, clinical studies using many ERBB1/ERBB2 inhibitors as single agents frequently did not demonstrate any form of tumor growth control.17 Exposure of tumor cells expressing a mutated active form of ERBB1, but generally not an overexpressed wild-type ERBB1, to kinase domain inhibitors results in growth arrest and tumor cell death.18,19 Over the course of many months exposure to kinase inhibitor(s), secondary mutations in the receptor kinase domain develop which render the receptor resistant to the kinase inhibitor. A more rapid mechanism of resistance to ERBB receptor inhibitors as single agents, prior to the development of secondary mutations, is the compensatory activation of growth factor receptors such as c-MET (+c-Src) and the IGF1R which can act in parallel to provide survival signaling.20–22 These receptors can provide a survival signal in their own right as receptor tyrosine kinases as well as causing trans-phosphorylation of inhibited ERBB receptors, thereby permitting the ERBB receptors to act as docking sites for e.g., RAS GTP exchange factors. We have found that resistance to Lapatinib in colon cancer cells is mediated by increased expression of mitochondrial and endoplasmic reticulum protective MCL-1 and BCL-XL proteins with reduced expression of pro-apoptotic BAX and mutation of p53.23
The BCL-2 family of proteins regulates the intrinsic/mitochondrial apoptosis pathway. Protective BCL-2 family proteins (BCL-2, BCL-XL, MCL-1, A1) associate via BH3 domains with pro-apoptotic family members including BAX and BAK. BAX and BAK, when released from protective BCL-2 proteins, can perturb the mitochondrial membrane forming pores that permit release of cytochrome c and AIF, leading ultimately to apoptosis. Tumor cells utilize a number of mechanisms to maintain viability, including loss of death receptor expression, e.g., CD95, by losing expression of pro-apoptotic BH3 domain proteins, e.g., BAX or by increasing expression of anti-apoptotic BCL-2 family members, e.g., MCL-1.24,25 In the case of protective BCL-2 family proteins, several clinically relevant small molecule inhibitors have been developed that specifically bind to the BCL-2 family protein, without altering expression of the protein and that block the binding of pro-apoptotic BH3 domain proteins, e.g., GX15-070 (Obatoclax).26,27 The drug-induced dissociation of BCL-2 protein from toxic BH3 domain protein results in greater levels of free BH3 domain protein that will facilitate mitochondrial dysfunction and promote the toxicity of other therapeutic agents.28,29
The present studies determined whether inhibition of BCL-2 family function using either CDK inhibitors to reduce protein expression or using Obatoclax to inhibit BH3 domain function, could promote tumor cell death.
Results
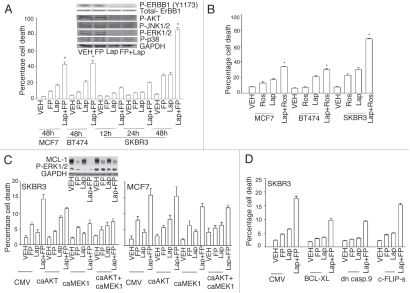
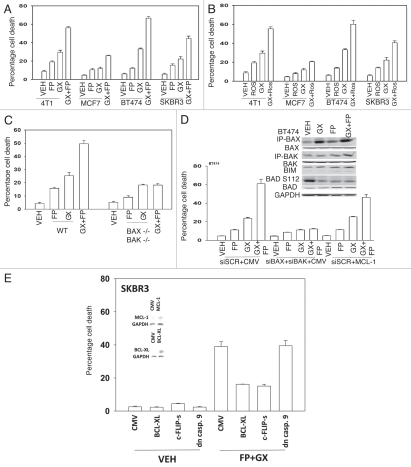
The impact of combined exposure of breast cancer cells to the CDK inhibitor flavopiridol and the ERBB1/ERBB2 inhibitor lapatinib was first investigated. In short-term cell viability assays simultaneous combined exposure of breast cancer cells to flavopiridol and lapatinib resulted in a greater than additive induction of short-term cell killing compared to either drug individually, which was synergistic as determined by Median Dose Effect analyses with Combination Index (CI) values consistently less than 1.00 (Fig. 1A and Table 1). These findings correlated with dephosphorylation of ERBB1, ERK1/2 and AKT. Parallel studies with another CDK inhibitor, roscovitine, generated data that was very similar to that generated using flavopiridol (Fig. 1B). Constitutive activation of MEK1 and of MEK1 and AKT, protected breast cancer cells from flavopiridol + lapatinib lethality that correlated with increased MCL-1 expression (Fig. 1C). Overexpression of either BCL-XL or of dominant negative caspase 9, but not c-FLIP-s, suppressed drug lethality (Fig. 1D). Lapatinib enhanced the rate of flavopiridol-induced MCL-1 depletion and overexpression of MCL-1 protected cells from flavopiridol + lapatinib lethality (Fig. 1E). Treatment of cells with lapatinib and flavopiridol enhanced BAX and BAK activation and knock down of BAX + BAK suppressed flavopiridol + lapatinib lethality (Fig. 1F). In colon cancer cells that were generated to be lapatinib resistant and that we had demonstrated was due to increased basal levels of MCL-1, flavopiridol partially circumvented lapatinib resistance (Fig. 1G).
Figure 1A–D.
Flavopiridol and lapatinib interact in a greater than additive manner to promote mammary tumor cell death in vitro. (A) MCF7, BT474 and SKBR3 were plated as in methods and 24 h after plating concurrently treated with vehicle control (VEH, DMSO), flavopiridol (FP, 50 nM) and/or lapatinib (Lap, 1 µM). Cell viability was determined in triplicate 48 h after drug exposure using trypan blue exclusion assays (±SEM, n = 3; *p < 0.05 greater than additive reduction in viability for the combined exposure compared to individual drug treatments). Inset part, SKBR3 cells were treated with flavopiridol and/or lapatinib and the phosphorylation of the indicated proteins determined 12 h after drug exposure following SDS PA GE and immunoblotting. (B) MCF7, BT474 and SKBR3 were plated as in methods and 24 h after plating concurrently treated with vehicle control (VEH, DMSO), roscovitine (Ros, 5 µM) and/or lapatinib (Lap, 1 µM). Cell viability was determined in triplicate 48 h after drug exposure using trypan blue exclusion assays (±SEM, n = 3; *p < 0.05 greater than additive reduction in viability for the combined exposure compared to individual drug treatments). (C) MCF7 and SKBR3 cells were infected with empty vector adenovirus (CMV) or viruses to express activated AKT (caAKT) or activated MEK1 (caMEK1). Twenty four h after infection cells were treated with vehicle control (VEH, DMSO), flavopiridol (FP, 50 nM) and/or lapatinib (Lap, 1 µM). Cell viability was determined in triplicate 48 h after drug exposure using trypan blue exclusion assays. Inset part, SKBR3 cells were infected with viruses and treated with drugs as indicated. The phosphorylation expression of the indicated proteins determined 12 h after drug exposure following SDS PA GE and immunoblotting. (D) SKBR3 cells were infected with empty vector adenovirus (CMV) or viruses to express BCL-XL, dominant negative caspase 9 or c-FLIP-s. Twenty four h after infection cells were treated with vehicle control (VEH, DMSO), flavopiridol (FP, 50 nM) and/or lapatinib (Lap, 1 µM). Viability was determined in triplicate 48 h after drug exposure using trypan blue exclusion assays (±SEM, n = 3).
Table 1.
Lapatinib and CDK inhibitors synergize to kill breast cancer cells
| Ros. (µM) | Lap. (µM) | Fa | CI |
| 2.5 | 0.5 | 0.36 | 0.55 |
| 5.0 | 1.0 | 0.39 | 0.49 |
| 7.5 | 1.5 | 0.49 | 0.27 |
| FP. (nM) | Lap. (µM) | Fa | CI |
| 25 | 0.5 | 0.35 | 0.27 |
| 50 | 1.0 | 0.46 | 0.27 |
| 75 | 1.5 | 0.61 | 0.17 |
SKBR3 cells were plated as single cells (250–1,500 cells/well) in sextuplicate and 12 h after this plating the cells were treated with vehicle (VEH, DMSO), Lapatinib (0.5–1.5 µM), flavopiridol (FP, 25–75 nM) or roscovitine (Ros, 2.5–7.5 µM), or with both drugs combined, as indicated at a fixed concentration ratio to perform median dose effect analyses for the determination of synergy. After drug exposure (24 h), the media was changed and cells cultured in drug free media for an additional 10–14 days. Cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. Colony formation data were entered into the Calcusyn program and combination index (CI) and Fraction Affected (Fa) values determined. A CI value of less than 1.00 indicates synergy.
