Abstract
Flowering plants have the unique ability to produce new organs continuously, for hundreds of years in some species, from stem cell populations maintained at their actively growing tips. The shoot tip is called the shoot apical meristem, and it acts as a self-renewing source of undifferentiated, pluripotent stem cells whose descendents become incorporated into organ and tissue primordia and acquire different fates. Stem cell maintenance is an active process, requiring constant communication between different regions of the shoot apical meristem to coordinate loss of stem cells from the meristem through differentiation with their replacement through cell division. Stem cell research in model plant systems is facilitated by the fact that mutants with altered meristem cell identity or accumulation are viable, allowing dissection of stem cell behavior by using genetic, molecular, and biochemical methods. Such studies have determined that in the model plant Arabidopsis thaliana stem cell maintenance information flows via a signal transduction pathway that is established during embryogenesis and maintained throughout the life cycle. Signaling through this pathway results in the generation of a spatial feedback loop, involving both positive and negative interactions, that maintains stem cell homeostasis. Stem cell activity during reproductive development is terminated by a temporal feedback loop involving both stem cell maintenance genes and a phase-specific flower patterning gene. Our current investigations provide additional insights into the molecular mechanisms that regulate stem cell activity in higher plants.
A major focus of present-day biological research is on the question of how stem cell fate is controlled during development. Plants make excellent model systems for studying this problem, because, unlike animals, plants maintain a reservoir of stem cells in their shoot and root apical meristems throughout their life spans. Thus plants provide an essentially endless supply of stem cells for study. Like animal stem cells, plant stem cells are self-renewing and have the potential to form many different organs and tissues. Stem cells in the shoot apical meristem (SAM) are the progenitors of all cells that make up stems, leaves, branches, and flowers. The root apical meristem is the source of all of the cells of the primary and lateral root system. Determining how stem cells are established and maintained in apical meristems is fundamental both for addressing the basic biological features of stem cell behavior and understanding many aspects of plant development. This article will discuss recent progress in our laboratory, and in numerous other laboratories, in unraveling the genetic regulatory mechanisms that maintain stem cell fate in the SAM of the model plant Arabidopsis thaliana.
Although mature plants have an enormous variety of forms, embryonic plant architecture is sufficiently limited that it is difficult to distinguish between embryos from different plant species based solely on morphological criteria. Likewise, a mature Arabidopsis embryo has a morphologically simple structure. It consists of an embryonic stem, two embryonic seed leaves called cotyledons, and a root apical meristem and a SAM at the basal and apical poles of the embryo, respectively. Because plants do not use cell migration or programmed cell death to achieve morphogenesis, the patterning of the embryo occurs mainly through symmetric and asymmetric cell divisions in different places and planes and through cell expansion (1). Plants also do not set aside a dedicated germ line during embryogenesis; instead, germ cells form independently during the generation of each flower from the SAM.
The elaboration of plant architecture occurs mainly postem-bryonically. After germination, the Arabidopsis SAM begins to initiate vegetative organs, the leaves, that form a basal rosette. After several weeks of vegetative development the plant undergoes the transition to flowering, during which the stem elongates and begins to produce secondary branches and flowers. The primary SAM at this stage is called the inflorescence, or flower-bearing, meristem. Flowers are formed from floral meristems, which are derived from the flanks of the SAM. Floral meristems have the same general structure and organization as SAMs. Each floral meristem produces a flower consisting of four types of floral organs: sepals, petals, stamens (male reproductive organs), and carpels, the latter of which fuse to form the gynoecium (the fruit) that encloses the seeds of the next generation. One important difference between a SAM and a floral meristem is that a SAM grows indefinitely and produces an unspecified number of lateral organs, whereas a floral meristem produces a specified number of organs and terminates in the formation of the central carpels.
The Arabidopsis SAM is patterned gradually during embryogenesis (2), culminating in the formation of a highly organized structure with overlapping functional domains (Fig. 1). The self-renewing stem cell pool is confined to the most apical, central portion of the meristem (3). Infrequent cell divisions in this region, which is called the central zone, causes displacement of daughter cells outward into the peripheral region, where they begin to divide more frequently (4). These more rapidly dividing peripheral zone cells begin to undergo differentiation and become incorporated into organ primordia on the meristem flanks. Cells in the central zone also divide downward into the interior of the meristem, a region called the rib zone that contributes to the meristem pith.
Fig. 1.
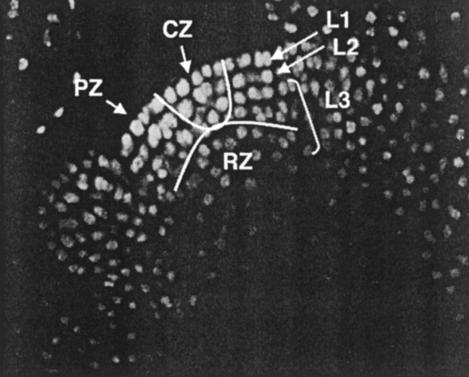
Organization of the Arabidopsis SAM. Confocal laser scanning microscope optical section through a WT flowering SAM and developing floral meristem. The section shows the cell layers (L1, L2, and L3) and the histologically defined domains. The central zone (CZ) lies at the apex of the SAM and harbors the stem cell reservoir. The surrounding peripheral zone (PZ) consists of progenitor cells for lateral organs, and the underlying rib zone (RZ) consists of progenitor cells for the core of the stem. The SAM has been stained with propidium iodide to visualize the cell nuclei. [Modified with permission from ref. 33 (Copyright 2000, Elsevier).]
The stem cell population consists of three clonally distinct cell layers (Fig. 1), as does the peripheral meristem region and each lateral organ primordium (3, 5). The epidermal layer, the L1, forms a single cell layer that remains distinct from the other layers because the cell divisions within it are always perpendicular to the plane of the meristem surface (anticlinal). The subepidermal L2 layer is also a single cell thick and divides exclusively in the anticlinal plane. The underlying L3 consists of all of the cells in the interior of the SAM, which divide in all planes. Yet despite the fact that cell divisions in the SAM are highly regular, mosaic analyses have demonstrated that, beyond the general maintenance of the clonal layers, cell lineage patterns are not fixed (6-8). Instead, the cells in the SAM rely almost entirely on positional cues to determine their correct fates (9), and cell fate and cell proliferation information must be communicated both within and between the different meristem layers for proper development to occur. Thus, as in animals, the local environment of the plant meristem provides a niche for the maintenance of the stem cell population.
