Abstract
The human disorder ataxia telangiectasia (AT), which is characterized by genetic instability and neurodegeneration, results from mutation of the ataxia telangiectasia mutated (ATM) kinase. The loss of ATM leads to cell cycle checkpoint deficiencies and other DNA damage signaling defects that do not fully explain all pathologies associated with A-T including neuronal loss. In addressing this enigma, we find here that ATM suppresses DNA double-strand break (DSB) repair by microhomology-mediated end joining (MMEJ). We show that ATM repression of DNA end-degradation is dependent on its kinase activities and that Mre11 is the major nuclease behind increased DNA end-degradation and MMEJ repair in A-T. Assessment of MMEJ by an in vivo reporter assay system reveals decreased levels of MMEJ repair in Mre11-knockdown cells and in cells treated with Mre11-nuclease inhibitor mirin. Structure-based modeling of Mre11 dimer engaging DNA ends suggests the 5′ ends of a bridged DSB are juxtaposed such that DNA unwinding and 3′–5′ exonuclease activities may collaborate to facilitate simultaneous pairing of extended 5′ termini and exonucleolytic degradation of the 3′ ends in MMEJ. Together our results provide an integrated understanding of ATM and Mre11 in MMEJ: ATM has a critical regulatory function in controlling DNA end-stability and error-prone DSB repair and Mre11 nuclease plays a major role in initiating MMEJ in mammalian cells. These functions of ATM and Mre11 could be particularly important in neuronal cells, which are post-mitotic and therefore depend on mechanisms other than homologous recombination between sister chromatids to repair DSBs.
Key words: ATM, Mre11, MRN complex, DNA degradation, double-strand break repair, microhomology-mediated end joining, PI-3-kinase-like kinases
Introduction
Deficiencies in mediators that participate in genomic maintenance are associated with several disorders characterized by genetic instability, cancer, abnormal development and neurodegeneration.1,2 Involved proteins frequently have dual tasks as both cell cycle checkpoint and DNA repair mediators. A key example is the ataxia telangiectasia mutated (ATM) protein. A defect in ATM results in ataxia telangiectasia (A-T), a human autosomal recessive disorder characterized by neurodegeneration, genetic instability, immunological malfunctions and a propensity for cancer development.3 ATM has an established role in activating checkpoints that halt cell cycle progression in response to a DNA double-strand break (DSB) in mammalian cells.4 However, the functions it may actively engage in during the repair of such a break have remained elusive. Although the focus of the literature has been on the two major pathways through which a DSB is repaired, non-homologous end-joining (NHEJ) and homologous recombination (HR), it has recently become evident that other “backup” or “alternative” pathways, such as microhomology-mediated end joining (MMEJ), exist.5–10 However, the mediators implicated in these pathways and their regulation require further elucidation.
We previously demonstrated that the fidelity of repairing a plasmid harboring a DSB was compromised in nuclear extracts from human ATM-deficient (A-T) cells.11 The mutation frequency was significantly higher in A-T nuclear extracts than in controls. Repaired plasmids harbored deletions spanning the repaired DSB site with rejoining occurring at sequences of microhomology (1–5 nucleotide repeats) flanking the break. This resulted in deletion of the sequences between the two sites of microhomology in addition to the loss of one microhomology site. This type of lesion is the product of repairing a break site via MMEJ, the alternative pathway of DSB repair. Genetic studies in yeast indicate that the Mre11 exonuclease has a major role in MMEJ.12,13 Biochemical studies performed with purified recombinant human Mre11 corroborate the ability of this nuclease to mediate MMEJ in vitro.14 Moreover, Mre11 was recently found to play a role in “alternative” pathways of DNA repair including MMEJ in mammalian cells.15,16 Nevertheless, the precise role played by Mre11 in MMEJ and the mediators that control this pathway in mammalian systems have not been clarified. Mre11 is a member of the mammalian MRN (Mre11-Rad50-Nbs1) complex that seems to play a role in recognition and sensing of a DSB.17 This complex rapidly migrates to DSB sites and recruits ATM.18 Through interacting with Nbs1, ATM undergoes trans-autophosphorylation and monomerization thus initiating a signaling cascade that results in activating DNA repair and cell cycle control machinery.19 Among ATM's substrates is the MRN complex itself; hence, this complex has functions upstream and downstream of ATM activation.20–22 An increased frequency of MMEJ in A-T nuclear extracts suggested that ATM suppresses this pathway, perhaps by hindering degradation of DNA ends at a break by an undefined mechanism.23
A major but poorly understood biological feature of the multi-system disorder syndrome A-T is neurodegeneration.3 This facet of the disease, unlike its other symptoms, is not readily explained by the cell cycle and DNA damage signaling deficiencies associated with an ATM malfunction. To better understand the cell biology underlying A-T, we characterize here the role of ATM in controlling MMEJ and the importance of the Mre11 nuclease. We find a key role for ATM's kinase activity in suppressing degradation at DNA ends and for the Mre11 nuclease in this degradation, which is consistent with structures and modeling of Mre11-DNA interactions. Moreover, we show that Mre11 participates in MMEJ in mammalian cells under regulation by ATM.
Results
DNA end-degradation is increased in kinase-dead (KD)-ATM nuclear extracts.
