Abstract
In interphase and mitosis, centrosomes play a major role in the spatial organization of the microtubule network. Alterations in centrosome number and structure are associated with genomic instability and occur in many cancers. Centrosome duplication is controlled by centriole replication. In most dividing animal cells, centrioles duplicate only once per cell cycle at a site adjacent to existing centrioles. The conserved protein kinase Polo-like kinase 4 (Plk4) has a key role in controlling centriole biogenesis. Overexpression of Plk4 drives centrosome amplification and is associated with tumorigenesis in flies. By contrast, haploinsufficiency of Plk4 promotes cytokinesis failure, leading to an increased incidence of tumors in mice. Recent studies have shown that Plk4 is a low abundance protein whose stability is linked to the activity of the enzyme. We discuss how this autoregulatory feedback loop acts to limit the damaging effects caused by too much or too little Plk4.
Key words: centrosome, centriole, polo-like kinase 4, Plk4, SAK, SCF, phosphodegron, β-TrCP, aneuploidy
Centrosomes are the major microtubule organizing centers of animal cells and play a particularly important role during mitosis where they organize the two opposite poles of the bipolar microtubule spindle apparatus upon which chromosomes are segregated. Although centrosomes are not strictly essential for the formation of the mitotic/meiotic spindle, whenever they are present they play a dominant role in guiding spindle formation.1,2 Extra copies of centrosomes frequently result in errors in spindle assembly that give rise to chromosome missegregation and the production of aneuploid daughter cells.3,4 Almost one hundred years ago, Theodor Boveri proposed that centrosome amplification can contribute to tumorigenesis.5 Since then supernumerary centrosomes have been reported in a variety of different tumor cells in vitro and in vivo and are a consistent feature of aneuploid tumors.6–13 However, despite the large body of circumstantial evidence linking extra centrosomes to the development of cancer, it remains unclear whether supernumerary centrosomes actively contribute to tumorigenesis or arise as a byproduct of cellular transformation.
Extra Centrosomes Promote Genomic Instability
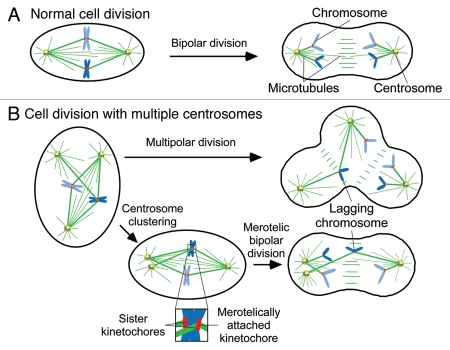
Given the critical role that centrosomes play in maintaining genomic integrity it is not surprising that their number is tightly regulated throughout the cell cycle. If centrosome duplication is not correctly coordinated cells may acquire extra copies of centrosomes, resulting in the formation of multi-polar spindles which can give rise to multi-polar divisions.3,4,14 Such divisions cause massive chromosome missegregation, leading to the subsequent production of highly aneuploid daughter cells that are typically inviable (Fig. 1). As a consequence, cells have evolved mechanisms to suppress multi-polar mitoses and hence prevent massive chromosome missegregation.6,7,15,16 In the majority of cases multiple centrosomes coalesce into two groups and form a bipolar spindle.17–19 While this allows cells to divide in a bipolar manner, the passage through a transient multi-polar intermediate prior to centrosome clustering increases the number of incorrect kinetochore-microtubule attachments, leading to chromosome missegregation3,4 (Fig. 1).
Figure 1.
Extra centrosomes promote chromosomal instability (CIN). (A) Cells with two centrosomes undergo normal bipolar divisions, allowing the chromosomes and centrosomes to be equally distributed in anaphase. (B) Cells possessing more than two centrosomes form multipolar spindles. If uncorrected, this can lead to a multipolar division giving rise to highly aneuploid and often inviable daughter cells. Centrosomes frequently coalesce into two groups prior to anaphase. While this allows the formation of a bi-polar spindle, the transition through a transient multipolar intermediate increases the frequency of incorrect, merotelic kinetochore-microtubule interactions leading to minor chromosome missegregation that is often compatible with cellular viability.