Mutations That Alter Stem Cell Fate
Our entrée into understanding the mechanism of plant stem cell maintenance has come from the analysis of Arabidopsis mutants that affect this process. Remarkably, loss-of-function shootmeristemless (stm) mutants, which lack a SAM and therefore contain no stem cells, survive embryogenesis and germinate normally (10), although they fail to produce any postembryonic lateral organs. Plants carrying loss-of-function mutations at the WUSCHEL (WUS) locus form an embryonic SAM, but it is not maintained after the production of a few lateral organs (11). These lateral organs often initiate ectopically from the center of the SAM, indicating that the stem cells at the shoot apex do not maintain their correct fate. New SAMs are established de novo at the base of the wus mutant cotyledons and leaves, but these meristems also terminate prematurely. This stop-and-start growth pattern, which culminates in a bushy or tousled-looking (wuschel in German) plant, continues through the floral phase. wus mutant inflorescence meristems produce a reduced number of flowers compared with WT plants, and most of the flowers terminate prematurely in a single stamen. Thus the wus mutant phenotypes define a gene whose WT function is required to promote shoot and floral meristem activity by maintaining the central stem cell reservoir.
WUS encodes a novel subtype of homeodomain protein that is nuclear localized and predicted to act as a transcription factor (12). WUS mRNA is first detected when embryos reach the 16-cell stage. At this time, WUS expression is confined to the four inner cells of the apical region. As embryogenesis proceeds, asymmetric cell divisions lead to an asymmetric distribution of WUS transcripts and the gradual localization of the WUS expression domain to the internal layers of the developing SAM. WUS mRNA is confined to a small group of cells in the internal layers of shoot, and later, floral meristems, and is not detected in either the epidermal or subepidermal layers of these tissues. The WUS expression domain is maintained by the activity of the FASCIATED1 (FAS1) and FAS2 genes, which encode components of chromatin assembly factor-1 (13). FAS1 and FAS2 may therefore promote stable WUS gene transcription by facilitating the appropriate chromatin conformation.
Arabidopsis mutants with phenotypes that are opposite those of wus phenotypes have also been characterized. Plants carrying loss-of-function mutations at the CLAVATA1 (CLV1), CLV2, or CLV3 loci generate many excess SAM cells and form enlarged meristems that grow as a ring or a strap instead of as a point. From this phenotype we infer that the WT function of the three CLV genes is to restrict meristem cell accumulation. This requirement for CLV gene activity begins during embryogenesis, because the SAMs of mature clv1 and clv3 embryos are already significantly enlarged relative to the SAMs of WT embryos and contain more stem cells (14, 15). Excess stem cells continue to accumulate throughout the life of clv mutant plants, so that by the transition to flowering clv shoot meristems are greatly enlarged and produce many more floral meristems than WT plants.
The floral meristems produced by clv mutants are also enlarged compared with the WT and contain excess stem cells. The enlarged floral meristems lead to an increase in the number of organs in clv flowers. WT flowers are arranged into four concentric rings of organs, called whorls. In Arabidopsis, organ number in each whorl is nearly invariant: the vast majority of WT flowers consist of four sepals, four petals, five to six stamens, and two fused carpels (16). Flowers of clv null mutants contain many additional organs, including up to seven or eight carpels that fuse to form a club-shaped fruit. It is from this phenotype that the mutants are named “clavata,” from the Latin term “clavatus” meaning club-like. In addition to generating extra organs, the floral meristems of clv1 and clv3 null mutants often contain a mass of proliferating stem cells that erupt through the gynoecium. This phenotype is further evidence that the CLV loci are required to prevent unrestricted stem cell amplification in the center of the floral meristem.
Genetic analysis has revealed that the three CLV genes and WUS interact to control stem cell fate during development. clv1 and clv3 null mutants have nearly indistinguishable phenotypes, and clv1 clv3 double mutants appear identical to either single mutant (15). Further, plants that are doubly heterozygous for clv1 and clv3 alleles display an enlarged meristem phenotype, indicating that each gene is sensitive to the dose of the other. clv2 null mutants have slightly weaker shoot and floral meristem phenotypes than clv1 and clv3 null mutants, and clv1 and clv3 are epistatic to clv2 with respect to those traits (17). However, clv2 mutants, unlike clv1 and clv3 mutants, also display phenotypes in nonmeristematic tissues, indicating that CLV2 functions more widely in development. Double mutants generated between wus null alleles and any clv allele have a wus mutant phenotype (11), revealing that wus is completely epistatic to clv1, clv2, and clv3. Thus all four genes act in the same genetic pathway to maintain the appropriate amount of stem cell accumulation in shoot and floral meristems.
A Short-Range Signaling Pathway Maintains Stem Cell Homeostasis
All three CLV genes have been cloned and found to encode components of a meristem signal transduction pathway. CLV1 encodes a leucine-rich repeat (LRR) receptor-like kinase (LRR-RLK) (18), one of 216 in the Arabidopsis genome (19). Members of this family contain multiple tandem extracellular LRRs, a transmembrane domain, and an intracellular serine/threonine kinase domain. The LRR-RLK family, which consists of 13 subclasses, is the largest class of receptor kinases in plants. The LRR-RLKs and other Arabidopsis RLKs form a monophyletic family that groups with Drosophila Pelle and other Pelle-like cytoplasmic kinases in phylogenetic analyses (19). Based on the fact that LRRs are considered to be protein-binding motifs, and that LRR-containing proteins in plants and animals are involved in protein-protein interactions and signal transduction (20), it has been proposed that the extracellular LRRs of the LRRRLKs bind protein or peptide ligands. The functions of a half-dozen Arabidopsis LRR-RLKs have been defined to date. They play roles in various plant signaling pathways, including those regulating different aspects of development (21-23), hormone signaling (24), and disease resistance (25).