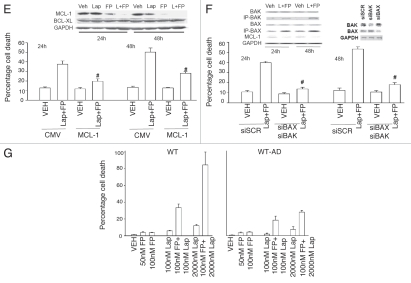
Figure 1E–G.
Flavopiridol and lapatinib interact in a greater than additive manner to promote mammary tumor cell death in vitro. (E) SKBR3 cells were transfected with empty vector plasmid of a plasmid to express MCL-1. Twenty four h after transfection cells were treated with vehicle control (VEH, DMSO), flavopiridol (FP, 50 nM) and/or lapatinib (Lap, 1 µM). Viability was determined in triplicate 48 h after drug exposure using trypan blue exclusion assays (±SEM, n = 3; #p < 0.05 less than corresponding empty vector drug treated cell killing). (F) SKBR3 cells were transfected with scrambled siRNA (siSCR) or siRNAs to knock down BAX and BAK (siBAX + siBAK). Twenty four h after transfection cells were treated with vehicle control (VEH, DMSO), flavopiridol (FP, 50 nM) and/or lapatinib (Lap, 1 µM). Viability was determined in triplicate 48 h after drug exposure using trypan blue exclusion assays (±SEM, n = 3; #p < 0.05 less than corresponding siSCR drug treated cell killing). Inset part, cells were treated with drugs and isolated 24 h and 48 h later. The activity of BAK and BAX was determined after immunoprecipitation of the conformationally active BAX and BAK proteins. (G) Parental wild-type (WT) and lapatinib adapted (WT-AD) HCT116 cells were treated with vehicle control (VEH, DMSO), flavopiridol (FP, 50-100 nM) and/or lapatinib (Lap, 100–200 nM). Viability was determined in triplicate 48 h after drug exposure using trypan blue exclusion assays (±SEM, n = 3).
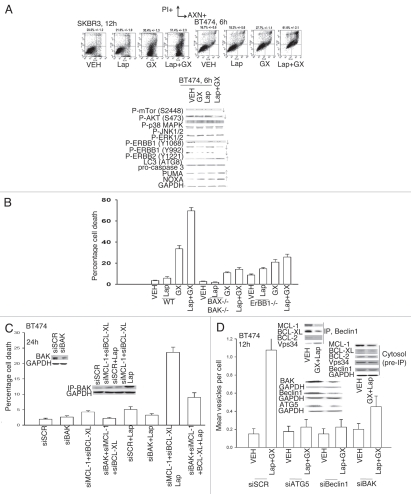
A number of BH3 domain inhibitor drugs are being explored in the clinic including the drug obatoclax (GX15-070 (GX)) that inhibits the protective function of BCL-2, BCL-XL and MCL-1 in terms of the abilities of these proteins to sequester toxic BH3 domain proteins such as BAX and BAK. Obatoclax enhanced lapatinib toxicity in a greater than additive fashion in short term and long term viability assays (Fig. 2A and Table 2). In BT474 breast cancer cells the lethal effects of obatoclax + lapatinib exposure correlated with loss of mTOR and AKT phosphorylation and increased expression of LC3, PUMA and NOXA. In transformed fibroblasts deletion of BAX+BAK or of ERBB1 suppressed the toxic interaction between lapatinib and obatoclax (Fig. 2B). Knock down of MCL-1 and BCL-XL expression enhanced lapatinib lethality in breast cancer cells and effect that was suppressed by concomitant knock down of BAK (Fig. 2C). This correlated with lapatinib + (MCL-1 + BCL-XL) knock down promoting BAK activation (Fig. 2C and inset).
Figure 2A–D.
Lapatinib and obatoclax interact to kill mammary carcinoma cells. (A) Human mammary carcinoma cells were treated with vehicle (DMSO), lapatinib (Lap, 2 µM), obatoclax (GX, 50 nM) or lapatinib + obatoclax. Floating and attached cells were isolated 6 h after exposure and cell viability determined using Annexin-PI flow cytometry assay (±SEM, n = 3). Lower blots: 6 h after drug treatment cells were isolated and blotting performed to determine protein expression/phosphorylation levels (n = 3). (B) Transformed mouse embryonic fibroblasts, wild-type (WT) or genetically deleted for the indicated proteins, were treated with vehicle (DMSO), lapatinib (Lap, 1 µM), obatoclax (GX, 50 nM) or lapatinib + obatoclax. Floating and attached cells were isolated 24 h after exposure and viability determined using trypan blue exclusion assay (±SEM, n = 3). (C) BT474 cells were plated and 12 h later as indicated transfected with siRNA molecules to reduce the expression of; control (siSCR); BCL-2 (siBCL-2); BCL-XL (siBCL-XL); or MCL-1 (siMCL). Forty eight h after transfection, cells were treated with vehicle (DMSO) or lapatinib (2 µM). Cells were isolated 24 h after serum starvation and cell viability determined in triplicate by trypan blue exclusion assay (±SEM n = 3). (D) BT474 cells 24 h after plating in glass chambered slides or in 60 mm dishes were transfected, as indicated in the part, with scrambled siRNA (siSCR); and siRNA molecules to knock down ATG5 (siATG5), Beclin1 (siBeclin1) and BAK (siBAK). Cells were co-transfected to express LC3-GFP. Twenty four h after transfection cells were treated with vehicle (VEH, DMSO) or with lapatinib (Lap, 2 µM) + obatoclax (GX, 50 nM). Six h after drug treatment cells in chamber slides were examined under a fluorescent microscope and the number of intense LC3-GFP staining vesicles per cell determined in 40 cells (n = 2; ±SEM). Inset: Transfected cells were treated with vehicle (VEH, DMSO) or with lapatinib (Lap, 2 µM) + obatoclax (GX, 50 nM) and 6 h after exposure cells were lysed and the association of Beclin1 with Vps 34, BCL-XL and MCL-1 determined after immunoprecipitation and SDS PA GE/blotting.
Table 2.
Obatoclax and lapatinib synergize to kill breast cancer cells
| GX (nM) | Lap. (µM) | Fa | CI |
| SKBR3 | |||
| 33.0 | 0.5 | 0.38 | 0.61 |
| 66.0 | 1.0 | 0.48 | 0.58 |
| 99.0 | 1.5 | 0.59 | 0.45 |
| BT474 | |||
| 15.0 | 0.5 | 0.28 | 0.67 |
| 30.0 | 1.0 | 0.54 | 0.57 |
| 45.0 | 1.5 | 0.82 | 0.50 |
SKBR3 and BT474 cells were plated as single cells (250–1,500 cells/well) in sextuplicate and 12 h after this plating the cells were treated with vehicle (VEH, DMSO), lapatinib (0.5–1.5 µM) or obatoclax (GX15-070, 33–99 nM or 15–45 nM as indicated), or with both drugs combined, as indicated at a fixed concentration ratio to perform median dose effect analyses for the determination of synergy. After drug exposure (24 h), the media was changed and cells cultured in drug free media for an additional 10–14 days. Cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. Colony formation data were entered into the Calcusyn program and combination index (CI) and Fraction Affected (Fa) values determined. A CI value of less than 1.00 indicates synergy.