A common feature of many tumor cells is high rates of chromosome gain or loss during mitosis, a phenomenon known as chromosomal instability (CIN).20 CIN is a major source of aneuploidy in human tumors and has long been proposed to have a causal role in cancer progression.21 The primary guard to protect against CIN is the mitotic checkpoint (also known as the spindle assembly checkpoint).22,23 This cell cycle control mechanism acts to delay the irreversible transition to anaphase until all chromosomes are correctly attached to the mitotic spindle. While weakening of the mitotic checkpoint in cells and tissues of mice causes aneuploidy and CIN,15 the mitotic checkpoint is rarely compromised in chromosomally unstable cancer cells.24–26 By contrast, elegant work has shown that supernumerary centrosomes are prevalent amongst CIN cells and are likely to form an important and frequent contributor to CIN.3,4
Centrosome Duplication is Tightly Coordinated with Cell Cycle Progression
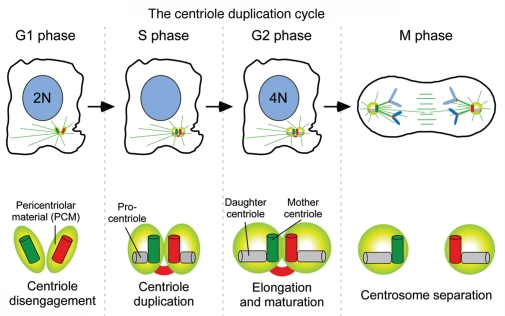
A centrosome is composed of two, barrel-shaped centrioles embedded in an amorphous proteinaceous matrix, known as the pericentriolar material (PCM) (Fig. 2). Like DNA, centrosome duplication is a semi-conservative process that occurs once, and only once, per cell cycle.27 G1 phase cells possess a single centrosome containing two centrioles (Fig. 2). In a normal somatic cell cycle centrosome duplication is first visible during early S-phase, when a new procentriole assembles next to the proximal end of each parental centriole. As the cell progresses through S and G2 phases, daughter centrioles elongate until they reach the length of their parents. At the G2-M phase transition the two centrosomes, each containing a parent and a daughter centriole, separate and instruct the formation of the two spindle poles. At anaphase, the microtubule spindle divides the centrosomes such that each incipient daughter cell will inherit one copy. Centrosomes and chromosomes are the only structures that are known to be precisely duplicated and partitioned equally during each cell division.
Figure 2.
The centriole replication cycle. G1 cells possess a single centrosome containing a pair of centrioles embedded in an amorphous pericentriolar material. In most somatic cells, centriole duplication occurs during S phase and is marked by the formation of procentrioles at the proximal end of each parental centriole. Procentrioles elongate until they reach the length of the parent centrioles in late G2. The procentriole and daughter centrioles are held in an orthogonal configuration and the two pairs of centrioles remain connected, functioning a single microtubule-organizing center until late G2. At the G2-M transition centrosome maturation occurs and the centrosomes separate and form opposite poles of the bipolar microtubule spindle. During mitosis the centrosomes are equally divided such that each incipient daughter cell inherits one centrosome. At the end of mitosis, centrioles lose their orthogonal configuration in a process known as disengagement. It has been proposed that this step may be required to license an additional round of centriole duplication in the next cell cycle.27
Since the centrioles recruit the PCM and determine the number of centrosomes in the cell, centrosome duplication is coupled to centriole replication. Canonical centriole duplication is triggered at a defined site on a parent centriole, which acts as a scaffold to concentrate key regulatory molecules required for centriole biogenesis.2 However, there are some well-characterized examples in specialized cell types where centriole formation can occur de novo without an existing centriole,28 illustrating that centriole formation can be a template-free, self-assembly process.29
Polo-Like Kinase 4 is a Key Regulator of Centriole Biogenesis
In flies and human, the protein kinase Plk4 localizes to the centriole and is a master regulator of centriole biogenesis.29–31 Plk4 is a distant member of the polo-like kinase family and plays an essential role in centriole duplication. In the absence of Plk4, Drosophila and human cells progressively lose centrioles through impaired duplication.30,31 Nevertheless, Plk4−/− mouse embryos do not arrest until embryonic day 7.5 and are thus, apparently capable of undergoing many divisions in the absence of Plk4.32 Since Plk4 turns over rapidly in cells,33 this suggests that either there is a large supply of maternal Plk4 transcript or that Plk4 is not necessary during early embryonic divisions.