CLV2 and CLV3 also encode members of large, plant-specific gene families. CLV2 encodes a receptor-like protein with 21 extracellular LRRs, a transmembrane domain, and a short cytoplasmic tail (26). There are ≈30 CLV2-like genes in the Arabidopsis genome, but their functions are unknown. CLV3 encodes a 96-aa predicted extracellular polypeptide (27) that is a founding member of the CLV3/ESR-related (CLE) protein family (28). The members of this family, which are found in many distantly related plant species, share common sequence elements including an amino-terminal signal peptide and a 14-aa region of homology at the carboxyl terminus. Two dozen CLE genes have been identified in Arabidopsis (28, 29). These genes are expressed in a wide range of tissues during development (29) and may function as ligands for RLKs in many plant signal transduction pathways.
The expression patterns of the CLV3 and CLV1 genes revealed important clues to their function. We found that, beginning during embryogenesis and continuing throughout the life of the plant, CLV3 marks the stem cell population in the SAM (27). CLV3 transcripts are detected predominantly in the superficial L1 and L2 cell layers at the apex of the meristem and in only a few underlying L3 cells. CLV3 mRNA is not found in peripheral meristem cells, mature stem tissues, or lateral organs. CLV3 is also expressed in the stem cell populations of all axillary shoot meristems and floral meristems. In floral meristems, CLV3 is detected until the stem cell population is consumed in the formation of the carpel primordia (see below), when CLV3 transcripts disappear from the flower. CLV1 mRNA is also induced during embryogenesis, at the same time as CLV3, and is likewise restricted to shoot and floral meristem cells (18). However, CLV1 transcripts are found predominantly in the L3 cells of the meristem, largely beneath the CLV3 expression domain, and are not detected in the L1 epidermal layer. Thus CLV1-expressing cells and CLV3-expressing cells are neighbors, suggesting that the two meristem domains may communicate with one another through a CLV3/CLV1 signal transduction pathway. CLV2 is also expressed in shoots and flowers based on RNA gel blot analysis (26), but the precise CLV2 expression domain in meristems has not been reported.
Important inroads have been made into determining the mechanism of CLV signal transduction. There is now solid biochemical evidence that CLV1 and CLV2 are bound together in a signaling complex (Fig. 2). The CLV1 kinase domain is capable of both autophosphorylation and transphosphorylation on serine residues, indicating that CLV1 is a bona fide receptor kinase (30, 31). CLV1, a predicted 105-kDa protein, is detected in size-fractionated meristem extracts in two different complexes: a 185-kDa complex and a 450-kDa complex (32). CLV1 and CLV3 are both components of the 450-kDa complex, whereas CLV3 is not detected in the 185-kDa complex. The 450-kDa complex does not form in clv3 mutants or clv1 mutants that lack an active kinase domain. The 450-kDa fraction is therefore likely to represent the active CLV receptor signaling complex, whereas the 185-kDa complex is inactive. Although specific antibodies for CLV2 have not yet been reported, its presence in the 450-kDa complex is inferred from the observation that this complex does not form in clv2 mutants although high levels of CLV1 and CLV3 transcripts are present (26). Mutations in CLV2 also reduce the accumulation of the 185-kDa complex, suggesting that CLV2 may also be a subunit of the inactive complex that forms a disulphide-linked heterodimer with CLV1.
Fig. 2.
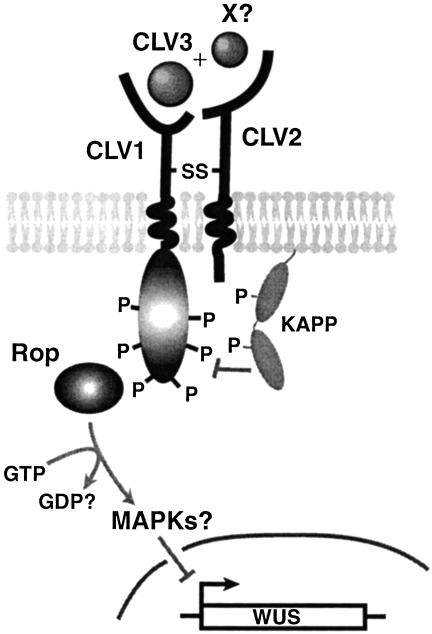
Schematic of the CLV signaling complex. The CLV1 LRR-RLK forms a heteromeric complex with the CLV2 receptor-like protein at the plasma membrane of interior SAM cells. Binding of the CLV3 polypeptide, possibly in association with another protein (X), is proposed to stimulate assembly of an active signaling complex that also contains a phosphatase (KAPP) and a Rho-like GTPase (Rop). The signal is relayed from the cytosol to the nucleus, potentially via a MAPK cascade, to limit WUS expression. P, phosphorylation site; SS, disulphide bond. [Reprinted with permission from ref. 68 (Copyright 2002, Annual Reviews, www.annualreviews.org).]
The active CLV signaling complex contains several additional cytosolic proteins. One is a type 2C kinase-associated protein phosphatase (KAPP), which interacts with several different LRR-RLKs (25, 34, 35). KAPP is expressed in Arabidopsis meristems in a broad region that encompasses the CLV1 expression domain (30), and KAPP protein binds to and dephosphorylates the phosphorylated form of the CLV1 kinase domain (30, 31). KAPP binds to the CLV1 kinase domain through its forkhead-associated (FHA) domain, and site-directed mutagenesis of conserved FHA residues abolishes the ability of KAPP to interact with RLKs (36). KAPP overexpression phenotypes mimic those of weak loss-of-function clv alleles (30), whereas reduction in KAPP transcription levels can suppress the clv1 mutant phenotype (31). These data are consistent with KAPP acting as a negative regulator of CLV signaling by dephosphorylating CLV1. AtSERK1, a plasma membrane-localized LRRRLK, was recently shown to become sequestered in intracellular vesicles when transiently coexpressed with KAPP (35). KAPP dephosphorylation of threonine residues in the AtSERK1 kinase domain A loop was found to play an active role in the receptor internalization. These results suggest that KAPP is an integral part of a endocytosis mechanism that internalizes AtSERK1, and possibly other LRR-RLKs such as CLV1, during receptor-mediated signaling.