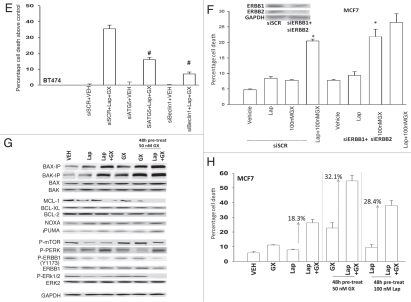
As lapatinib + obatoclax exposure was increasing the levels of the autophagy regulator LC3 in breast cancer cells and because we had previously noted a similar effect in colon cancer cells, we investigated in breast cancer cells the role of autophagy in the lethality of this drug combination. Lapatinib + obatoclax exposure of BT474 cells increased the numbers of autophagic vesicles per cell (Fig. 2D). Increased autophagy was dependent on expression of Beclin1, ATG5 or of BAK. Lapatinib + obatoclax exposure promoted increased association of Beclin1 with Vps34 and decreased association of the protein with BCL-XL and MCL-1 (Fig. 2D, upper blots). Knock down of either ATG5 or Beclin1 protected BT474 cells from the lethal effects of the drug combination (Fig. 2E). In agreement with lapatinib acting in an ontarget fashion to inhibit ERBB receptor signaling, knock down of ERBB1 and ERBB2 enhanced obatoclax toxicity in MCF7 cells; toxicity in the absence of ERBB1 + ERBB2 was not further enhanced by lapatinib exposure (Fig. 2F).
Figure 2E–H.
Lapatinib and obatoclax interact to kill mammary carcinoma cells. (E) BT474 cells 24 h after plating were transfected with scrambled siRNA (siSCR, 20 nM) or transfected with siRNAs to knock down ATG5 (siATG5) or Beclin1 (siBeclin1). Twenty four h after transfection, cells were treated with vehicle (VEH, DMSO) or lapatinib (Lap, 2 µM) and obatoclax (GX, 50 nM). Three h after exposure cells were isolated and viability determined by Annexin-PI staining (±SEM, n = 2). #p < 0.05 less than corresponding value in siSCR cells. (F) MCF7 cells were transfected with scrambled siRNA (siSCR) or siRNA molecules to knock down ERBB1 (siERBB1) and ERBB2 (siERBB2). Twenty four h after transfection, cells were treated with vehicle (VEH, DMSO) or lapatinib (Lap, 2 µM) and obatoclax (GX, 50 nM). Twelve h after exposure cells were isolated and viability determined by Annexin-PI staining (±SEM, n = 2). *p < 0.05 greater than corresponding value in vehicle cells. (G) MCF7 cells were pre-treated with either vehicle (DMSO), lapatinib (100 nM) or obatoclax (50 nM) for 48 h as indicated. Portions of cells were isolated. The remaining portions of cells were then treated with vehicle (DMSO), lapatinib (100 nM) and/or obatoclax (50 nM) in the indicated combinations for 12 h followed by isolation of cells. The activity of BAK and BAX under each treatment condition was determined after immunoprecipitation of the conformationally active BAX and BAK proteins. The expression of other proteins and the phosphorylation of these proteins was in cell lysates not undergoing IP. (H) MCF7 cells in triplicate were pre-treated with either vehicle (DMSO), lapatinib (100 nM) or obatoclax (50 nM) for 48 h. Cells were then treated with vehicle (DMSO), lapatinib (100 nM) and/or obatoclax (50 nM) in the indicated combinations for 24 h. After exposure cells were isolated and viability determined by trypan blue staining (±SEM, n = 2).
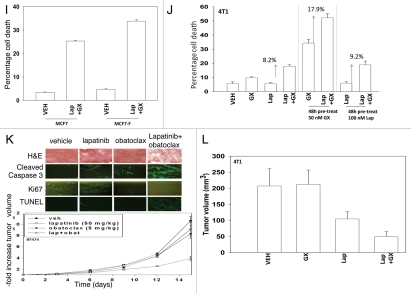
Pre-treatment of MCF7 cells with lapatinib or with obatoclax enhanced basal levels of BAX and BAK activity and pre-treatment reduced expression of protective BCL-2 family proteins (Fig. 2G). Combined exposure to both drugs promoted PKR-like endoplasmic reticulum kinase (PERK) activation, indicative of an elevated ER stress response with concomitant suppression of translation. Pre-treatment of MCF7 cells with lapatinib or with obatoclax significantly enhanced the toxicity of the drug combination compared to a simple continuous exposure to both drugs without any drug pre-treatment (Fig. 2H). Fulvestrant resistant MCF7 cells were more sensitive to lapatinib and obatoclax toxicity than parental estrogen sensitive MCF7 cells (Fig. 2I). In 4T1 mammary tumors we noted in a similar manner to sequence dependent apoptosis promoting effects of pre-treatment with obatoclax but in this cell line not with lapatinib (Fig. 2J). Combined exposure of orthotopic established BT474 human mammary carcinoma xenograft tumors to lapatinib and obatoclax significantly reduced tumor growth below that of tumors treated with either individual agent, and this suppression of tumor growth correlated with profound disruption of tumor cyto-architecture as judged using H&E staining, increased cleavage of pro-caspase 3 and abolition of Ki67 staining (Fig. 2K). Similar growth suppression data were observed in 4T1 mammary tumors growing in the fat pads of syngeneic immune competent mice (Fig. 2L).
Figure 2I–L.
Lapatinib and obatoclax interact to kill mammary carcinoma cells. (I) MCF7 (parental) and MCF7 fulvestrant resistant cells were then treated with vehicle (DMSO), lapatinib (100 nM) and/or obatoclax (50 nM) in the indicated combinations for 24 h. After exposure cells were isolated and viability determined by trypan blue staining (±SEM, n = 2). (J) Mouse mammary carcinoma 4T1 cells in triplicate were pre-treated with either vehicle (DMSO), lapatinib (100 nM) or obatoclax (50 nM) for 48 h. Cells were then treated with vehicle (DMSO), lapatinib (100 nM) and/or obatoclax (50 nM) in the indicated combinations for 24 h. After exposure cells were isolated and viability determined by trypan blue staining (±SEM, n = 2). (K) Lower Graph: BT474 xenograft tumors were established in the mammary fat pad of an athymic mouse and tumors permitted to form to ∼100 mm3. Animals were treated with lapatinib and/or obatoclax as indicated and tumor volume determined every 2–3 days (±SEM, n = 2). Upper: immuno-histochemistry of BT474 tumors 16 days after cessation of drug treatment. (L) 4T1 mouse mammary tumor cells were injected into the 4th mammary fat pad of a Balb/c mouse (1 − 107). Seven days after injection, mice were treated with lapatinib and/or obatoclax. The tumor volumes presented were calculated 14 days after the start of drug exposure (±SEM, n = 2).
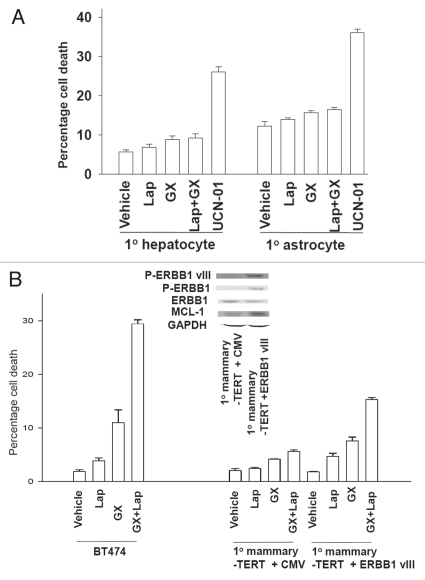
Lapatinib and obatoclax exposure did not kill primary rodent hepatocytes or primary human astrocytes (Fig. 3A and B). However, transfection of primary mammary epithelial cells expressing hTERT with a plasmid to express activated ERBB1 vIII resulted in increased expression of MCL-1 and increased cell killing following lapatinib + obatoclax exposure (Fig. 3B).
Figure 3.