While reduced levels of Plk4 impair centriole duplication, overexpression of the kinase overrides the mechanism that normally limits centriole replication, leading to the concurrent formation of multiple daughter centrioles in a single cell cycle.31,34,35 This demonstrates that there is no structural limitation to the formation of multiple daughter centrioles, but rather the number of centrioles produced during each cell cycle is limited by the level of active Plk4.
Plk4 Misregulation Promotes Tumorigenesis
The production of multiple centrioles ultimately leads to the formation of extra centrosomes. Consequently, overexpression of Plk4 in Drosophila gave rise to flies possessing supernumerary centrosomes in ∼60% of somatic cells.19 Many of these cells initially formed multipolar spindles, but these spindles ultimately become bipolar. Consequently, flies with extra centrosomes are overtly normal and fertile and display only a modest increase in aneuploidy. Transplanted larval brain tissue from animals with extra centrosomes was able to initiate tumorigenesis in wild type host flies,19 providing the first direct causative link between centrosome amplification and tumorigenesis. However, fruit flies are not normally affected by cancer, and therefore, it will be important to extend these exciting findings to investigate the long-term effect of centrosome amplification in mammalian tumors models.
Surprisingly, Plk4 heterozygosity also leads to centrosome amplification, abnormal spindle formation and aneuploidy, but through a mechanism that does not involve centriole overduplication. Plk4+/− murine embryonic fibroblasts exhibit a high frequency of cytokinesis failure leading to the production of tetraploid daughter cells with twice the normal centrosome content.36 Exactly how Plk4 acts to control cytokinesis remains to be established. There is compelling evidence that the uncontrolled proliferation of tetraploid cells acts as a catalyst to promote further genetic instability and tumorigenesis.15,37 Consistently, Plk4+/− cells spontaneously immortalize in culture and are capable of forming tumors when injected into immunocompromised mice.36 Moreover, Plk4 heterozygous mice are prone to the development of spontaneous lung and liver tumors and loss of Plk4 heterozygozity occurs frequently in human hepatoma.32,36,37 Together, these studies demonstrate that Plk4 has the unusual property of acting both as a haploinsufficient tumor suppressor and an oncogene. Clearly, controlling the level of Plk4 is of great importance for the cell and the organism.
Plk4 Levels are Tightly Controlled
The level and activity of Plk4 must be carefully controlled in space and time to ensure proper co-ordination of centriole duplication. Indeed, Plk4 expression is controlled transcriptionally, with Plk4 mRNA increasing in late G1 and reaching a maximal level during mitosis.33 The C-terminus of Plk4 also contains three putative PEST domains, which are commonly associated with reduced protein stability.38 Consistently, endogenous Plk4 is a low abundance protein, while overexpressed Plk4 has a short half-life of 2–3 hours in cells.33 Regulated proteolysis is a common mechanism for downregulating active kinases and in many cases involves targeted destruction of kinases through the ubiquitin-proteasome pathway.39 Several studies have shown that Plk4 is ubiquitinated and destroyed by the 26S proteasome33,40–43 and moreover, chemical inhibition of the proteasome promotes centriole overduplication in a Plk4 dependent manner.44
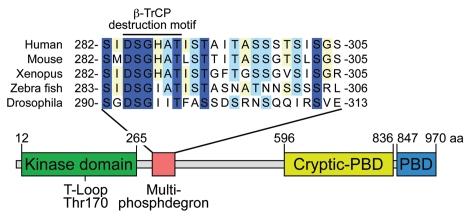
Several components of the ubiquitination-degradation machinery have been found to localize to the centrosome, including components of the SKP1-CUL1-F-Box (SCF) E3 ubiquitin ligase.40,42,45 The SCF complex is an E3 ubiquitin ligase comprised of 3 core subunits (Skp1, Cullin 1 and Rbx1) and an interchangeable F-box protein responsible for substrate recognition.46 F-box proteins often associate with substrates phosphorylated at specific sites in “phosphodegron” motifs. Plk4 possess a conserved phosphodegron binding site for the F-box protein β-TrCP (Fig. 3). Consistent, with a role in controlling Plk4 stability, depletion of Slimb (the Drosophila homologue of vertebrate β-TrCP) or knockout of β-TrCP in mouse cells causes an increase in centrosome number.47–49 Two recent studies demonstrated that Slimb associates with Plk4 in Drosophila cells and mutation of the Slimb binding site increases Plk4 stability leading to centriole overduplication.41,42 Nevertheless, the kinase responsible for regulating phosphorylation of Plk4 in the β-TrCP/Slimb degron was not identified.