An additional cytoplasmic component of the active CLV complex, identified through coimmunoprecipitation experiments (32), is a member of the plant-specific Rop subfamily of Rho GTPases (37). Members of the Rho GTPase family, which includes CDC42, Rac, and Rho, are key cytosolic components of cell surface-receptor signaling cascades that regulate cytoskeletal organization, polarized cell growth, and many other cellular processes in animals and fungi (38-40). Arabidopsis contains at least 10 Rop GTPases (41, 42), some of which have defined roles in mediating tip growth in pollen tubes and root hairs (37, 43) and others of which act in various developmental processes (44). It is not yet clear which Rop protein(s) participates in CLV signaling, because the antibody used in the biochemical experiments cross-reacts with multiple Rop isoforms (32).
How is the signal relayed downstream of the CLV complex? Although the exact mechanism is currently unknown, a model has been proposed based on precedents in other systems (32). Rho/Rac GTPases are members of the Ras superfamily of cytoplasmic signal transduction molecules, many of which relay signals through a mitogen-activated protein kinase (MAPK) pathway. The Arabidopsis genome contains multiple genes encoding MAPK, MAPKK, and MAPKKK components of this pathway (45). A complete MAPK cascade has recently been reported (46) to function downstream of the Arabidopsis flagellin receptor FLS2, which encodes an LRR-RLK (25), in innate immune responses. Based on this paradigm, once the CLV1 kinase domain becomes activated via phosphorylation, it might associate with and activate the Rop GTPase, which in turn could activate a MAPK cascade that ultimately leads to changes in target gene expression. Functionally redundant MAPK signaling proteins were discovered at several steps in the FLS2 pathway, and several of the components of the FLS2 cascade may also function in other pathways (46). If this is a common theme for other LRR-RLK signaling pathways, it could account for the lack of MAPK cascade mutants identified to date in the CLV signaling pathway.
What is the biological basis for signaling through the CLV complex in meristematic cells? Genetic and molecular evidence has led to the conclusion that the CLV signaling pathway is part of a negative feedback loop that controls stem cell fate in the Arabidopsis shoot apex. Our first insight came from an experiment designed to determine what happens to CLV3 expression in the absence of the CLV1 receptor kinase. Because clv1 mutant phenotypes are very similar to clv3 mutant phenotypes, we tested the possibility that the clv1 phenotypes might be caused by a reduction in either the domain or level of CLV3 expression. In contrast to our prediction, we observed that CLV3 was expressed at very high levels in a greatly enlarged domain in clv1 SAMs (27). This result indicates that CLV3 expression is not reduced in clv1 mutant plants, but instead that CLV1 normally acts to restrict the number of CLV3-expressing cells in WT meristems. Similarly, the CLV1 expression domain is enlarged in clv3 mutant plants. In fact, in clv1, clv2, or clv3 mutants the expression domains of CLV1 and CLV3 enlarge coordinately (17, 27). From these data we inferred the presence of two opposing stem cell regulatory pathways. The expansion of the CLV expression domains would be normally controlled by a positive, stem cell-promoting pathway, which in turn would be negatively regulated by the stem cell-restricting CLV pathway. The stem cell-promoting pathway would then also promote the expression of the CLV genes by causing the enlargement of the cell populations that express them.
We were able to separate these two pathways genetically by expressing CLV3 constitutively in Arabidopsis plants under the control of the cauliflower mosaic virus 35S promoter. Transgenic 35S::CLV3 plants expressing high levels of CLV3 mRNA germinated normally, but stopped initiating organs after producing a few leaves (47). These results indicate that stem cells are not correctly maintained when CLV3 is constitutively expressed at high levels. They further demonstrate that CLV3 is sufficient to restrict stem cell accumulation throughout the SAM. Flower meristems could be produced by lines expressing lower levels of constitutive CLV3. Such meristems failed to initiate the inner organ types, again demonstrating an inability to maintain stem cells. The level of CLV3 protein therefore appears to be the critical factor that determines the amount of stem cells that are produced in both shoot and floral meristems. In addition, when the 35S::CLV3 transgene was introduced into the clv1 or clv2 mutant background, the resulting plants constitutively expressed CLV3 at high levels but exhibited the typical clv mutant phenotype. Thus CLV3 signaling requires functional CLV1 and CLV2 proteins, and the terminal meristem phenotypes observed in the transgenic 35S::CLV3 plants result from enhanced CLV3 signaling through the CLV1 receptor complex.
WUS is a key target of regulation by the CLV signal transduction pathway, as determined by examining its expression pattern in clv3 mutant and CLV3 overexpressing plants (47). In WT plants, WUS mRNA is restricted to the internal layers of shoot and floral meristems, beneath the CLV3 domain, and is not detected in either the epidermal or subepidermal layers. Transgenic CLV3 overexpression lines phenocopy the wus mutant phenotype, suggesting that WUS activity is down-regulated in response to signaling through the CLV complex. Consistent with this hypothesis, the WUS expression domain expands laterally and into the subepidermal layer of clv3 mutant meristems (47). Conversely, WUS mRNA is not detected in the arrested meristems of 35S::CLV3 plants, indicating that one outcome of enhanced CLV signaling is a severe reduction in the level of WUS transcription. WUS therefore appears to respond directly, but negatively, to the level of CLV3 signaling: a high level of CLV3 reduces the size of the WUS expression domain, whereas a low level allows it to expand.
The positive, stem cell-promoting pathway depends on activity of the WUS gene. Driving WUS expression ectopically in all initiating organ primordia on the flanks of the SAM results in seedlings that form normal cotyledons and an apex that consists entirely of undifferentiated meristematic cells (48). CLV3 mRNA, which in WT plants is found only at the very apex of the meristem, can be detected on the periphery of this mound of cells. This result indicates that WUS misexpression is sufficient to induce CLV3 transcription and stem cell fate. Based on these data, WUS is proposed to enable the cells that express it to act as an “organizing center” that confers stem cell fate on the overlying cells of the meristem (12). WUS is therefore a key component of the stem cell-promoting pathway, which functions to control the stem cell-restricting CLV pathway by regulating CLV3 transcription. The regulation of CLV3 by WUS is probably indirect, because the WUS expression domain does not overlap with the CLV3 domain and there is no evidence that WUS protein moves out of the cells in which it is made.