Lapatinib and obatoclax therapy does not kill non-transformed cells; primary mammary epithelial cells are sensitized to drug toxicity by overexpression of an activated ERBB1 mutant. (A) Primary mouse hepatocytes and primary human fetal astrocytes were treated with vehicle (VEH, DMSO) or lapatinib (Lap, 2 µM) and obatoclax (GX, 50 nM); or with 250 nM UCN-01. Twelve h after exposure cells were isolated and viability determined by Annexin-PI staining (±SEM, n = 2). (B) Primary mammary epithelial cells stably expressing hTERT were transiently transfected with an empty vector plasmid or with a plasmid to express ERBB1 vIII. Forty eight h after transfection cells were treated with vehicle (VEH, DMSO) or lapatinib (Lap, 2 µM) and obatoclax (GX, 50 nM). Twelve h after exposure cells were isolated and viability determined by Annexin-PI staining (±SEM, n = 2). Parallel data using BT474 cells is also presented for comparison. Inset: Cells were isolated 48 h after transfection and the phosphorylation/expression of the indicated proteins measured by immunoblotting.
We next determined if obatoclax and flavopiridol that directly inhibit and downregulate expression, respectively, of the function of MCL-1, also interacted to kill breast cancer cells. Flavopiridol enhanced obatoclax toxicity in a greater than additive fashion in short term and long term viability assays (Fig. 4A and Table 3). Similar data were obtained using the structurally dissimilar CDK inhibitor roscovitine (Fig. 4B). In transformed fibroblasts deletion of BAX + BAK suppressed the toxic interaction between lapatinib and obatoclax (Fig. 4C). Knock down of BAX + BAK expression suppressed drug combination lethality in breast cancer cells, whereas overexpression of MCL-1 only modestly protected cells from drug toxicity (Fig. 4D). Obatoclax enhanced BAX activity that was increased by flavopiridol; flavopiridol permitted obatoclax to enhance BAK activation (Fig. 4D, upper inset part). Overexpression of BCL-XL which was overexpressed to a much higher level than that of MCL-1 in Figure 4D more potently suppressed flavopiridol and obatoclax toxicity (Fig. 4E and and inset blots). Expression of dominant negative caspase 9 but not of c-FLIP-s also suppressed flavopiridol and obatoclax combination toxicity.
Figure 4.
CDK inhibitors enhance obatoclax toxicity in breast cancer cells. (A) Human and rodent mammary carcinoma cells were treated with vehicle control (VEH, DMSO), flavopiridol (FP, 50 nM) and/or obatoclax (GX, 50 nM). Viability was determined in triplicate 24 h after drug exposure using trypan blue exclusion assays (±SEM, n = 3). (B) Human and rodent mammary carcinoma cells were treated with vehicle control (VEH, DMSO), roscovitine (Ros, 50 nM) and/or obatoclax (GX, 50 nM). Viability was determined in triplicate 24 h after drug exposure using trypan blue exclusion assays (±SEM, n = 3). (C) Transformed mouse embryonic fibroblasts, wild-type (WT) or genetically deleted for the indicated proteins, were treated with vehicle (DMSO), (VEH, DMSO), flavopiridol (FP, 50 nM) and/or obatoclax (GX, 50 nM). Floating and attached cells were isolated 24 h after exposure and viability determined using trypan blue exclusion assay (±SEM, n = 3). (D) BT474 cells were transfected with scrambled siRNA (siSCR), empty vector plasmid (CMV), siRNAs to knock down BAX and BAK (siBAX + siBAK) or with a plasmid to express MCL-1. Twenty four h after transfection cells are treated with vehicle (DMSO), (VEH, DMSO), flavopiridol (FP, 50 nM) and/or obatoclax (GX, 50 nM). Floating and attached cells were isolated 12 h after exposure and viability determined using trypan blue exclusion assay (±SEM, n = 3). Inset: 6 h after drug treatment cells were isolated and blotting performed to determine protein expression/phosphorylation/activity levels (n = 3). (E) SKBR3 cells were infected with empty vector adenovirus (CMV) or viruses to express BCL-XL, dominant negative caspase 9 or c-FLIP-s. Twenty four h after infection cells were treated with vehicle (DMSO), (VEH, DMSO), flavopiridol (FP, 50 nM) and/or obatoclax (GX, 50 nM). Floating and attached cells were isolated 12 h after exposure and viability determined using trypan blue exclusion assay (±SEM, n = 3).
Table 3.
CDK inhibitors and obatoclax interact in a synergistic fashionto kill breast cancer cells
| FP. (nM) | GX (nM) | Fa | CI |
| 4T1 | |||
| 10.0 | 10.0 | 0.62 | 0.32 |
| 17.5 | 17.5 | 0.72 | 0.32 |
| 25.0 | 25.0 | 0.78 | 0.39 |
| MCF7 | |||
| 10.0 | 10.0 | 0.36 | 0.49 |
| 17.5 | 17.5 | 0.52 | 0.57 |
| 25.0 | 25.0 | 0.68 | 0.63 |
MCF7 and 4T1 cells were plated as single cells (250–1,500 cells/well) in sextuplicate and 12 h after this plating the cells were treated with vehicle (VEH, DMSO), flavopiridol (10–25 nM) or Obatoclax (GX15-070, 10–25 nM), or with both drugs combined, as indicated at a fixed concentration ratio to perform median dose effect analyses for the determination of synergy. After drug exposure (24 h), the media was changed and cells cultured in drug free media for an additional 10–14 days. Cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. Colony formation data were entered into the Calcusyn program and combination index (CI) and Fraction Affected (Fa) values determined. A CI value of less than 1.00 indicates synergy.
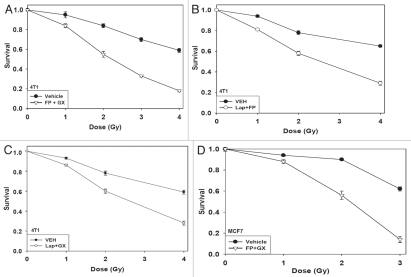
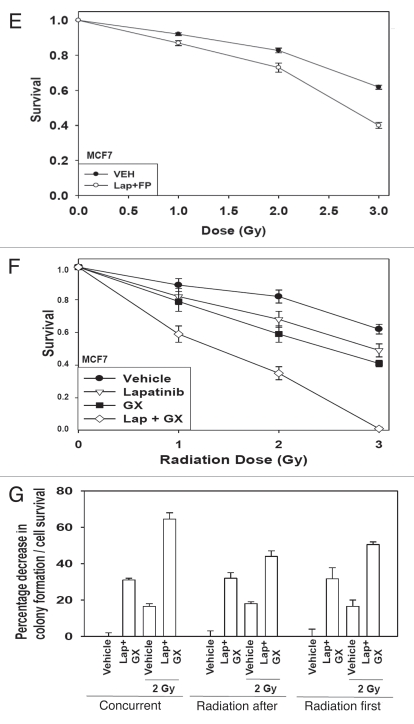
Radiotherapy is a primary therapeutic modality for breast cancer and is used in conjunction with a variety of chemotherapies. Treatment of 4T1 rodent and MCF7 human breast cancer cells with flavopiridol and obatoclax radiosensitized breast cancer cells (Fig. 5A and D). Treatment of cells with lapatinib and flavopiridol radiosensitized breast cancer cells (Fig. 5B and E). Treatment of cells with lapatinib and obatoclax radiosensitized breast cancer cells (Fig. 5C and F). Finally, we determined whether there was a schedule dependency for radiosensitization by lapatinib and obatoclax treatment. Concurrent drug and radiation exposure provided a greater radiosensitizing effect than irradiation either prior to or following drug treatment (Fig. 5G). Collectively, the data in this manuscript demonstrate that inhibition of MCL-1 function renders breast cancer cells susceptible to mitochondrial dysfunction and tumor cell death and in parallel increases mammary carcinoma cell radiosensitivity.
Figure 5A–D.