ATM suppresses degradation at DNA ends by an ATP dependent function that is inhibited by the PI3-kinase-like kinase (PIKK) inhibitors caffeine and wortmannin.23 Also, the addition of pre-phosphorylated ATM to an ATM-deficient nuclear extract does not restore DNA end-stability to levels observed in control extracts (wtATM+). These data suggest that the kinase activity of ATM may be required for suppression of DNA end-degradation. To test this possibility, we expressed kinase-dead (KD)-ATM in both A-T and control (wtATM+) fibroblasts. In this form of ATM two key catalytic amino acid residues in the kinase active site are substituted (Asp2870→Ala, Asn2875→Lys) rendering the kinase inactive.24
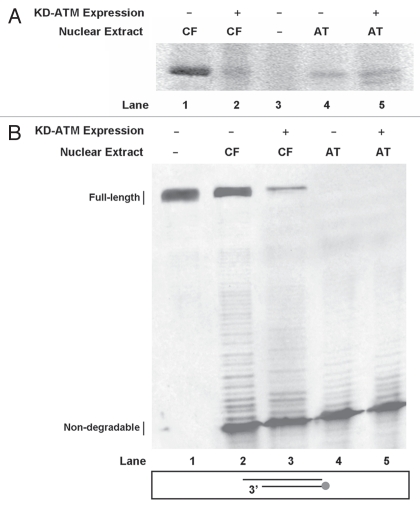
The kinase status of ATM in the KD-ATM expressing cells was assessed by testing the ability of ATM immunoprecipitated from nuclear extracts to phosphorylate PHAS-I (Fig. 1A). Nuclear extracts were prepared and ATM was then immunoprecipitated by mouse anti-ATM that was later bound to rProtein G agarose beads. The beads from the immunoprecipitation reactions were incubated with PHAS-I and (γ-32P)ATP. As previously reported,24,25 the expression of KD-ATM in control (wtATM+) cells imparts a dominant negative effect on these cells; we observed an 80% reduction in phosphorylation of the PHAS-I substrate in nuclear extracts from KD-ATM-expressing control cells compared to their non-expressing counterparts (Fig. 1A; compare Lane 1 to Lane 2). This reduction was down to levels seen in extracts from A-T cells and A-T cells expressing KD-ATM. As expected, the expression of KD-ATM in A-T cells did not alter the inability of extracts from these cells to phosphorylate PHAS-I. Residual phosphorylation of PHAS-I in nuclear extracts from A-T cells and cells expressing KD-ATM can be attributed to a kinase that non-specifically binds to the employed rProtein G agarose beads.
Figure 1.
Enhanced DNA degradation after expression of kinase dead (KD)-ATM. (A) KD-ATM expression tested by assessing 32p incorporation into PHAS-I. Nuclear extracts from the ATM+ control fibroblast (CF) cell line WI-38VA13, ATM-deficient (AT) cell line AT5BIVA and corresponding cells transfected with the KD-ATM expression plasmid were prepared. ATM was immunoprecipitated from these extracts and the precipitates were employed in a kinase assay in the presence of PHAS-I as a substrate and (γ-32P) ATP. A reaction containing PHAS-I and (γ-32P)ATP in the absence of a nuclear extract (Lane 3) was included as control. Reactions were separated on a 12% SDS-PAGE and the gel was exposed to a storage phosphor screen which was then scanned. (B) 5′Cy3-labeled Template degradation products following gel separation. A duplex DNA substrate with a 5′Cy3-labeled Template was incubated with nuclear extracts from WI-38VA13 (CF), AT5BIVA (AT) and respective cell lines expressing KD-ATM. A reaction with duplex only (Lane 1) in the absence of nuclear extract was included as control.
To test the role of ATM's kinase function in controlling nuclease activity at DNA ends, nuclear extracts were prepared from KD-ATM-expressing cells and their respective controls were tested for their ability to sustain DNA end-stability (Fig. 1B). We employed a DNA end-processing assay to measure degradation of 3′ DNA ends (Template) in the various extracts.23 We detected a 90% decrease in full-length product in A-T nuclear extracts when compared to controls (Fig. 1B; compare Lane 4 to Lane 2). Furthermore, expression of KD-ATM in the controls led to a 60% decrease in detection of full-length product and an increase in the levels of degradation products (Fig. 1B; compare Lane 2 to Lane 3). No marked differences were observed between extracts from A-T cells and A-T cells expressing KD-ATM. This increase in degradation in extracts from control cells expressing KD-ATM suggests that ATM engages via its kinase activity in suppressing the degradation of DNA ends.
DNA degradation is decreased in Mre11-immunodepleted nuclear extracts.
The dependence of DNA stability on the kinase activity of ATM suggested that this PIKK regulates the activities of nucleases participating in end-degradation by phosphorylating downstream mediators. One candidate nuclease that may be regulated by ATM is the MRN nuclease complex composed of Mre11 nuclease, Rad50 ATPase/adenylate kinase and Nbs1, which mediates interactions with multiple proteins including ATM.26 ATM-dependent phosphorylation of Mre11,27 Rad50,28 and Nbs120,21 in response to DNA damage has been described.
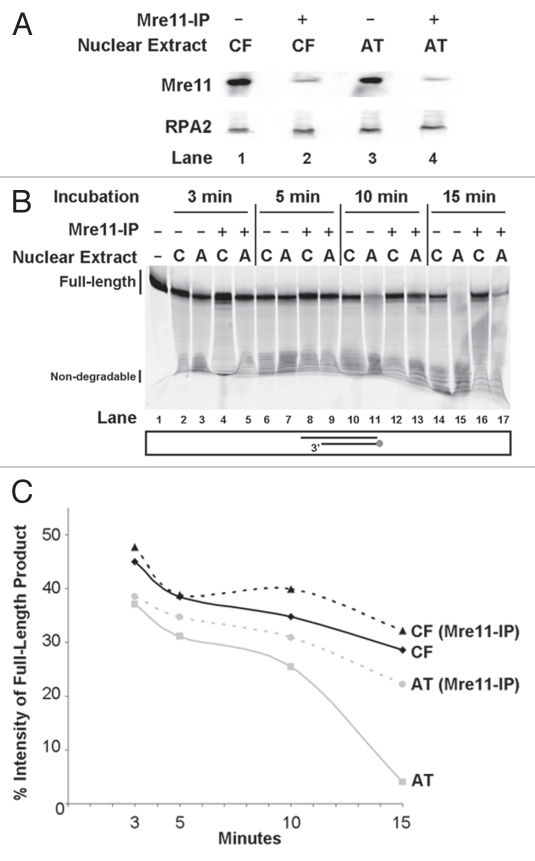
To investigate the possible role of Mre11 in the elevated degradation levels observed in our A-T extracts, we immunodepleted Mre11 from A-T and control nuclear extracts. An 80–90% decrease in Mre11 levels was detectable in extracts immunodepleted with an Mre11 antibody when compared to non-depleted extracts (Fig. 2A).
Figure 2.