Figure 3.
Structural features of human Plk4. Plk4 possesses an N-terminal kinase domain, a C-teminal polo-box domain (PBD) and cryptic polo-box domain (Cryptic-PBD). The multi-phosphodegron is required to destabilize kinase active Plk4. The multi-phosphodegron contains a conserved β-TrCP binding motif and is rich in potential phosphorylation sites. The conserved Threonine in the T-loop activation domain is marked.
Plk4 Kinase Activity Autoregulates Protein Stability
There are several examples in which the sustained activation of protein kinases promotes their irreversible downregulation though ubiquitin-targeted proteolysis.39 To investigate how the stability of Plk4 is regulated in mammalian cells, we created stable cell lines expressing kinase active and kinase dead versions of Plk4 from the same genomic locus. Interestingly, we found that kinase inactive Plk4 accumulated to >10 fold higher levels than the active kinase, suggesting that kinase active Plk4 stimulates its own breakdown.43 This was confirmed by engineering an analogue sensitive allele of Plk4 whose activity could be specifically inhibited with a bulky ATP analogue:50 addition of the drug caused Plk4 protein stability to dramatically increase. Expression of kinase active Plk4 was also able to promote the destabilization of kinase inactive Plk4, indicating Plk4 is capable of intermolecular destabilization. Concentrating Plk4 at the centriole may thus, aid Plk4's ability to promote its own destruction.
An attractive explanation for how Plk4 regulates its own stability is that Plk4 self-phosphorylates the β-TrCP phosphodegron and targets itself for destruction. Surprisingly, however, mutation of the phosphodegron to abolish phosphorylation only modestly stabilized Plk4, despite the fact β-TrCP binding was largely abolished.43 In vitro analysis of Plk4 autophosphorylation sites revealed a densely phosphorylated region located just downstream of the Plk4 kinase domain43 (Fig. 3). Strikingly, this 24 amino acid region encompasses the entire β-TrCP phosphodegron and lies within one of the PEST domains previously shown to destabilize Plk4.38 Consistent with a role in controlling protein stability, deletion of this 24 amino acid region dramatically stabilized Plk4 without affecting kinase activity, demonstrating this region integrates Plk4 activity with the protein's instability.43 Using a series of Plk4 mutants defective in phosphorylation at specific sites within this 24 amino acid region, we set out to establish if a combination of phosphorylation events is required for Plk4 destruction. Plk4 was progressively stabilized as more phosphorylation sites in the 24 amino acid region were abolished, indicating Plk4 destruction depends upon phosphorylation at multiple sites within this region. We propose that this 24 amino acid region functions as a “multi-phosphodegron” that promotes the destruction of Plk4 after the bulk addition of phosphates.
The bulk addition of phosphates to the multi-phosphodegron would create a substantial change in electrostatic charge that may enhance docking to E3 ligases. Since counteracting phosphatases would act to remove phosphates from this region, a requirement for multi-site phosphorylation creates a higher threshold of Plk4 activity that would be required to degrade the protein. This may provide a temporal delay that allows Plk4 to phosphorylate the substrates required for centriole duplication before it promotes its own degradation.