Radiosensitization of mammary carcinoma cells by drug combinations that target MCL-1. (A) Single 4T1 cells plated in sextuplicate and 12 h after plating were treated with vehicle or flavopiridol (FP, 50 nM) and obatoclax (GX, 50 nM). Cells were irradiated 1 h after drug treatment. Media was changed 48 h after irradiation and colonies permitted to form in drug free media for an additional ∼14 days. Data are the means of data points from two separate studies (12 data points) ±SEM. (B) Single 4T1 cells plated in sextuplicate and 12 h after plating were treated with vehicle or flavopiridol (FP, 50 nM) and lapatinib (Lap, 1 µM). Cells were irradiated 1 h after drug treatment. Media was changed 48 h after irradiation and colonies permitted to form in drug free media for an additional ∼14 days. Data are the means of data points from two separate studies (12 data points) ±SEM. (C) Single 4T1 cells plated in sextuplicate and 12 h after plating were treated with vehicle or lapatinib (Lap, 1 µM) and obatoclax (GX, 50 nM). Cells were irradiated 1 h after drug treatment. Media was changed 48 h after irradiation and colonies permitted to form in drug free media for an additional ∼14 days. Data are the means of data points from two separate studies (12 data points) ±SEM. (D) Single MCF7 cells plated in sextuplicate and 12 h after plating were treated with vehicle or flavopiridol (FP, 50 nM) and obatoclax (GX, 50 nM). Cells were irradiated 1 h after drug treatment. Media was changed 48 h after irradiation and colonies permitted to form in drug free media for an additional ∼14 days. Data are the means of data points from two separate studies (12 data points) ±SEM.
Figure 5E–G.
Radiosensitization of mammary carcinoma cells by drug combinations that target MCL-1. (E) Single MCF7 cells plated in sextuplicate and 12 h after plating were treated with vehicle or flavopiridol (FP, 50 nM) and lapatinib (Lap, 1 µM). Cells were irradiated 1 h after drug treatment. Media was changed 48 h after irradiation and colonies permitted to form in drug free media for an additional ∼14 days. Data are the means of data points from two separate studies (12 data points) ±SEM. (F) Single MCF7 cells plated in sextuplicate and 12 h after plating were treated with vehicle or lapatinib (Lap, 1 µM) and obatoclax (GX, 50 nM). Cells were irradiated 1 h after drug treatment. Media was changed 48 h after irradiation and colonies permitted to form in drug free media for an additional ∼14 days. Data are the means of data points from two separate studies (12 data points) ±SEM. (G) Single BT474 cells plated in sextuplicate and 12 h after plating were: (a) irradiated followed 24 later by a 48 h treatment with vehicle or lapatinib (Lap, 1 µM) and obatoclax (GX, 50 nM); (b) treated with vehicle or lapatinib (Lap, 1 µM) and obatoclax (GX, 50 nM) for 48 h then irradiated 24 h after drug exposure; (c) treated with vehicle or lapatinib (Lap, 1 µM) and obatoclax (GX, 50 nM) and cells were irradiated 1 h after drug treatment and media was changed 48 h after irradiation. Under all conditions, colonies after irradiation/drug exposure were permitted to form in drug free media for an additional ∼14 days. Data are the means of data points from two separate studies (12 data points) ±SEM.
Discussion
The studies described herein were designed to explore the mechanisms by which the protective actions of the mitochondrial protein MCL-1 could be subverted, thereby promoting breast cancer cell death.
CDK inhibitors flavopiridol or roscovitine and the ERBB1/2 inhibitor lapatinib, administered at relatively low, potentially clinically relevant concentrations, interact to kill mammary carcinoma cells in vitro. Cell killing correlated with loss of MCL-1 expression and was dependent on activation of the pro-apoptotic BH3 domain proteins BAX and BAK; overexpression of MCL-1 suppressed drug-induced cell killing. As a more direct approach to inhibit MCL-1 we made use of the BH3 domain inhibitor obatoclax that inhibits MCL-1 sequestration of toxic pore forming proteins, such as BAX and BAK. Obatoclax enhanced lapatinib toxicity. Again, cell killing correlated with activation of BAK. Finally, as both CDK inhibitors and obatoclax directly and independently, target MCL-1 function, we determined whether such agents interacted to kill breast cancer cells. Obatoclax and CDK inhibitors synergized to kill breast cancer cells in a BAX and BAK dependent fashion; overexpression of MCL-1 weakly suppressed drug-induced lethality. Radiotherapy is a mainstay in the treatment of breast cancer patients. Our findings revealed that all three drug combinations targeted towards inhibiting MCL-1 resulted in enhanced breast cancer cell radiosensitization. Collectively, our data validates the hypothesis that inhibiting the ability of MCL-1 to protect breast cancer cells from apoptosis could have therapeutic utility.
The mechanisms by which flavopiridol and roscovitine downregulate expression of anti-apoptotic proteins may be multifactorial. For example, flavopiridol, by inhibiting the pTEFb transcription complex, can act as a transcriptional repressor, and can block the transcription of short-lived proteins including MCL-1 (reviewed in ref. 22). Deletion of BAX and BAK function modestly suppressed flavopiridol toxicity but abolished the potentiation of obatoclax or lapatinib lethality. Such findings are in accord with previous studies indicating that loss of these multi-domain BCL-2 family members protects cells from diverse noxious stimuli.24,25 In clinical trials using a 72 h infusion schedule, the predicted free plasma concentrations of flavopiridol were found to be approximately 10% of the total amount of infused drug, with peak free plasma concentrations in the 25–80 nM range (reviewed in ref. 5, 24 and 25). These drug levels caused significant toxicities in patients with modest apparent benefit in terms of tumor control. Hence, based on patient performance and tumor response rates, alternate schedules of flavopiridol infusion were explored, with the rate of drug administration being increased in many trials to 1 h–24 h, achieving similar free flavopiridol concentrations with objective clinical responses being noted. More recently, a novel loading and 4 hr flavopiridol infusion schedule has been described which results in higher and more sustained plasma flavopiridol concentrations.
Lapatinib is approved for treatment of breast cancer patients in combination with the thymidylate synthase inhibitor capecitabine. Stable plasma lapatinib concentrations in excess of 2 µM have been reported in patients with this value being increased at least 2–3 fold with repeated dosing and ingestion of the drug with food.37–39 The half life of the drug in human plasma is ∼24 h (cf 2–3 hours in rodents) and once bound lapatinib slowly dissociates from ERBB1 and ERBB2.37–39 Lapatinib treatment reduced ERK1/2 activity and facilitated flavopiridolinduced suppression of MCL-1 levels and expression of constitutively active MEK1 partially maintained MCL-1 levels in flavopiridol treated cells and suppressed drug lethality; the protective effect of activated MEK1 was greater than that induced by activated AKT. SKBR3 and BT474 cells overexpress ERBB2 and BT474 and MCF7 cells express a mutant active PI3K protein, and as a result of these genetic alterations all of these cells have been argued to be more dependent on AKT signaling for growth and cell survival than the MEK-ERK pathway.40 In contrast to other systems where we have observed BAX/BAK dependent tumor cell killing that was associated with JNK and/or p38 MAPK signaling, CDK inhibitor + lapatinib toxicity was apparently not dependent on the JNK or p38 MAPK pathways to promote the activation of the toxic BH3 domain proteins.30
Knock down of MCL-1 and BCL-XL enhanced lapatinib toxicity in breast cancer cells; this is similar to our prior observations in colon cancer cells.36 Inhibition of BCL-2 family protein function using the small molecule BH3 domain antagonist obatoclax, a drug that is entering phase II trials, enhanced lapatinib toxicity in multiple breast cancer cell lines. Several drugs designed to inhibit protective BCL-2 family function are presently undergoing clinical evaluation including ABT-263 and AT-101 (R + gossypol).26–28 ABT-263 inhibits only BCL-2 and BCL-XL, whereas AT-101 is claimed, like obatoclax, to inhibit BCL-2, BCL-XL and MCL-1. In lung cancer cells addicted for survival to mutant active ERBB1 signaling that inhibition of BCL-2/BCL-XL using ABT-737 enhances gefitinib toxicity and that in other tumor cell types ERBB1 inhibitor toxicity is mediated via mitochondrial dysfunction.26–29 Our in vitro findings not only demonstrated that lapatinib and obatoclax synergized to kill breast cancer cells but that pre-treatment with either obatoclax or lapatinib enhanced basal activity levels of BAX and BAK which facilitated subsequent drug combination toxicity. Our in vivo findings demonstrated that lapatinib and obatoclax interacted to suppress mammary tumor growth. Collectively, these findings in combination with our own in the present manuscript argue that one useful approach to sensitize breast cancer cells to ERBB1 inhibitors is to inhibit the function of protective BCL-2 family proteins.