Decreased DNA degradation in nuclear extracts after Mre11 immunodepletion. (A) Following Mre11 immunodepletion of WI-38VA13 (CF) and AT5BIVA (AT) nuclear extracts, supernatants were subjected to SDS-PAGE, transferred to a membrane and then probed for Mre11. RPA2 was assayed for as a loading control. (B) 5′Cy3-labeled Template degradation products following gel separation. A duplex DNA substrate with a 5′Cy3-labeled Template was incubated with nuclear extracts from WI-38VA13 (CF), AT5BIVA (AT) and respective nuclear extracts that were Mre11-immunodepleted. Degradation was allowed to proceed for 3, 5, 10 or 15 min. A reaction with duplex only (Lane 1) in the absence of nuclear extract was included as control and incubated for 15 min. (C) Plot for % intensities of Full-length products. Full-length products in (A) were quantified and expressed as percent intensity (% intensity = (product intensity/total intensity) × 100).
Immunodepleted extracts were assayed for DNA end-processing by following a time-course of Template degradation (Fig. 2B and C). A slight increase in detection of full-length products at the various time-points tested was noted in the control extracts after Mre11 immunodepletion. In stark contrast, immunodepletion of Mre11 from A-T nuclear extracts resulted in a large increase in full-length products. The maximal difference was noted after 15 minutes of incubating the DNA substrate in the nuclear extracts (Fig. 2B; compare Lane 15 to Lane 17). At that time point, a 6-fold increase in full-length products was seen in Mre11-immunodepleted A-T nuclear extracts when compared to non-depleted A-T extracts. Furthermore, a 1.12-fold increase in full-length product level was detected in depleted control extracts compared to respective non-depleted ones (Fig. 2B; compare Lane 14 to Lane 16). In A-T extracts, degradation of DNA is more extensive than observed in control extracts. Immunodepletion of Mre11 from A-T extracts prevents the excessive degradation.
Treatment with the Mre11 nuclease inhibitor mirin reduces DNAdegradation in nuclear extracts.
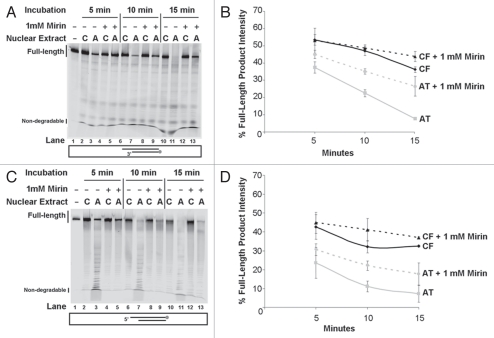
We wished to test if immunodepleting Mre11 may have also immunoprecipitated other mediators that are directly responsible for the detected degradation and interact with Mre11. To examine this possibility, we used the Mre11 nuclease inhibitor Z-5-(4-hydroxybenzylidene)-2-imino-1,3-thiazolidin-4-one, more commonly know as Mirin.29 This inhibitor suppresses DNA degradation by purified Mre11 in vitro and inhibits cellular pathways mediated by the nuclease in vivo.30 Nuclear extracts from A-T and control cells were pre-incubated with Mirin prior to employing them in an end-processing assay.
Mirin-treated extracts were first tested for the degradation of the DNA template in a short oligonucleotide duplex at various time-points (Fig. 3A and B). As observed with the Mre11-immunodepletion experiments, improved full-length product detection, and decreased DNA 3′ end-degradation, was minimal in Mirin-treated control extracts. Addition of Mirin to control extracts resulted in only a 1.2-fold increase in full-length products observed after 15 min of incubation (Fig. 3A; compare Lane 10 to Lane 12). Yet, at the same 15 min time point, in Mirin-treated A-T nuclear extracts the amount of full-length products observed was 3.5-fold higher than that in non-treated A-T nuclear extracts (Fig. 3A; compare Lane 11 to Lane 13). The difference seen between Mirin-treated A-T and control extracts demonstrates the tighter regulation of nuclease function when ATM is present. Enhancement of full-length products in A-T extracts after preincubation with the Mre11 nuclease inhibitor was less than that observed after Mre11-immunodepletion. This may indicate that immunodepletion was incomplete or alternatively that accessory nucleases that may participate in DNA degradation co-immunoprecipitate with Mre11.
Figure 3.
Reduced duplex DNA degradation in Mirin-treated nuclear extracts. (A) 5′Cy3-labeled Template degradation products following gel separation. A duplex DNA substrate with a 5′Cy3-labeled Template was incubated with nuclear extracts from WI-38VA13 (CF), AT5BIVA (AT) and respective nuclear extracts treated with 1 mM Mirin. Degradation was allowed to proceed for 5, 10 or 15 min. A reaction with duplex only (Lane 1) in the absence of nuclear extract was included as control and incubated for 15 min. (B) Plot for % intensities of Full-length products. Full products in (A) were quantified and expressed as percent intensity (% intensity = (product intensity/total intensity) × 100). (C) 3′Cy3sp-labeled Top Strand degradation products following gel separation. A duplex DNA substrate with a 3′Cy3sp-labeled Top Strand was incubated with nuclear extracts from WI-38VA13 (CF), AT5BIVA (AT) and respective nuclear extracts treated with 1 mM Mirin. Degradation was allowed to proceed for 5, 10 or 15 min. A reaction with duplex only (Lane 1) in the absence of nuclear extract was included as control and incubated for 15 min. (D) Plot for % intensities of Full-length products. Full products in (A) were quantified and expressed as percent intensity (% intensity = (product intensity/total intensity) × 100).
Top Strand (5′-end) degradation in Mirin-treated and non-treated extracts was also monitored at various time-points (Fig. 3C and D). The level of full-length products seen was slightly higher in the Mirin-treated control extracts than in nontreated controls, with a 1.14-fold increase at the 15 min time point (Fig. 3C; compare Lane 10 to Lane 12). The level of full-length product observed in A-T nuclear extracts treated with the Mre11 nuclease inhibitor was 2.5-fold higher than in non-treated A-T nuclear extracts (Fig. 3C; compare Lane 11 to Lane 13). Mre11 preferentially acts as a 3′ to 5′ exonuclease on 3′ recessed strands and has endonuclease functions on single-stranded loops.31 Consequently, the decrease in 5′ end degradation after treating nuclear extracts with Mirin may be indicative of inhibition of a direct Mre11 activity on that strand or an indirect inhibition of an accessory nuclease that acts on that end. One possibility is that mediator-facilitated single-stranded loop structures may form on one strand thus allowing direct endonuclease degradation by Mre11. Another possibility is that by inhibiting Mre11 with Mirin, the degradation of the 3′ end was hindered thus slowing down the kinetics of 5′ end-degradation by another nuclease.
shRNA knock-down of Mre11 decreased DNA degradation in nuclear extracts.