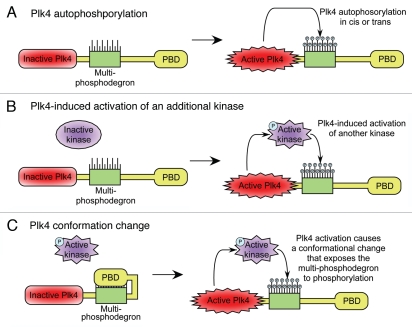
In vitro Plk4 is able to autophosphorylate several sites within the multi-phosphodegron, suggesting that kinase active Plk4 may act directly to promote its own destruction43 (Fig. 4). A recent report identified a site in the multi-phosphodegron (serine 305) as a major Plk4 autophosphorylation site in vitro and in vivo.51 Although preventing phosphorylation of this site alone does not alter Plk4 stability in cells, proteasome inhibition caused a marked increase in S305-phosphorylated Plk4, indicating that phosphorylation of S305 occurs concurrently with other sites in the multi-phosphodegron that have an additive role in promoting Plk4 destruction. However, it also remains possible that Plk4 could regulate its own destruction indirectly (Fig. 4). For example, Plk4 may activate an as yet unidentified kinase that in-turn phosphorylates the multi-phosphodegron. Alternatively, the activation of Plk4 may promote a conformational change that exposes the previously masked multi-phosphodegron to phosphorylation by other kinases. Regardless of the exact mechanism, placing Plk4 stability under direct control of the proteins activity creates an autoregulatory feedback loop that allows Plk4 to self-limit kinase activity by promoting its own destruction by the proteasome. This is likely to be important in preventing Plk4 from accumulating to dangerous levels in the cell. Interestingly, the levels of the centriole component SAS-6 are also controlled by proteolysis. SAS-6 is a substrate for the APCcdh1 E3 ligase and targeted for degradation during late mitosis.52 Overexpression of SAS-6 in human cells leads to the formation of multiple daughter centrioles in a single cell cycle. Proteolysis may therefore, be a general mechanism to limit the availability of centriole components and prevent centriole overduplication.
Figure 4.
Model for how Plk4 kinase activity autoregulates protein stability. (A) Kinase active Plk4 autophosphorylates the multi-phosphodegron. Phosphorylation promotes Plk4's ubiquitination by an E3 ligase and subsequent destruction by the proteasome. (B) Kinase active Plk4 phosphorylates and activates an additional kinase. This kinase subsequently phosphorylates the multi-phosphodegron leading to Plk4's downregulation. (C) Activation of Plk4 induces a conformational shift in the protein that exposes the previously masked multi-phosphodegron to phosphorylation by other kinases.
Conclusions and Future Prospects
The regulation of Plk4 levels is a clear example of biological self-control that likely contributes to limiting normal centriole duplication to once per cell cycle. A remaining question however, is how Plk4 is able to promote centriole replication at a defined period in the cell cycle. It is probable that other levels of control also act to precisely control Plk4 activity. For example, the recruitment of Plk4 to the centriole may control the phosphorylation of substrates51 or Plk4 activity may be regulated through phosphorylation of the T-loop threonine by other cell cycle regulated kinases.53 A more detailed appreciation of Plk4's ability to orchestrate centriole replication may aid our understanding as to the origins of centrosome amplification in cancer and provide opportunities to develop therapies that target cancer cells possessing extra centrosomes.
Acknowledgements
This work was supported by a grant (GM29513) from the National Institutes of Health to D.W. Cleveland, who receives salary support from the Ludwig Institute for Cancer Research. A.J.H. was supported by a European Molecular Biology Organization (EMBO) Long-Term Fellowship.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12184
References
- 1.Debec A, Sullivan W, Bettencourt-Dias M. Centrioles: active players or passengers during mitosis? Cell Mol Life Sci. 2010;67:2173–2194. doi: 10.1007/s00018-010-0323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nigg EA, Raff JW. Centrioles, centrosomes and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 3.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome missegregation in cancer cells. PLoS ONE. 2009;4:6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boveri T. Zur Frage der Entstenhung Maligner Tumoren. Jena: Gustav Fischer Verlag; 1914. (Ger). [Google Scholar]
- 6.Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- 7.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 8.Ghadimi BM, Sackett DL, Difilippantonio MJ, Schrock E, Neumann T, Jauho A, et al. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- 9.Kronenwett U, Huwendiek S, Castro J, Ried T, Auer G. Characterisation of breast fine-needle aspiration biopsies by centrosome aberrations and genomic instability. Br J Cancer. 2005;92:389–395. doi: 10.1038/sj.bjc.6602246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci USA. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, et al. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- 13.Yamamoto Y, Matsuyama H, Furuya T, Oga A, Yoshihiro S, Okuda M, et al. Centrosome hyperamplification predicts progression and tumor recurrence in bladder cancer. Clin Cancer Res. 2004;10:6449–6455. doi: 10.1158/1078-0432.CCR-04-0773. [DOI] [PubMed] [Google Scholar]