Thus in WT Arabidopsis plants, stem cell maintenance is mediated by short-range signaling events. The CLV3 signal is likely to be released from the stem cells at the apex of the meristem and bind a CLV1/CLV2 receptor complex in underlying cells. Signaling through the CLV pathway limits WUS activity by restricting its expression to a narrow domain of cells in the deeper layers of the meristem. Constitutive signaling through CLV3 enhances this negative pathway, causing down-regulation of WUS and complete loss of stem cells. Disruption of the negative pathway in clv mutants causes the WUS expression domain to expand laterally and upward into the subepidermal cell layer. Activity of the positive pathway, mediated by WUS, promotes expression of CLV3 and maintenance of the stem cell domain. This mutual regulation, involving both positive and negative interactions, provides a feedback system required to maintain an appropriately sized stem cell reservoir throughout Arabidopsis development.
This negative feedback loop elegantly compensates for the departure of cells from the meristem during organ formation. As stem cell daughters enter the transition zone and become incorporated into organ primordia, the decrease in stem cell number leads to a reduction in the level of the CLV3 signal. The drop in negative signaling through the CLV pathway causes the expansion of the WUS-expressing cell population. This, in turn, triggers the amplification of the stem cell population via the stem cell-promoting pathway, until the level of CLV3 produced by the stem cells rises sufficiently to restrict further expansion of the WUS domain and equilibrium is once again attained.
Signaling Via CLV3
One major question to be addressed is how the CLV1/CLV2 receptor complex is activated by CLV3. Based on its sequence, CLV3 is predicted to be exported through the secretory pathway to the extracellular space by the default pathway for soluble plant proteins. CLV3 has been shown to act in a cell nonautonomous fashion (27), but the expression data do not exclude the possibility that activation of the CLV pathway occurs intracellularly within those few cells in the interior of the SAM that express both CLV3 and the receptor complex. In collaboration with Enrique Rojo, Valentina Kovaleva, and Natasha Raikhel at the University of California, Riverside, we devised several assays to determine whether the CLV3 protein is secreted and whether secretion is required for its function in vivo (49).
To test the prediction that the CLV3 protein is secreted, we used two translational fusions of CLV3 to the amino terminus of β-glucuronidase, one full length and the other lacking the putative signal peptide, in transient subcellular localization assays. The fusion protein lacking the signal peptide was detected in the cytoplasm, whereas the full-length CLV3-GUS fusion protein was detected only in the extracellular space. Thus the CLV3 protein is exported through the secretory pathway, and the presence of its signal peptide is required for its extracellular localization. Additional transient assays, in which full-length CLV3-GUS was cobombarded together with a nuclear-localized protein as a marker for transformed cells, show that CLV3 is capable of moving beyond the cells in which it is made (V.K.S. and J.C.F., unpublished data). We confirmed that these results were relevant in vivo by overexpressing full-length and truncated CLV3-GFP fusion proteins in clv3 mutant plants. We found that plants transformed with the full-length 35S::CLV3-GFP fusion protein rescued the clv3 mutant phenotype and showed GFP florescence only in the extracellular space. Plants transformed with the truncated version of CLV3 fused to GFP did not rescue the clv3 phenotype and showed GFP florescence only in the cytoplasm. Based on these data, we conclude that CLV3 is a secreted protein both in vitro and in planta.
Next, we asked whether CLV3 secretion is essential for its activity in shoot and floral meristems. We tested whether preventing the CLV3 protein from reaching the cell surface would block its function in restricting stem cell accumulation (Fig. 3). Overexpression constructs were generated in which CLV3 was tagged with a vacuolar sorting signal to direct the protein to the vacuole rather than the extracellular space (Vac constructs). We also generated several overexpression constructs in which CLV3 was tagged with a mutated vacuolar sorting signal that forces the protein into the secretory pathway (Sec constructs). When these constructs were introduced into clv3 null mutant or WT plants, several phenotypic classes were distinguished. Most of the Sec-containing plants showed complete rescue of the clv3 phenotype or had a gain-of-function phenotype, whereas the majority of Vac-containing plants had little or no rescue of the phenotype. A few Vac-containing plants showed a gain-of-function phenotype, which could be attributed to saturation of the vacuolar sorting pathway by high levels of CLV3.
Fig. 3.
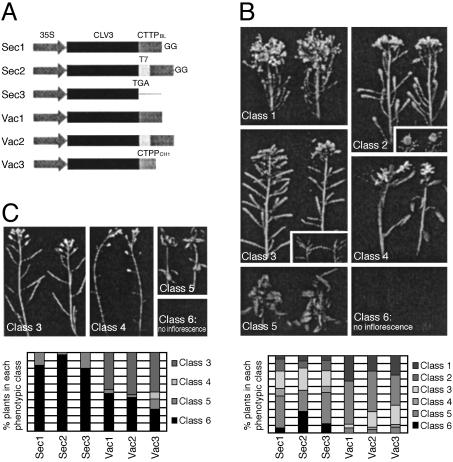
Targeting to the vacuole blocks the activity of CLV3. (A) Scheme of the fusion constructs. The three Vac constructs contain fusions of CLV3 to the C-terminal vacuolar sorting signal from barley lectin (ctVSSBL) or tobacco chitinase A (ctVSSCH). The ctVSSBL with two additional Gly residues (GG), which no longer functions as a vacuolar-sorting signal, was attached at the carboxyl terminus of CLV3 (Sec1) or CLV3-T7 (Sec2). Sec3 contains the full-length CLV3 protein with no additional tag. (B) clv3-2 plants transformed with the fusion constructs. Primary transformants were grouped into six phenotypic classes according to the severity of the meristem phenotype. Class 1 plants showed no transgene activity and resembled untransformed clv3-2 plants. Class 6 plants showed the strongest transgene activity and exhibited a gain-of-function phenotype in which the SAM of each plant terminated prematurely before flowering. Plants in the other classes fell between these two extremes. The graph at the bottom shows for each construct the percentage of transformed plants recovered in each phenotypic class. (C) WT plants transformed with the fusion constructs. Primary transformants were grouped into classes 3, 4, 5, and 6. The graph at the bottom shows for each construct the percentage of transformed plants recovered in each phenotypic class. [Reprinted with permission from ref. 49 (Copyright 2002, American Society of Plant Biologists).]