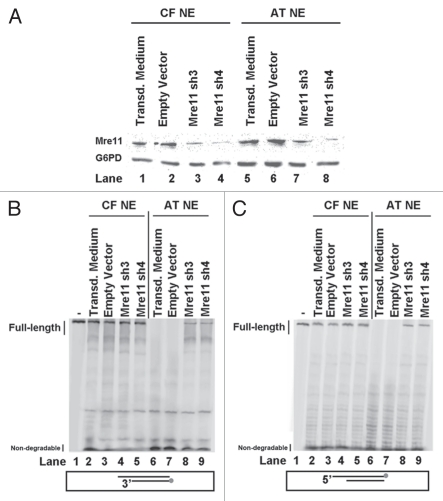
To further test the role of Mre11 nuclease in degration of DNA ends at breaks, Mre11 levels were knocked down by transducing A-T and control fibroblast cells with lentiviral particles encoding shRNA for Mre11. Mre11 levels were decreased by ∼80% (Fig. 4A). Nuclear extracts from knocked down cells were prepared and tested for DNA end-degradation.
Figure 4.
Reduced DNA degradation in nuclear extracts after Mre11 knockdown. (A) Immunoblot for Mre11 in nuclear extracts after Mre11 knockdown. Nuclear extracts from WI-38VA13 (CF) and AT5BIVA (AT) were prepared. Extracts were also prepared from respective cells in which Mre11 was knocked down by transduction with lentivirus encoding Mre11 shRNA (Mre11 sh3 and Mre11 sh4). The extracts were subjected to SDS-PAGE, transferred to a membrane and then probed for Mre11. Nuclear extracts from cells plated in virus-free transduction medium (Lanes 1 and 5) or transduced with an empty vector (Lanes 2 and 6) were included as controls. G6PD was probed for as a loading control. (B) 5′Cy3-labeled Template degradation products following gel separation. A duplex DNA substrate with a 5′Cy3-labeled Template was incubated with nuclear extracts from WI-38VA13 (CF) and AT5BIVA (AT) cells. The duplex was also incubated with respective nuclear extracts from cells in which Mre11 was knocked down by transduction with lentivirus encoding Mre11 shRNA (Mre11 sh3 or Mre11 sh4). Nuclear extracts from cells plated in virus-free transduction medium (Lanes 2 and 6) or transduced with an empty vector (Lanes 3 and 7) were included as controls. A reaction with duplex only (Lane 1) in the absence of nuclear extract was included as control. (C) 3′Cy3sp-labeled Top Strand degradation products following gel separation. A duplex DNA substrate with a 3′Cy3sp-labeled Top Strand was incubated with nuclear extracts from WI-38VA13 (CF) and AT5BIVA (AT) cells. The duplex was also incubated with respective nuclear extracts from cells in which Mre11 was knocked down by transduction with lentivirus encoding Mre11 shRNA (Mre11 sh3 or Mre11 sh4). Nuclear extracts from cells plated in virus-free transduction medium (Lanes 2 and 6) or transduced with an empty vector (Lanes 3 and 7) were included as controls. A reaction with duplex only (Lane 1) in the absence of nuclear extract was included as control.
The degradation of both Template (Fig. 4B) and Top Strand (Fig. 4C) were assessed. Full-length products after incubation with nuclear extracts from Mre11-knocked down control fibroblasts for 15 min were 1.2-fold higher than the non-treated controls (Fig. 4B and C; compare Lanes 1 and 2 to Lanes 4 and 5). Knocking down Mre11 in A-T fibroblasts resulted in a 5–6 fold rescue of full-length products for both strands of the duplex DNA substrate (Fig. 4B and C; compare Lanes 6 and 7 to Lanes 8 and 9). Residual Mre11 is potentially responsible for our inability to fully-rescue full-length product stability. The amount of full-length Template in Mre11-knockdown A-T nuclear extracts was around 10% less than in non-treated control extracts (Fig. 4B; compare Lanes 2 and 3 to Lanes 8 and 9). On the other hand, the amount of full-length Top Strand in Mre11-knockdown A-T nuclear extracts was about 30% less than in non-knockdown control extracts (Fig. 4C; compare Lanes 2 and 3 to Lanes 8 and 9). Notably, the increase of full-length products observed after Mre11 knockdown was similar to that seen after Mre11 immunodepletion, but higher than after nuclease inhibition with Mirin.
Mre11-immunodepleted leads to deficient MMEJ in nuclear extracts.
To further assess the role played by Mre11 in the increased MMEJ levels observed in A-T extracts we used the SupF22 plasmid system (Fig. 5).11 The repair of a restriction-digested SupF22 plasmid by MMEJ in nuclear extracts can be detected by a blue-white transformed bacterial colony screen (Fig. 5). We reported that the highest MMEJ levels observed with linearized SupF22 plasmids repaired in A-T extracts occurred with plasmids harboring breaks with blunt ends or recessed 5′ overhangs. Based on this observation, plasmids with these types of ends were selected to examine the contribution of Mre11 to MMEJ. Data obtained from two independent experiments assessing the two different types of breaks extended our previous findings of elevated microhomology-dependent repair in A-T extracts (Table 1). The frequency of MMEJ revertants in A-T nuclear extracts was 5–10 fold higher than that observed with controls. Immunodepletion of Mre11 resulted in a drop in the frequency of MMEJ in both types of extracts. Results from two independent experiments revealed an average of a three-fold decrease in MMEJ repair of plasmids with blunt ends in A-T nuclear extracts and in control extracts after immunodepletion of Mre11. A similar three-fold decrease was observed in the MMEJ repair of plasmids with recessed 5′ overhangs in Mre11-immunodepleted A-T nuclear extracts; a seven-fold decrease in MMEJ was observed with plasmids with recessed 5′ overhangs in Mre11-immunodepleted control extracts. This decrease in MMEJ levels was not the result of an effect of Mre11-immunodepletion on rejoining efficiency in the nuclear extracts. Comparable rejoining efficiencies, as indicated by transformation efficiencies, were obtained from both Mre11-immunodepleted and non-depleted extracts.