- 14.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 15.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimini D. Merotelic kinetochore orientation, aneuploidy and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat Cell Biol. 2008;10:748–751. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 19.Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 21.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 22.Taylor SS, Scott MI, Holland AJ. The spindle checkpoint: a quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 2004;12:599–616. doi: 10.1023/B:CHRO.0000036610.78380.51. [DOI] [PubMed] [Google Scholar]
- 23.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 24.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Holland AJ, Cleveland DW. Beyond genetics: surprising determinants of cell fate in antitumor drugs. Cancer Cell. 2008;14:103–105. doi: 10.1016/j.ccr.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsou MF, Stearns T. Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol. 2006;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol Cells. 2009;27:135–142. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- 30.Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 31.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 32.Ko MA, Rosario CO, Hudson JW, Kulkarni S, Pollett A, Dennis JW, et al. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- 33.Fode C, Binkert C, Dennis JW. Constitutive expression of murine Sak-a suppresses cell growth and induces multinucleation. Mol Cell Biol. 1996;16:4665–4672. doi: 10.1128/mcb.16.9.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Rosario CO, Ko MA, Haffani YZ, Gladdy RA, Paderova J, Pollett A, et al. Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci USA. 2010;107:6888–6893. doi: 10.1073/pnas.0910941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macmillan JC, Hudson JW, Bull S, Dennis JW, Swallow CJ. Comparative expression of the mitotic regulators SAK and PLK in colorectal cancer. Ann Surg Oncol. 2001;8:729–740. doi: 10.1007/s10434-001-0729-6. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita Y, Kajigaya S, Yoshida K, Ueno S, Ota J, Ohmine K, et al. Sak serine-threonine kinase acts as an effector of Tec tyrosine kinase. J Biol Chem. 2001;276:39012–39020. doi: 10.1074/jbc.M106249200. [DOI] [PubMed] [Google Scholar]
- 39.Lu Z, Hunter T. Degradation of activated protein kinases by ubiquitination. Annu Rev Biochem. 2009;78:435–475. doi: 10.1146/annurev.biochem.013008.092711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korzeniewski N, Zheng L, Cuevas R, Parry J, Chatterjee P, Anderton B, et al. Cullin 1 functions as a centrosomal suppressor of centriole multiplication by regulating polo-like kinase 4 protein levels. Cancer Res. 2009;69:6668–6675. doi: 10.1158/0008-5472.CAN-09-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli M, Zhang W, Laue E, et al. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 42.Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duensing A, Liu Y, Perdreau SA, Kleylein-Sohn J, Nigg EA, Duensing S. Centriole overduplication through the concurrent formation of multiple daughter centrioles at single maternal templates. Oncogene. 2007;26:6280–6288. doi: 10.1038/sj.onc.1210456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freed E, Lacey KR, Huie P, Lyapina SA, Deshaies RJ, Stearns T, et al. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 1999;13:2242–2257. doi: 10.1101/gad.13.17.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wojcik EJ, Glover DM, Hays TS. The SCF ubiquitin ligase protein slimb regulates centrosome duplication in Drosophila. Curr Biol. 2000;10:1131–1134. doi: 10.1016/s0960-9822(00)00703-x. [DOI] [PubMed] [Google Scholar]
- 48.Murphy TD. Drosophila skpA, a component of SCF ubiquitin ligases, regulates centrosome duplication independently of cyclin E accumulation. J Cell Sci. 2003;116:2321–2332. doi: 10.1242/jcs.00463. [DOI] [PubMed] [Google Scholar]
- 49.Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, et al. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 50.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 51.Sillibourne JE, Tack F, Vloemans N, Boeckx A, Thambirajah S, Bonnet P, et al. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol Biol Cell. 2009;21:547–561. doi: 10.1091/mbc.E09-06-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swallow CJ, Ko MA, Siddiqui NU, Hudson JW, Dennis JW. Sak/Plk4 and mitotic fidelity. Oncogene. 2005;24:306–312. doi: 10.1038/sj.onc.1208275. [DOI] [PubMed] [Google Scholar]