Based on our findings combining CDK inhibitors and lapatinib and obatoclax and lapatinib we determined whether the drug combination of CDK inhibitors and obatoclax caused a greater than additive killing of breast cancer cells. CDK inhibitors and obatoclax interacted in a synergistic fashion to kill cells that was associated with the drug combination, but not the individual agents, promoting activation of BAK. Knock down of BAK and BAX abolished drug combination lethality whereas overexpression of MCL-1 or of BCL-XL had only a weak protective effect (Unpublished data). The lack of MCL-1 or BCL-XL having a protective effect against CDK inhibitor + obatoclax lethality was indicative that obatoclax within the drug combination directly inhibited the toxic BH3 protein sequestering function and that overexpression of the protective BCL-2 family protein could not block the action of this drug.
In all instances, the primary mode by which tumor cells in this manuscript were induced to die after drug combination exposure required mitochondrial dysfunction. Individually, lapatinib, CDK inhibitors and obatoclax all have been shown to promote radiosensitization by mechanisms as diverse as inhibition of NFκB; suppression of cyto-protective protein expression and the generation of ROS and autophagy.41–43 In addition to causing DNA damage, one well recognized route of ionizing radiation-induced cell killing is also by causing mitochondrial dysfunction and promoting cytochrome c release into the cytosol.44 All three drug combinations that targeted MCL-1 function enhanced breast cancer cell radiosensitivity. The precise mechanisms by which each drug combination enhances radiosensitivity will need to be explored in a future manuscript.
In summary, the data in this manuscript demonstrates that multiple drug combinations which target MCL-1 function and/or expression kill breast cancer cells in vitro. A primary mode of drug combination lethality is due to the untethering and activation of BAK. Future studies will be required to validate whether our in vitro and in vivo discoveries translate into effective therapies for breast cancer.
Materials and Methods
Materials.
Phospho-/total-ERK1/2, Phospho-/total-JNK1/2, Phospho-/total-p38 MAPK, Anti-S473 AKT and total AKT antibodies were purchased from Cell Signaling Technologies (Worcester, MA). Lapatinib was supplied by Glaxo Smith Kline (Collegeville, PA) and Obatoclax by GeminX (Malvern, PA). Flavopiridol and roscovitine were purchased from Enzo Life Sciences (Plymouth Meeting, PA). Trypsin-EDTA, RPMI medium, penicillin-streptomycin were purchased from GIBCOBRL (GIBCOBRL Life Technologies, Grand Island, NY). The activated MEK1 EE adenovirus was kindly provided by Dr. J. Moltken (University of Cincinnati, Cincinnati, OH). BAX/BAK-/-,BIM-/- and BID-/- fibroblasts were kindly provided by Dr. S. Korsmeyer (Harvard University, Boston, MA). ERBB1-/- MEFs were supplied by Dr. J. Grandis (University of Pittsburgh, Pittsburgh, PA). ATG5-/- MEFs were supplied by Dr. M. Czaja (Yeshiva University, New York, NY). Mammary carcinoma cells and TERT transfected normal mammary epithelial cells were from the ATCC and also from Dr. Kenneth P. Nephew (Indiana University, Bloomington, IN) and Dr. A. Larner (Virginia Commonwealth University, Richmond, VA). The plasmid to express ERBB1 vIII was from Addgene. The plasmid to express MCL-1 was from Dr. Steven Grant (VCU). Reagents and the detailed performance of all experimental procedures were as described references 23 and 30–36.
Methods.
Culture and in vitro exposure of cells to drugs. Tumor cells and fibroblasts were cultured at 37°C (5% (v/v CO2)) in vitro using RPMI supplemented with 10% (v/v) fetal calf serum. In vitro drug treatments were from 100 mM stock solutions of each drug and the maximal concentration of Vehicle (DMSO) in media was 0.02% (v/v). For colony formation assays, cells were plated at low density (250–2,000 cells per dish) and 12 h after plating, cells were treated with the drugs in the order stated and at the concentrations stated in the Figure/Figure legend. Ten-14 days after exposure, plates were washed in PBS, fixed with methanol and stained with a filtered solution of crystal violet (5% w/v). After washing with tap water, the colonies were counted both manually (by eye) and digitally using a ColCountTM plate reader. Data presented is the arithmetic mean (±SEM) from both counting methods from multiple studies.
Cell treatments, SDS-PAGE and western blot analysis. Cells were treated with drugs, as indicated in the Figure legend. For SDS PAGE and immunoblotting, cells were lysed in either a nondenaturing lysis buffer and prepared for immunoprecipitation or in whole-cell lysis buffer (0.5 M Tris-HCl, pH 6.8, 2% SDS, 10% ycerol, 1% β-mercaptoethanol, 0.02% bromophenol blue) and the samples were boiled for 30 min. After immunoprecipitation, samples were boiled in whole cell lysis buffer. The boiled samples were loaded onto 10–14% SDS-PAGE and electrophoresis was run overnight. Proteins were electrophoretically transferred onto 0.22 µm nitrocellulose and immunoblotted with various primary antibodies against different proteins. All immunoblots were visualized using a Li-Cor Odyssey Infra Red Imager.
Recombinant adenoviral vectors; infection in vitro. We generated and purchased previously described recombinant adenoviruses to modulate protein expression and to express constitutively activated and dominant negative AKT and MEK1 proteins, dominant negative caspase 9 and BCL-XL (Vector Biolabs, Philadelphia, PA). Cells were infected with these adenoviruses at an approximate m.o.i. of 50. Cells were further incubated for 24 hours to ensure adequate expression of transduced gene products prior to drug exposures.
Detection of cell death by trypan blue and flow cytometery assays. Cells were harvested by trypsinization with Trypsin/EDTA for ∼10 min at 37°C. As some apoptotic cells detached from the culture substratum into the medium, these cells were also collected by centrifugation of the medium at 1,500 rpm for 5 min. The pooled cell pellets were resuspended and mixed with trypan blue dye. Trypan blue stain, in which blue dye incorporating cells were scored as being dead was performed by counting of cells using a light microscope and a hemacytometer. Five hundred cells from randomly chosen fields were counted and the number of dead cells was counted and expressed as a percentage of the total number of cells counted. Alternatively, the Annexin V/propidium iodide assay was carried to determine cell viability out as per the manufacturer's instructions (BD PharMingen) using a Becton Dickinson FACScan flow cytometer (Mansfield, MA).
Morphological detection of apoptosis by wright giemsa assays. Morphological assessment of apoptosis was performed as follows; cells were harvested by trypsinization with Trypsin/EDTA for ∼10 min at 37°C. As some apoptotic cells detached from the culture substratum into the medium, these cells were also collected by centrifugation of the medium at 1,500 rpm for 5 min. The pooled cell pellets were resuspended and a fraction of the suspension was centrifuged in a cytospinner (Cytospin 3, Shandon Inc., Pittsburgh, PA). For Wright Giemsa staining, the slides were fixed and stained in Diff-Quik7 Stain Set (Dade Diagnostics of P.P. Inc., Aguada, PR USA), according to the manufacturer's instruction and viewed under a light microscope. Nuclear and total cellular morphology was evaluated. Giemsa staining was used to identify total cell numbers and total numbers of apoptotic and non-apoptotic manifestations of cell killing. Five hundred cells from several randomly chosen fields were counted and the number of apoptotic cells was counted and expressed as a percentage of the total number of cells counted.