Figure 5.
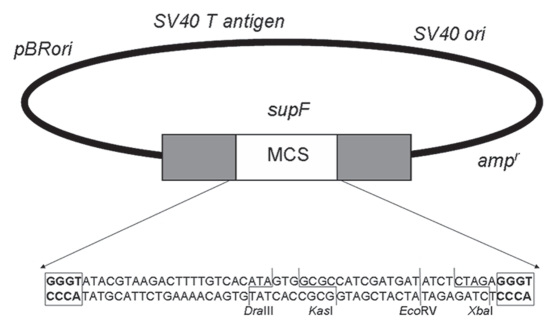
Organization of the SupF22 plasmid. The SupF22 plasmid contains the ampicilin resistance gene (bla) and the pBR327 ori from πAN7, the ori and T antigen coding region from SV40 and the IG region of M13. This plasmid also harbors the SupF gene, which encodes an amber suppressor tyrosyl-tRNA, interrupted by a multicloning site flanked by two GGGT sequences (bold in box frames). This renders the amber suppressor gene non-functional. Repair by MMEJ of a SupF22 plasmid linearized within the multicloning site and utilizing the aforementioned GGGT microhomologies results in deletion of the intervening multicloning site along with one GGGT repeat. This restores the functionality of the amber suppressor tRNA in the plasmid, which is then used to transform the indicator MBM7070 bacterial strain. This strain harbors an amber mutation within the lacZ gene. Reversion of the mutated SupF gene to a functional one by MMEJ is therefore scored as blue colony formation with the indicator strain on agar plates containing IPTG and X-gal. Restriction endonuclease sites within the multicloning site are indicated.
Table 1.
MMEJ frequency after repair of linearized SupF22 plasmids in Mre11-immunodepleted nuclear extracts
| Blunt ends (EcoRV) | Recessed 5′ ends (DraIII) | |||||||
| Nuclear extract | Mre11-immunodepletion | Exp. # | Trans. effi. (104) | Ratio of blue:total colonies | MMEJ freq. (%) | Trans. effi. (104) | Ratio of blue:total colonies | MMEJ freq. (%) |
| Control | − | 1 | 9.0 | 6:6808 | 0.088 | 20.9 | 35:13828 | 0.25c |
| 2 | 7.0 | 10:6272 | 0.16 | 13.9 | 16:11792 | 0.14c | ||
| + | 1 | 2.7 | 1:4316 | 0.023 | 15.6 | 4:11820 | 0.034 | |
| 2 | 11.1 | 5:6596 | 0.076 | 14.8 | 2:9160 | 0.022 | ||
| A-T | − | 1 | 15.1 | 98:9270 | 1.06a | 25.6 | 205:18300 | 1.12d |
| 2 | 29.8 | 143:16792 | 0.85a | 33.6 | 240:23732 | 1.01d | ||
| + | 1 | 17.2 | 36:12800 | 0.28b | 21.2 | 61:16780 | 0.36e | |
| 2 | 16.4 | 35:11420 | 0.31b | 50.1 | 66:23484 | 0.28e | ||
For blunt ends: p < 0.001; Fisher's Exact test of MMEJ frequency in aA-T versus control nuclear extracts and bA-T versus Mre11-imunodepleted A-T nuclear extracts. For recessed 5′ ends: p < 0.001; Fisher's Exact Test of MMEJ frequency in ccontrol versus Mre11-imunodepleted control nuclear extracts, dA-T versus control nuclear extracts and eA-T versus Mre11-imunodepleted A-T nuclear extracts.
MMEJ is suppressed in vivo after a reduction in Mre11 treatment with mirin or shRNA knock down.
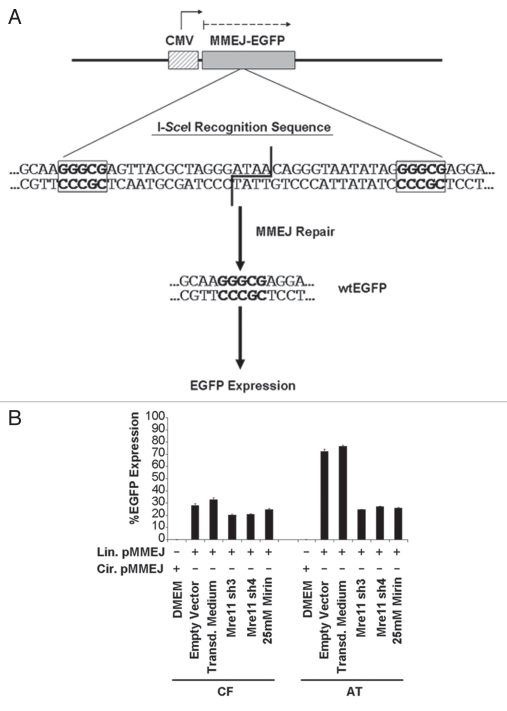
To assess the importance of the Mre11 nuclease in MMEJ and the regulation of this pathway by ATM in vivo, we used the pMMEJ plasmid (Fig. 6A), which harbors an insertion of 35 bases within the ORF of a wtEGFP gene. This insertion creates an I-SceI megaendonuclease recognition site flanked by two 5 bp microhomologies. The repair of an I-SceI linearized pMMEJ plasmid by MMEJ results in reconstitution of the wtEGFP gene and consequently allows expression of EGFP. Repair through MMEJ, as indicated by %EGFP expression levels (Fig. 6B), was 2.5-fold higher in A-T cell than in controls. Treatment of control fibroblasts with Mirin resulted in a 1.2-fold decrease in MMEJ levels and similar results were obtained with Mre11 knockdown. On the other hand, treatment of A-T cells with Mirin or knocking down Mre11 in that background resulted in a three-fold decrease in MMEJ, down to levels seen in control fibroblasts. Residual MMEJ levels may be due to incomplete inhibition of Mre11 or an Mre11-independent MMEJ pathway. Results from flow cytometry were corroborated with detection of EGFP expression in tested cells on a western immunoblot (Fig. 7).