To localize the CLV3 protein in the different transgenic lines, we performed immunogold labeling with an antibody-directed, T7-tagged CLV3 protein. CLV3 protein was detected in the cell wall of Sec-containing plants, but not in Vac-containing plants that failed to rescue the clv3 phenotype. CLV3 was also detected in the cell wall of Vac-containing plants that displayed a gain-of-function phenotype. This observation confirmed that the vacuolar sorting pathway became saturated in these lines, and thus the excess CLV3 protein was secreted and able to interact with the CLV1 complex. Therefore we conclude from this work that CLV3 functions as a secreted protein that activates the CLV stem cell signaling pathway in the extracellular space.
A Temporal Feedback Loop Terminates Stem Cell Activity in Flowers
Flowers are produced from floral meristems, which are derived from the SAM and likewise harbor a stem cell reservoir that provides cells for the generation of the floral organs. Each floral meristem forms as a primordium on the flank of the SAM. As it grows the floral meristem initiates three concentric rings of organs (sepals, petals, and stamens) and then the stem cells are consumed in the formation of the carpels that comprise the central gynoecium. Thus unlike SAMs, in which the stem cell population is maintained indefinitely to provide progeny cells for continuous organogenesis, floral meristems ultimately terminate stem cell production to permit the female reproductive organs to differentiate in the center of the flower.
Floral meristem activity is regulated by several overlapping sets of regulatory factors. The CLV signaling pathway is active at the floral meristem apex until carpel initiation, when CLV1, CLV3, and WUS all are down-regulated to undetectable levels. Floral meristem identity is conferred by transcription factors such as LEAFY (LFY) and APETALA1 (AP1) (50, 51), which can convert shoot meristems into floral meristems (52, 53). The floral meristem identity genes induce the transcription of flower-specific homeotic genes in overlapping spatial domains of the floral meristem (54, 55). Activity of the floral homeotic gene products in four different combinations then specifies the identity of the sepals, petals, stamens, and carpels (56).
The floral homeotic gene AGAMOUS (AG) encodes a MADS-domain transcription factor (57) that plays key roles in both floral organ identity specification and stem cell termination. ag mutations cause the transformation of stamens into petals and replacement of the central carpels with an entirely new flower (58). The resulting ag flowers consist entirely of sepals and petals and resemble shoots because they retain a stem cell population at their apex (59) and continue to produce organs indefinitely. AG overexpression has the opposite effect: instead of maintaining indeterminate growth, the SAMs of 35S::AG plants terminate in a solitary flower (60). Thus AG is required to terminate stem cell activity in the center of the developing flower and is sufficient to convert a shoot meristem into a floral meristem.
ag mutants and wus mutants have opposite flower phenotypes, and genetic experiments demonstrate that these two genes play antagonistic roles in regulating floral meristem activity. WUS expression persists in ag mutant flowers even after the formation of many organs (59, 61). AG is therefore a negative regulator of WUS transcription and, consequently, of stem cell maintenance. Stem cell termination is restored in ag wus double mutant flowers, which resemble wus single mutant flowers, indicating that the capability of ag flowers to continuously produce new organs depends on the ectopic activity of WUS. Thus prolonged WUS expression is sufficient to permit indefinite floral meristem activity, and a key role of AG is to down-regulate WUS and terminate stem cell maintenance in the center of the developing flower.
The expression of AG is regulated by the floral meristem identity pathway and also the stem cell signaling pathway (Fig. 4). AG is initially transcribed in the center of developing floral meristems in the cells that will ultimately become specified as stamens and carpels, and AG expression in these organs persists until the late stages of flower development (57). AG is directly activated by LFY (55), which binds to regulatory elements in the second intron of the gene. However, LFY protein is distributed throughout the floral meristem (62), and thus does not confer region-specific AG induction. This function is provided by WUS, which is expressed in the center of floral meristems in a subset of cells that eventually express AG. Although wus mutant flowers do not display homeotic organ transformations they lack all carpels and most stamens, the organ types that are specified by AG. In addition, misexpression of WUS in flowers causes the formation of ectopic stamens and carpels as a consequence of ectopic AG activation. Recent reports have shown that WUS is also a direct activator of AG, acting through a homeodomain protein consensus binding site in the second intron (59, 61). These sites are adjacent to the LFY binding sites, although the two proteins bind DNA independently (61). Thus LFY provides the flower specificity and WUS provides the regional specificity for AG induction in the central region of floral meristems.
Fig. 4.
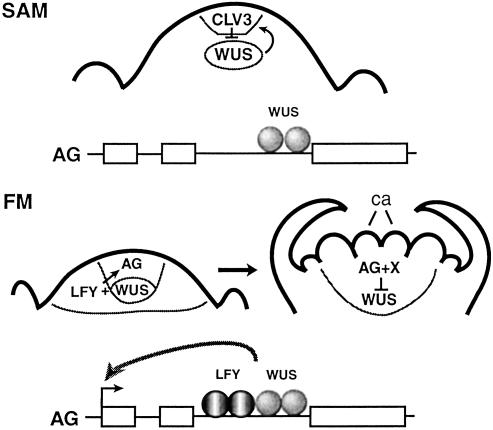
Temporal feedback loop regulating stem cell termination in determinate floral meristems. (Upper) Schematic of an indeterminate SAM, showing the interaction between CLV3 and WUS in their respective domains (gray circles). In the SAM, LFY is absent and AG expression is not induced. (Lower) Schematic of a determinate floral meristem over time. LFY is present throughout the young floral meristem. Both LFY and WUS bind to enhancer sequences and cooperate to induce AG transcription in the center of the developing flower. At the time of carpel (ca) initiation, AG and an additional factor (X) repress WUS expression to terminate stem cell activity. [Reprinted with permission from ref 68 (Copyright 2002, Annual Reviews, www.annualreviews.org).]
A third gene that appears to be involved in AG activation in Arabidopsis floral meristems is ULTRAPETALA (ULT). We have shown that mutations in ULT cause shoot and floral meristem enlargement, leading to the production of extra flowers from the SAMs and extra floral organs from the floral meristems (63). ult mutant floral meristems also produce some additional organs in the center of the flower before terminating, reminescent of ag mutants. AG activation is delayed in the very center of ult floral meristems (63), whereas overexpression of ULT leads to premature and ectopic AG induction (C.C. and J.C.F., unpublished data). Thus ULT, which encodes a member of a plant-specific family of novel proteins (C.C. and J.C.F., unpublished data), is required under normal circumstances for the correct timing of AG induction in floral meristems.