Plasmid transfection. Plasmid DNA (0.5 µg/total plasmid transfected) was diluted into 50 µl of RPMI growth media that lacked supplementation with FBS or with penicillin-streptomycin. Lipofectamine 2000 reagent (1 µl) (Invitrogen, Carlsbad, CA) was diluted into 50 µl growth media that lacked supplementation with FBS or with penicillin-streptomycin. The two solutions were then mixed together and incubated at room temperature for 30 min. The total mixture was added to each well (4 well glass slide or 12-well plate) containing 200 µl growth media that lacked supplementation with FBS or with penicillinstreptomycin. The cells were incubated for 4 h at 37oC, after which time the media was replaced with RPMI growth media containing 5% (v/v) FBS and 1x pen-strep.
Animal studies. For studies with human mammary carcinoma cells, athymic Nu/Nu mice (8 week old, female) were obtained from the NCI and were irradiated (1.5 Gy) 48 h prior to injection of animals into the 4th mammary fat pad with 1.0 × 107 BT474 cells. Tumors of ∼100 mm3 grew over the following month. Animals were segregated into tumor volumes of approximate equivalent mean tumor size and standard error. The animals were administered vehicle diluent (cremophore), lapatinib (50 mg/kg), obatoclax (5 mg/kg) or the drug combination by oral gavage once every day for 4 days. Tumor volumes are measured every two-three days. For studies with mouse mammary tumor cells Balb/c mice (8 week old, female) were obtained from the NCI and animals injected into the 4th mammary fat pad with 1.0 × 107 4T1 cells. Five days after implantation the animals were administered vehicle diluent (cremophore), lapatinib (100 mg/kg, BID), obatoclax (5 mg/kg, daily) or the drug combination by oral gavage for 5 days followed by two days of rest followed by another 5 days of treatment. The volumes of the tumors in each group were calculated on the day after the final drug treatment.
Immunohistochemistry and staining of fixed tumor sections. Post sacrifice, tumors were fixed in OCT compound (Tissue Tek); cryostat sectioned (Leica) as 12 µm sections. Nonspecific binding was blocked with a 2% (v/v) Rat Sera, 1% (v/v). Bovine Sera, 0.1% (v/v) Triton X100, 0.05% (v/v) Tween-20 solution then sections were stained for cell signaling pathway markers: anti-Ki67; anti-cleaved caspase 3. For staining of sectioned tumors, primary antibodies were applied overnight, sections washed with phosphate buffer solution and secondary antibodies applied for detection. Apoptotic cells with double stranded DNA breaks were detected using the Upstate TUNEL Apototic Detection Kit according to the manufacturer's instructions. Slides were applied to high powered light/confocal microscopes (Zeiss LSM 510 Meta-confocal scanning microscope; Zeiss HBO 100 microscope with Axio Cam MRm camera) at the indicated magnification in the Figures/Figure legends. The proliferation zone, which included both tumor and normal peritoneal tissue, was usually selected as the site of interest, within 2 mm of, or juxtaposed to leading edge of the tumor. Data shown are representative slides from several sections from the same tumor with multiple tumors (from multiple animals; and multiple experiments) having been examined (n = at least 3–6 animals-tumors).
Data analysis. Comparison of the effects of various treatments was performed using one way analysis of variance and a two tailed Student's t-test. Differences with a p value of <0.05 were considered statistically significant. Experiments shown are the means of multiple individual points from multiple experiments (±SEM). Characterization of synergistic and antagonistic interactions in cells exposed to a range of drug concentrations administered at a fixed ratio of one drug to another was done using median dose effect analysis in conjunction with a commercially available software program (CalcuSyn, Biosoft, Ferguson, MO).
Acknowledgements
This work was funded; to P.D. from The Goodwin Foundation, PHS grants (R01-DK52825, P01-CA104177, R01-CA150214), Department of Defense Award (W81XWH-10-1-0009). These studies were also funded in part by a grant from The Jim Valvano “V” foundation. P.D. is the holder of the Universal Inc., Professorship in Signal Transduction Research.
Abbreviations
- ERK
extracellular regulated kinase
- MEK
mitogen activated extracellular regulated kinase
- PI3K
phosphatidyl inositol 3 kinase
- FP
flavopiridol
- GX
obatoclax
- FLIP
flice inhibitory protein
- ca
constitutively active
- dn
dominant negative
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/13273
References
- 1.Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, et al. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- 2.De Azevedo WF, Jr, Mueller-Dieckmann HJ, Schulze-Gahmen U, Worland PJ, Sausville E, Kim SH. Structural basis for specificity and potency of a flavonoid inhibitor of human CDK2, a cell cycle kinase. Proc Natl Acad Sci USA. 1996;93:2735–2740. doi: 10.1073/pnas.93.7.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wesierska-Gadek J, Krystof V. Selective cyclin-dependent kinase inhibitors discriminating between cell cycle and transcriptional kinases: future reality or utopia? Ann N Y Acad Sci. 2009;1171:228–241. doi: 10.1111/j.1749-6632.2009.04726.x. [DOI] [PubMed] [Google Scholar]
- 4.Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 5.Senderowicz AM. Flavopiridol: the first cyclin-dependent kinase inhibitor in human clinical trials. Investig New Drugs. 1999;17:313–320. doi: 10.1023/a:1006353008903. [DOI] [PubMed] [Google Scholar]
- 6.Carlson BA, Dubay MM, Sauville EA, Brizuela L, Worland PJ. Flavopiridol induces G1 arrest with inhibition of cyclin-dependent kinase (CDK)2 and CDK4 in human breast carcinoma cells. Cancer Res. 1996;56:2973–2978. [PubMed] [Google Scholar]
- 7.Parker BW, Kaur G, Nieves-Neira W, Taimi M, Kohlhagen G, Shimizu T, et al. Early induction of apoptosis in hematopoietic cell lines after exposure to flavopiridol. Blood. 1998;91:458–465. [PubMed] [Google Scholar]
- 8.Gojo I, Zhang B, Fenton RG. The cyclin-dependent kinase inhibitor flavopiridol induces apoptosis in multiple myeloma cells through transcriptional repression and downregulation of Mcl-1. Clin Cancer Res. 2002;11:3527–3538. [PubMed] [Google Scholar]
- 9.Rosato RR, Almenara JA, Yu C, Grant S. Evidence of a functional role for p21WAF1/CIP1 downregulation in synergistic antileukemic interactions between the histone deacetylase inhibitor sodium butyrate and flavopiridol. Mol Pharmacol. 2004;65:571–581. doi: 10.1124/mol.65.3.571. [DOI] [PubMed] [Google Scholar]
- 10.Takada Y, Aggarwal BB. Flavopiridol inhibits NFκB activation induced by various carcinogens and inflammatory agents through inhibition of IκBα kinase and p65 phosphorylation: abrogation of cyclin D1, cyclooxygenase-2 and matrix metalloprotease-9. J Biol Chem. 2004;279:4750–4759. doi: 10.1074/jbc.M304546200. [DOI] [PubMed] [Google Scholar]
- 11.Almenara J, Rosato RR, Grant S. Synergistic induction of mitochondrial damage and apoptosis in human leukemia cells by flavopiridol and the histone deacetylase inhibitor suberoylanilide hydroxamic acid (Vorinostat) Leukemia. 2002;16:1331–1343. doi: 10.1038/sj.leu.2402535. [DOI] [PubMed] [Google Scholar]
- 12.Gao N, Dai Y, Rahmani M, Dent P, Grant S. Contribution of disruption of the nuclear factorkappaB pathway to induction of apoptosis in human leukemia cells by histone deacetylase inhibitors and flavopiridol. Mol Pharmacol. 2004;66:956–963. doi: 10.1124/mol.104.002014. [DOI] [PubMed] [Google Scholar]