Figure 6.
Suppression of in vivo MMEJ after Mirin-treatment or knocking down Mre11. (A) Schematic representation of the pMMEJ plasmid used to assess in vivo MMEJ repair. The pMMEJ plasmid was derived from the pEGFP-C3 plasmid by an insertion of 35 bp within the ORF of the wtEGFP gene. This creates an I-SceI megaendonuclease recognition site flanked by two 5 bp microhomologies. The repair of a linearized pMMEJ plasmid by MMEJ reconstitutes the wtEGFP gene and allows expression of EGFP. (B) WI-38VA13 (CF) and AT5BIVA (AT) cells were transfected with either linear or circular pMMEJ and with a mCherry transfection control plasmid. Repair was allowed and cells were analyzed for fluorescent protein expression by flow cytometry. Cells treated with 25 mM Mirin or in which Mre11 was knocked down by transduction with lentivirus encoding Mre11 shRNA (Mre11 sh3 or Mre11 sh4) were also analyzed. Cells transduced with empty vector or virus-free transduction medium were included as controls. Repair through MMEJ is represented by %EGFP expression; %EGFP expression = (number of cells expressing both EGFP and mCherry/number of cells expressing mCherry) × 100.
Figure 7.
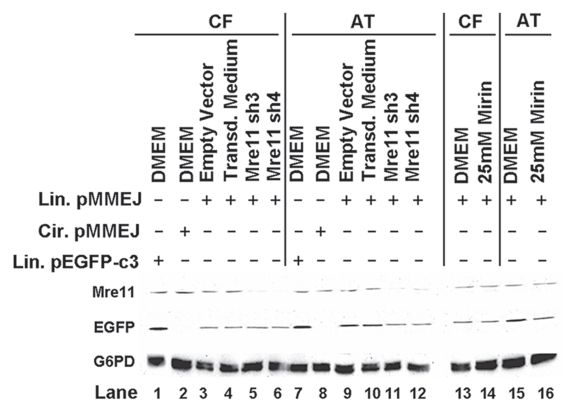
Western immunoblot for Mre11 and EGFP after MMEJ repair in Mirin-treated or Mre11-knockdown cells. WI-38VA13 (CF) and AT5BIVA (AT) cells were transfected with either linear or circular pMMEJ. Repair was allowed and cell extracts were then prepared. Cells treated with 25 mM Mirin or in which Mre11 was knocked down by transduction with lentivirus encoding Mre11 shRNA (Mre11 sh3 and Mre11 sh4) were also analyzed. Cells transduced with empty vector (Lanes 2 and 9) or virus-free transduction medium (Lanes 4 and 10) were included as controls. Transfection with linear pEGFP-c3 was used as an EGFP expression control (Lanes 1 and 7). Cell extracts were subjected to SDS-PAGE, transferred to a membrane and then probed for Mre11 and EGFP. G6PD was assayed for as a loading control.
Discussion
Neurodegeneration is a prominent pathologic feature of A-T3 that is not well explained by the cell cycle checkpoint deficiencies and other DNA damage signaling defects associated with an ATM malfunction. We show here that ATM suppresses an error-prone pathway of DNA double-strand break repair referred to as microhomology-mediated end joining (MMEJ). This pathway results in deletion of sequences surrounding the break site and rejoining at short regions of homology. Incorrect modulation of MMEJ may be the basis for the pathobiological mechanisms leading to neuronal degeneration in A-T. Neurocytes are postmitotic and therefore depend on mechanisms other than homologous recombination between sister chromatids to repair DSBs. One such mechanism is MMEJ which can lead to loss of significant genetic information if not kept in check. The accumulation of genetic damage could ultimately lead to loss of function and/or death of neuronal cells. In addition, the misregulation of error-prone repair discussed herein likely contributes to the genetic instability and propensity for leukemias and lymphomas observed in A-T.
Repair events via MMEJ are associated with disease processes resulting in chromosomal translocations and tumor-formation.32–36 We show here that DNA degradation and consequent microhomology-directed break rejoining involved in MMEJ are highly dependent on Mre11 and in particular on its nuclease function. Both Mre11 knockdown and immunodepletion resulted in a decreased level of DNA substrate degradation. This was prominent in ATM-deficient nuclear extracts which have a marked elevation in nuclease activity and in MMEJ. Treating these extracts with Mirin, an inhibitor of Mre11 nuclease activity,30 also resulted in decreased substrate degradation, but not to the extent seen with Mre11 knock down or immunodepletion. This may indicate incomplete inhibition of Mre11 or the participation of one or more accessory nucleases that require the physical presence of Mre11 at the DNA end but whose kinetics are less dependent on Mre11 nuclease function.
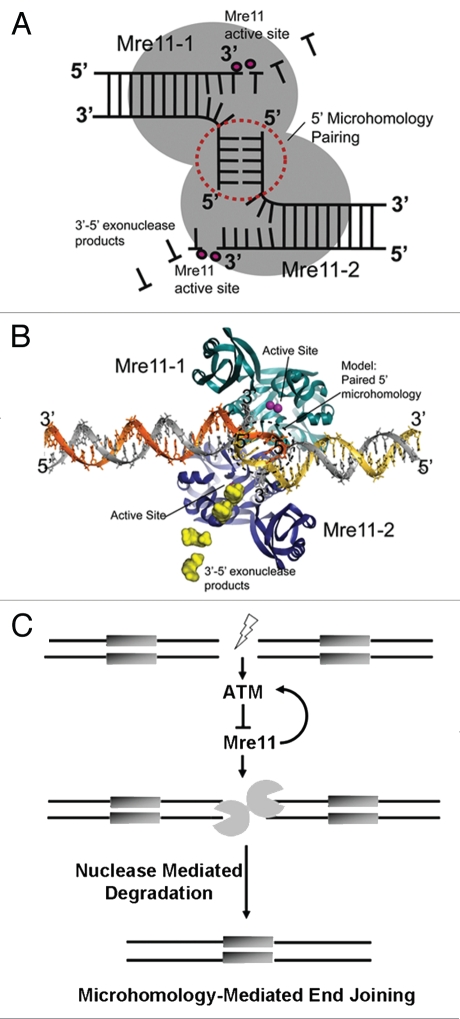
Immunodepletetion of Mre11 from nuclear extracts resulted in decreased levels of linearized plasmid rejoining in these extracts via MMEJ. Moreover, using an in vivo reporter assay system we observed a decrease in MMEJ levels after Mre11 knockdown and after treating cells with Mirin. This direct demonstration of Mre11 involvement as a nuclease in MMEJ in vivo in a mammalian system is consistent with recent studies.15,16 Overexpression of Mre11 was demonstrated to increase the frequency of deletions at DNA ends whereas a nuclease deficient mutant Mre11 failed to cause a similar increase. Previous reports have indicated that purified Mre11 degrades DNA in vitro up to regions of microhomology, where it stalls.14 Consistent with biochemical analyses, recent Mre11-DNA complex crystal structures suggest that Mre11 DNA bridging may provide the requisite synaptic DNA architectures for DNA end pairing and degradation reactions regulating MMEJ (Fig. 8A and B).37 We modeled the structure of an Mre11 dimer directly engaging DNA ends and the model shows how the 5′ ends of a bridged DSB are placed into close juxtaposition by the dimeric protein scaffold. In this model, Mre11 complex DNA unwinding and 3′–5′ exonuclease activities may collaborate to facilitate simultaneous pairing of extended 5′ termini and exonucleolytic degradation of the 3′ ends in MMEJ (Fig. 8A and B).