Thus the termination of stem cell activity in floral meristems is mediated by a temporal negative feedback loop requiring both flower patterning and stem cell maintenance genes. As in SAMs, WUS promotes maintenance of the stem cell domain in floral meristems to permit the generation of the full complement of floral organs. In addition, WUS and the flower-specific factor LFY bind independently to AG regulatory sequences and cooperate to direct AG transcription in the center of developing floral meristems, with ULT providing temporal specificity. Once activated, AG feeds back to repress WUS, thus terminating stem cell activity and allowing the differentiation of the central cells into carpel primordia. Unlike the other target of WUS induction, CLV3, AG does not require the constant presence of WUS to maintain its expression and consequently AG transcription persists even after WUS has been switched off.
Conclusions and Final Thoughts
Long-term maintenance of a stem cell population is critical for the particular developmental habit of plants, that is, continuous organ formation to achieve maximal growth under constantly changing environmental conditions. The past 10 years have produced a quantum leap in our understanding of plant stem cell activity from a descriptive to a mechanistic level. We and others have demonstrated that stem cell fate in shoot and floral meristems is controlled by a signal transduction pathway consisting of the CLV receptor complex and the WUS transcription factor. We have shown that this pathway consists of both plant-specific proteins and those with similarity to animal proteins, and that activation of the pathway occurs extracellularly. We have also determined that the CLV/WUS pathway functions as a homeostatic feedback loop that elegantly corrects for fluctuations in stem cell number that occur as a consequence of organogenesis. Finally, studies in several labs are providing insights into how a temporal negative feedback loop terminates stem cell activity to permit the correct patterning of flowers during the reproductive phase of plant development.
Many of the principles of stem cell regulation elucidated in Arabidopsis are also likely to apply to other plants, even those distantly related to the rose family of which Arabidopsis is a member. Mutants with clv-like fasciated meristem phenotypes have been identified in a number of plant species (64-66), and the cloning of some of the affected genes reveals that the CLV signaling pathway may be conserved among higher plants (65, 67). The challenges for the future will be to characterize all of the components of the CLV signaling pathway, including the targets of WUS transcriptional regulation, to obtain a comprehensive understanding of how the output of this pathway controls stem cell fate and to integrate stem cell signaling into an overall picture of meristem activity.
Acknowledgments
Funding for this work has been provided by National Science Foundation Grant IBN 0110667 and U.S. Department of Agriculture Current Research Information System Grant 5335-21000-016-00D.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Regenerative Medicine,” held October 18-22, 2002, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: SAM, shoot apical meristem; LRR, leucine-rich repeat; LRR-RLK, LRR receptor-like kinase; KAPP, kinase-associated protein phosphatase; MAPK, mitogen-activated protein kinase; CLV, Clavata; WUS, Wuschel; AG, Agamous; LFY, Leafy; ULT, Ultrapetala.
References
- 1.Meyerowitz, E. M. (1997) Cell 88, 299-308. [DOI] [PubMed] [Google Scholar]
- 2.Long, J. A. & Barton, M. K. (1998) Development (Cambridge, U.K.) 125, 3027-3035. [DOI] [PubMed] [Google Scholar]
- 3.Steeves, T. A. & Sussex, I. M. (1989) Patterns in Plant Development (Cambridge Univ. Press, New York).
- 4.Laufs, P., Grandjean, O., Jonak, C., Kieu, K. & Traas, J. (1998) Plant Cell 10, 1375-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyndon, R. F. (1998) The Shoot Apical Meristem: Its Growth and Development (Cambridge Univ. Press, Cambridge, U.K.).
- 6.Poethig, R. S., Coe, E. H. J. & Johri, M. M. (1986) Dev. Biol. 117, 392-404. [Google Scholar]
- 7.Irish, V. F. & Sussex, I. M. (1992) Development (Cambridge, U.K.) 115, 745-753. [Google Scholar]
- 8.Furner, I. J. & Pumfrey, J. E. (1992) Development (Cambridge, U.K.) 115, 755-764. [Google Scholar]
- 9.Stewart, R. N. (1978) in The Clonal Basis of Development, eds. Subtelny, S. & Sussex, I. M. (Academic, New York), pp. 117-124.
- 10.Barton, M. K. & Poethig, R. S. (1993) Development (Cambridge, U.K.) 119, 823-831. [Google Scholar]
- 11.Laux, T., Mayer, K. F. X., Berger, J. & Jurgens, G. (1996) Development (Cambridge, U.K.) 122, 87-96. [DOI] [PubMed] [Google Scholar]
- 12.Mayer, K. F. X., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G. & Laux, T. (1998) Cell 95, 805-815. [DOI] [PubMed] [Google Scholar]
- 13.Kaya, H., Shibahara, K.-I., Taoka, K.-I., Iwabuchi, M., Stillman, B. & Araki, T. (2000) Cell 104, 131-142. [DOI] [PubMed] [Google Scholar]
- 14.Clark, S. E., Running, M. P. & Meyerowitz, E. M. (1993) Development (Cambridge, U.K.) 119, 397-418. [DOI] [PubMed] [Google Scholar]
- 15.Clark, S. E., Running, M. P. & Meyerowitz, E. M. (1995) Development (Cambridge, U.K.) 121, 2057-2067. [Google Scholar]
- 16.Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. (1990) Plant Cell 2, 755-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kayes, J. M. & Clark, S. E. (1998) Development (Cambridge, U.K.) 125, 3843-3851. [DOI] [PubMed] [Google Scholar]
- 18.Clark, S. E., Williams, R. W. & Meyerowitz, E. M. (1997) Cell 89, 575-585. [DOI] [PubMed] [Google Scholar]