- 13.Pytel D, Sliwinski T, Poplawski T, Ferriola D, Majsterek I. Tyrosine kinase blockers: new hope for successful cancer therapy. Anticancer Agents Med Chem. 2009;9:66–76. doi: 10.2174/187152009787047752. [DOI] [PubMed] [Google Scholar]
- 14.Carter CA, Kelly RJ, Giaccone G. Small-molecule inhibitors of the human epidermal receptor family. Expert Opin Investig Drugs. 2009;18:1829–1842. doi: 10.1517/13543780903373343. [DOI] [PubMed] [Google Scholar]
- 15.McCubrey JA, Milella M, Tafuri A, Martelli AM, Lunghi P, Bonati A, et al. Targeting the Raf/MEK/ERK pathway with small-molecule inhibitors. Curr Opin Investig Drugs. 2008;9:614–630. [PubMed] [Google Scholar]
- 16.Benavente S, Huang S, Armstrong EA, Chi A, Hsu KT, Wheeler DL, Harari PM. Establishment and Characterization of a Model of Acquired Resistance to Epidermal Growth Factor Receptor Targeting Agents in Human Cancer Cells. Clin Cancer Res. 2009;15:1585–1592. doi: 10.1158/1078-0432.CCR-08-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hida T, Ogawa S, Park JC, Park JY, Shimizu J, Horio Y, Yoshida K. Gefitinib for the treatment of non-small-cell lung cancer. Expert Rev Anticancer Ther. 2009;9:17–35. doi: 10.1586/14737140.9.1.17. [DOI] [PubMed] [Google Scholar]
- 18.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinibsensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 20.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Q, Esteva FJ. Cross-talk between the ErbB/HER family and the type I insulin-like growth factor receptor signaling pathway in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:485–498. doi: 10.1007/s10911-008-9107-3. [DOI] [PubMed] [Google Scholar]
- 22.Mueller KL, Hunter LA, Ethier SP, Boerner JL. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68:3314–3322. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin AP, Miller A, Emad L, Rahmani M, Walker T, Mitchell C, et al. Lapatinib resistance in HCT116 cells is mediated by elevated MCL-1 expression and decreased BAK activation and not by ERBB receptor kinase mutation. Mol Pharmacol. 2008;74:807–822. doi: 10.1124/mol.108.047365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant S, Dent P. Simultaneous interruption of signal transduction and cell cycle regulatory pathways: implications for new approaches to the treatment of childhood leukemias. Curr Drug Targets. 2007;8:751–759. doi: 10.2174/138945007780830764. [DOI] [PubMed] [Google Scholar]
- 25.Grant S. Is the focus moving toward a combination of targeted drugs? Best Pract Res Clin Haematol. 2008;21:629–637. doi: 10.1016/j.beha.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azmi AS, Mohammad RM. Non-peptidic small molecule inhibitors against Bcl-2 for cancer therapy. J Cell Physiol. 2009;218:13–21. doi: 10.1002/jcp.21567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15:1126–1132. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cragg MS, Harris C, Strasser A, Scott CL. Unleashing the power of inhibitors of oncogenic kinases through BH3 mimetics. Nat Rev Cancer. 2009;9:321–326. doi: 10.1038/nrc2615. [DOI] [PubMed] [Google Scholar]
- 29.Li R, Zang Y, Li C, Patel NS, Grandis JR, Johnson DE. ABT-737 Synergizes with Chemotherapy to Kill Head and Neck Squamous Cell Carcinoma Cells Via a NOXA-Mediated. Mol Pharmacol. 2009;75:1231–1239. doi: 10.1124/mol.108.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park MA, Zhang G, Mitchell C, Rahmani M, Hamed H, Hagan MP, et al. Mitogen-activated protein kinase kinase 1/2 inhibitors and 17-allylamino-17-demethoxy-geldanamycin synergize to kill human gastrointestinal tumor cells in vitro via suppression of c-FLIP-s levels and activation of CD95. Mol Cancer Ther. 2008;7:2633–2648. doi: 10.1158/1535-7163.MCT-08-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park MA, Yacoub A, Sarkar D, Emdad L, Rahmani M, Spiegel S, et al. PERK-dependent regulation of MDA-7/IL-24-induced autophagy in primary human glioma cells. Autophagy. 2008;4:513–515. doi: 10.4161/auto.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park MA, Yacoub A, Rahmani M, Zhang G, Hart L, Hagan MP, et al. OSU-03012 stimulates PKR-like endoplasmic reticulum-dependent increases in 70 kDa heat shock protein expression, attenuating its lethal actions in transformed cells. Mol Pharmacol. 2008;73:1168–1184. doi: 10.1124/mol.107.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park MA, Zhang G, Martin AP, Hamed H, Mitchell C, Hylemon PB, et al. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide-dependent CD95 and PERK activation. Cancer Biol Ther. 2008;7:1648–1662. doi: 10.4161/cbt.7.10.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang G, Park MA, Mitchell C, Walker T, Hamed H, Studer E, et al. Multiple cyclin kinase inhibitors promote bile acid-induced apoptosis and autophagy in primary hepatocytes via p53-CD95-dependent signaling. J Biol Chem. 2008;283:24343–24358. doi: 10.1074/jbc.M803444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell C, Park MA, Zhang G, Yacoub A, Curiel DT, Fisher PB, et al. Extrinsic pathway- and cathepsin-dependent induction of mitochondrial dysfunction are essential for synergistic flavopiridol and vorinostat lethality in breast cancer cells. Mol Cancer Ther. 2007;6:3101–3112. doi: 10.1158/1535-7163.MCT-07-0561. [DOI] [PubMed] [Google Scholar]
- 36.Martin AP, Mitchell C, Rahmani M, Nephew KP, Grant S, Dent P. Inhibition of MCL-1 enhances lapatinib toxicity and overcomes lapatinib resistance via BAK-dependent autophagy. Cancer Biol Ther. 2009;8:2084–2096. doi: 10.4161/cbt.8.21.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yip AY, Tse LA, Ong EY, Chow LW. Survival benefits from lapatinib therapy in women with HER2-overexpressing breast cancer: a systematic review. Anticancer Drugs. 2010;21:487–493. doi: 10.1097/CAD.0b013e3283388eaf. [DOI] [PubMed] [Google Scholar]
- 38.Tevaarwerk AJ, Kolesar JM. Lapatinib: a small-molecule inhibitor of epidermal growth factor receptor and human epidermal growth factor receptor-2 tyrosine kinases used in the treatment of breast cancer. Clin Ther. 2009;31:2332–2348. doi: 10.1016/j.clinthera.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 39.Henson ES, Johnston JB, Los M, Gibson SB. Clinical activities of the epidermal growth factor receptor family inhibitors in breast cancer. Biologics. 2007;1:229–239. [PMC free article] [PubMed] [Google Scholar]
- 40.Zoppoli G, Moran E, Soncini D, Cea M, Garuti A, Rocco I, et al. Ras-induced resistance to lapatinib is overcome by MEK inhibition. Curr Cancer Drug Targets. 2010;10:168–175. doi: 10.2174/156800910791054211. [DOI] [PubMed] [Google Scholar]
- 41.Kimple RJ, Vaseva AV, Cox AD, Baerman KM, Calvo BF, Tepper JE, et al. Radiosensitization of epidermal growth factor receptor/HER2-positive pancreatic cancer is mediated by inhibition of Akt independent of ras mutational status. Clin Cancer Res. 2010;16:912–923. doi: 10.1158/1078-0432.CCR-09-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newcomb EW, Lymberis SC, Lukyanov Y, Shao Y, Schnee T, Devitt M, et al. Radiation sensitivity of GL261 murine glioma model and enhanced radiation response by flavopiridol. Cell Cycle. 2006;5:93–99. doi: 10.4161/cc.5.1.2271. [DOI] [PubMed] [Google Scholar]
- 43.Pan J, Cheng C, Verstovsek S, Chen Q, Jin Y, Cao Q. The BH3-mimetic GX15-070 induces autophagy, potentiates the cytotoxicity of carboplatin and 5-fluorouracil in esophageal carcinoma cells. Cancer Lett. 2010 28;293:167–174. doi: 10.1016/j.canlet.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Yang G, Wu L, Chen S, Zhu L, Huang P, Tong L, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs radiation-induced bystander effect. Br J Cancer. 2009;100:1912–1916. doi: 10.1038/sj.bjc.6605087. [DOI] [PMC free article] [PubMed] [Google Scholar]