Figure 8.
Mre11-mediated microhomology-mediated end-joining and its regulation by ATM. (A) Schematic for Mre11 dimer-mediated MMEJ. The Mre11 homodimer is a symmetric structure that binds two DNA ends. 5′ termini are paired at the interface of the dimer, whereas 3′ ends are degraded by Mre11 3′–5′ exonuclease activity in Mre11 active site (B) Structure-based model for an Mre11 5′-microhomology end joining complex based on the Mre11-DNA synaptic complex (PDB ID 3DSC). The 5 bp 5′ microhomology (circled dotted line) was modeled by extending the 5′ termini of DNAs observed in the X-ray structure. 5′ MMEJ pairing could occur in the space between crystallographically defined DNA interaction sites. (C) Model delineating the role of ATM in suppression of error-prone DSB repair via microhomology-mediated end joining. ATM is activated upon the formation of a DSB. Then, its kinase activity maintains end-stability at a break through suppression of Mre11-dependent microhomology-directed DNA degradation and subsequent end joining.
Other mediators also have been associated with this error-prone repair pathway and these include the BLM helicase,38 PARP-1, XRCC1, DNA ligase III,39 the FEN1 endonuclease, DNA polymerase ε 40 and the Exo1 exonuclease.41 Whether these mediators are involved in the Mre11-depedent pathway merits investigation. Given the complexity of DSB repair in mammalian cells it is possible that more than one pathway leads to the formation of MMEJ products.
Based upon the key roles identified here for ATM and Mre11, the regulation of MMEJ and suppression of error-prone repair can be better explored. ATM deficiency leads to an increase in MMEJ in vitro.11 Here we extend this observation in vivo and reveal that Mre11 is the target of this regulation. This increase in MMEJ is not simply due to a reduction in the ability to rejoin DNA ends in the absence of appropriate ATM function, as nuclear extracts from both A-T and control cells are equally efficient in rejoining DNA ends.11 The ATM kinase activity was necessary for repression of DNA degradation by Mre11. Thus, inhibition of degradation and MMEJ by ATM may be through phosphorylation of the nuclease complex. ATM phosphorylates Nbs1 in response to DNA damage,20,21 and ATM-dependent phosphorylation of Mre11,27 and Rad50,28 was reported. Structurally, the Nbs1 connects directly to the Mre11 adjacent to its ATM interaction motif, so the mechanical linkage of Nbs1 to both ATM and Mre11 is established structurally.42 Furthermore, the connection of Rad50 to Mre11 suggests ATP driven conformational change may help open the DNA for Mre11 processing in ways dependent upon the state of Rad50.43 Notably, examination of MMEJ in yeast revealed roles for both Mre11 and Tel1, the yeast ATM homologue, in promoting MMEJ.12,13 The observation that Tel1 promotes MMEJ in yeast, but ATM suppresses MMEJ in mammalian cells may indicate the divergence of these two proteins and of their roles. It may also demonstrate pathway differences between yeast and mammalian systems. The yeast Dnl4 (DNA ligase 4), for example is required for MMEJ, whereas its mammalian homologue, LIG4, is dispensable for the pathway.44–47
Together our results and prior observations provide an integrated understanding of the critical roles of both ATM and Mre11 nuclease in MMEJ. We conclude that a DSB leads to activation of ATM via the MRN complex. ATM then regulates, through its kinase activities, the degradation and rejoining of DNA ends by Mre11. In particular, it suppresses error-prone MMEJ repair of the DSB by inhibiting thomology-directed DNA degradation mediated by the Mre11 nuclease (Fig. 8C).
Materials and Methods
Cell culture and preparation of nuclear extracts.
AT5BIVA, a SV40-transformed A-T fibroblast cell line, was obtained from the Coriell Cell Repository (Coriell Institute of Medical Research, Camden, NJ). The SV40-transformed human lung fibroblast cell line, WI38VA13, was obtained from ATCC (American Type Culture Collection, Manassas, VA) and was used as a control cell line for AT5BIVA. Maintenance of cell cultures and preparation of nuclear extracts were performed as previously described.23
Transient kinase dead (KD)-ATM expression.
The KD-ATM expression plasmid24 was a generous gift from Dr. M. Kastan (St. Jude Children's Research Hospital, Memphis, TN). Cells grown on 150 mm plates in DMEM supplemented with 10% FBS were allowed to reach a confluency of 60%. The medium was replaced with DMEM and then cells were transfected with 26 µg of the KD-ATM plasmid or the pEGFP-C3 (Clontech, Palo Alto, CA) transfection control plasmid. Lipofectin (Sigma, St. Louis, MO) was used as a transfection reagent according to manufacturer protocols. After an overnight incubation with the transfection medium at 37°C in 5% CO2, the medium was replaced with DMEM containing 10% FBS and cells were then incubated for 48 hr to allow expression. Growth medium was then replaced with DMEM containing 10% FBS and supplemented with 1.5 mg/ml Geneticin to select for KD-ATM expressing cells. After 48 hr, cells were washed and harvested for preparing nuclear extracts as previously described.23
Assessment of kinase dead (KD)-ATM expression.