- 19.Shiu, S. H. & Bleecker, A. B. (2001) Proc. Natl. Acad. Sci. USA 98, 10763-10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobe, B. & Deisenhofer, J. (1994) Trends Biochem. Sci. 19, 415-421. [DOI] [PubMed] [Google Scholar]
- 21.Torii, K. U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R. F. & Komeda, Y. (1996) Plant Cell 8, 735-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao, D.-Z., Wang, G.-F., Speal, B. & Ma, H. (2002) Genes Dev. 16, 2021-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clay, N. K. & Nelson, T. (2002) Plant Cell 14, 2707-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J. & Chory, J. (1997) Cell 90, 929-938. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Gomez, L. & Boller, T. (2000) Mol. Cell 5, 1-20. [DOI] [PubMed] [Google Scholar]
- 26.Jeong, S., Trotochaud, A. E. & Clark, S. E. (1999) Plant Cell 11, 1925-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fletcher, J. C., Brand, U., Running, M. P., Simon, R. & Meyerowitz, E. M. (1999) Science 283, 1911-1914. [DOI] [PubMed] [Google Scholar]
- 28.Cock, J. M. & McCormick, S. (2001) Plant Physiol. 126, 939-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma, V. K., Ramirez, J. & Fletcher, J. C. (2003) Plant Mol. Biol. 51, 415-425. [DOI] [PubMed] [Google Scholar]
- 30.Williams, R. W., Wilson, J. M. & Meyerowitz, E. M. (1997) Proc. Natl. Acad. Sci. USA 94, 10467-10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone, J. M., Trotochaud, A. E., Walker, J. C. & Clark, S. E. (1998) Plant Physiol. 117, 1217-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trotochaud, A. E., Hao, T., Wu, G., Yang, Z. & Clark, S. E. (1999) Plant Cell 11, 393-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fletcher, J. C. & Meyerowitz, E. M. (2000) Curr. Opin. Plant Biol. 3, 23-30. [DOI] [PubMed] [Google Scholar]
- 34.Stone, J. M., Collinge, M. A., Smith, R. D., Horn, M. A. & Walker, J. C. (1994) Science 266, 793-795. [DOI] [PubMed] [Google Scholar]
- 35.Shah, K., Russinova, E., Gadella, T. W. J., Jr., Willemse, J. & de Vries, S. C. (2002) Genes Dev. 16, 1707-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, J., Smith, G. P. & Walker, J. C. (1999) Proc. Natl. Acad. Sci. USA 96, 7821-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, H., Wu, G., Ware, D., Davis, K. R. & Yang, Z. (1998) Plant Physiol. 118, 407-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chant, J. & Stowers, L. (1995) Cell 81, 1-4. [DOI] [PubMed] [Google Scholar]
- 39.Nagata, K.-I. & Hall, A. (1996) BioEssays 18, 529-531. [Google Scholar]
- 40.Settleman, J. (2001) Dev. Cell 1, 321-331. [DOI] [PubMed] [Google Scholar]
- 41.Lin, Y., Wang, Y., Zhu, J. & Yang, Z. (1996) Plant Cell 8, 293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winge, P., Brembu, T. & Bones, A. M. (1997) Plant Mol. Biol. 35, 483-495. [DOI] [PubMed] [Google Scholar]
- 43.Jones, M. A., Shen, J.-J., Fu, Y., Li, H., Yang, Z. & Grierson, C. S. (2002) Plant Cell 14, 763-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, H., Shen, J.-J., Zheng, Z.-L., Lin, Y. & Yang, Z. (2001) Plant Physiol. 126, 670-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirt, H. (1997) Trends Plant Sci. 2, 11-15. [Google Scholar]
- 46.Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W.-L., Gomez-Gomez, L., Boller, T., Ausubel, F. M. & Sheen, J. (2002) Nature 415, 977-983. [DOI] [PubMed] [Google Scholar]
- 47.Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M. & Simon, R. (2000) Science 289, 617-619. [DOI] [PubMed] [Google Scholar]
- 48.Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F. X., Jurgens, G. & Laux, T. (2000) Cell 100, 635-644. [DOI] [PubMed] [Google Scholar]
- 49.Rojo, E., Sharma, V. K., Kovaleva, V., Raikhel, N. V. & Fletcher, J. C. (2002) Plant Cell 14, 969-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F. & Meyerowitz, E. M. (1992) Cell 69, 843-859. [DOI] [PubMed] [Google Scholar]
- 51.Mandel, M. A., Gustafson-Brown, C., Savidge, B. & Yanofsky, M. F. (1992) Nature 360, 273-277. [DOI] [PubMed] [Google Scholar]
- 52.Weigel, D. & Nilsson, O. (1995) Nature 377, 495-500. [DOI] [PubMed] [Google Scholar]
- 53.Mandel, M. A. & Yanofsky, M. F. (1995) Nature 377, 522-524. [DOI] [PubMed] [Google Scholar]
- 54.Parcy, F., Nilsson, O., Busch, M. A., Lee, I. & Weigel, D. (1998) Nature 395, 561-566. [DOI] [PubMed] [Google Scholar]
- 55.Busch, M. A., Bomblies, K. & Weigel, D. (2000) Science 285, 585-587. [DOI] [PubMed] [Google Scholar]
- 56.Coen, E. S. & Meyerowitz, E. M. (1991) Nature 353, 31-37. [DOI] [PubMed] [Google Scholar]
- 57.Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A. & Meyerowitz, E. M. (1990) Nature 346, 35-39. [DOI] [PubMed] [Google Scholar]
- 58.Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. (1989) Plant Cell 1, 37-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lenhard, M., Bohnert, A., Jurgens, G. & Laux, T. (2001) Cell 105, 805-814. [DOI] [PubMed] [Google Scholar]
- 60.Mizukami, Y. & Ma, H. (1997) Plant Cell 9, 393-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R. & Weigel, D. (2001) Cell 105, 793-803. [DOI] [PubMed] [Google Scholar]
- 62.Sessions, A., Yanofsky, M. F. & Weigel, D. (2000) Science 289, 779-781. [DOI] [PubMed] [Google Scholar]
- 63.Fletcher, J. C. (2001) Development (Cambridge, U.K.) 128, 1323-1333. [DOI] [PubMed] [Google Scholar]
- 64.Emerson, R. A. (1912) Am. Breed. Assoc. Rep. 8, 385-399. [Google Scholar]
- 65.Stuurman, J., Jaggi, F. & Kuhlemeier, C. (2002) Genes Dev. 16, 2213-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacArthur, J. W. (1928) Genetics 13, 410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taguchi-Shiobara, F., Yuan, Z., Hake, S. & Jackson, D. (2001) Genes Dev. 15, 2755-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fletcher, J. C. (2002) Annu. Rev. Plant Biol. 53, 45-66. [DOI] [PubMed] [Google Scholar]