To verify the expression of KD-ATM, ATM was immunoprecipitated from nuclear extracts and the precipitates tested in a kinase assay as previously described with some modification.24 To immunoprecipitate ATM, rProtein G agarose beads (Invitrogen Corporation, Carlsbad, CA) were preblocked overnight in IP buffer (50 mM Tris Cl pH 7.5, 300 mM NaCl, 10% Glycerol, 1% Triton-X, 1 mM PMSF, 1 mM DTT) containing 3% BSA and supplemented with Complete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics GmbH, Mannheim, Germany) used according to manufacturer recommendations. Nuclear lysates were precleared by incubation with the pre-blocked rProtein G agarose beads to eliminate kinases that bind non-specifically to the beads. Pre-blocked beads (20 µl) were drained from the pre-blocking buffer and added to the nuclear extracts (50 µg). Reactions were mixed by rotation at 4°C for 30 min, centrifuged at 5,000 rpm for 1 min, and the beads were discarded. Pre-clearing was repeated three times. Mouse anti-ATM (Sigma, St. Louis, MO) (2.5 µg) was added and the reactions were mixed by rotation overnight at 4°C. Then, pre-blocked beads (20 µl) were added and the reactions were mixed by rotation at 4°C for 3 hr. Beads were collected by centrifugation at 5,000 rpm for 1 min, washed three times with 200 µl IP buffer, once with 200 µl Tris/LiCl buffer (100 mM Tris Cl pH 7.5, 0.5 M LiCl) and twice with 200 µl kinase buffer (10 mM HEPES pH 7.5, 50 mM NaCl, 10 mM MgCl2, 10 mM MnCl2, 5 µM ATP). To perform the kinase assay, following the final wash, beads were resuspended in 20 µl of kinase buffer in the presence of 1 µg of PHAS-I (Stratagene, La Jolla, CA) and 3.33 pmol of (γ-32P)ATP. Reaction products were separated on 12% denaturing-polyacrylamide gels. Gels were dried and exposed to a storage phosphor screen (GE Healthcare, Princeton, NJ) for phosphoimage analysis. The exposed screen was scanned with a Typhoon 9410 Variable Mode Imager (GE Healthcare, Princeton, NJ) and analyzed using ImageQuant v5.2 software (Molecular Dynamics, Amersham Biosciences, Princeton, NJ).
Mre11 immunodepletion.
To prepare Mre11-immunodepleted nuclear extracts, 50 µg of nuclear extract were added to 1 µg of rabbit anti-Mre11 (Abcam, Inc., Cambridge, MA) and mixed by rotation at 4°C overnight. Then, 30 µl of protein G magnetic beads (New England Biolabs, Ipswich, MA) were added to the protein-antibody complexes and mixed by rotation for 3 hr at 4°C. The magnetic beads were removed from the extracts by separation on a magnetic rack and the immunodepleted supernatants were collected.
Knockdown of Mre11 expression.
To knockdown Mre11, cells at a confluency of 50% were transduced with lentiviral particles encoding shRNA for Mre11 in DMEM containing Polybrene (Sigma, St. Louis, MO) at a concentration of 8 µg/ml. Two shRNA sequences from the MISSION™ TRC shRNA Target Set NM_005591 (Sigma, St. Louis, MO) were used. The two plasmids used had Sigma reference numbers TRCN0000039868, referred to as Mre11 sh3 in the text, and TRCN0000039872, referred to as Mre11 sh4. Cells were incubated with the virus particles overnight, medium was replaced with DMEM containing 10% FBS and the cells were tested for expression or employed in subsequent experiments 48 hours later.
Immunoblotting.
Samples (20 µg) were incubated at 100°C for 5 min in Laemmli sample buffer and then electrophoresed on 12% SDS-polyacrylamide gels. Proteins were transferred to Trans-Blot Medium nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA), probed and then visualized with the SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology, Inc., Milwaukee, WI). The FluorChem system (Alpha Innotech Corporation, San Leandro, CA) was used for gel documentation. The Mre11 (1:70,000) primary antibody was obtained from Abcam, Inc., (Cambridge, MA). The GFP (1:1,000) primary antibody was obtained from Santa Cruz Biotechnology, Inc., (Santa Cruz, CA). The G6PD (1:10,000) primary antibody was purchased from GeneTex, Inc., (Irvine, CA).
Duplex oligonucleotide substrates.
Duplex DNA substrates (Table 2) were generated as previously described23 and employed in the DNA end-processing assay to assess degradation in the nuclear extracts tested. To examine 3′ recessed strand degradation a 71 nt Template strand harboring a Cy3 moiety at its 5′ end was hybridized to a 76 nt Top Strand. The 5′ end of the 71 nt Template and the 3′ end of the 76 nt. Top Strand were protected from nuclease mediated degradation by phosphorothioate bonds linking the last six bases at those ends. To examine 5′ end-degradation, a 50 nt Top Strand harboring a Cy3sp moiety at its 3′ end was hybridized to a 45 nt Template. The 3′ end of the 50 nt Top Strand and the 5′ end of the 45 nt Template were protected from nuclease mediated degradation by phosphorothioate bonds linking the last six bases at those ends.
Table 2.
Oligonucleotides used to assess DNA degradation
| Name | Length (nt) | Sequence |
| Top strand | 76 | 5′ACCCAG AGC TCG GTA CCC GGG GAT CCT CTA GAG TCG ACC TGC AGG CAT GCA AGC TTG GCA CTG GCC GTC GTT TTA C |
| Cy3 Template | 71 | 3′CTC GAG CCA TGG GCC CCT AGG AGA TCT CAG CTG GAC GTC CGT ACG TTC GAA CCG TGA CCG GCA GCA AAA TG/Cy3 |
| Cy3Sp Top Strand | 50 | 5′ACCCAA GTC GAC CTG CAG GCA TGC AAG CTT GGC ACT GGC CGT CGT TTT AC/Cy3sp |
| Template | 45 | 3′TCA GCT GGA CGT CCG TAC GTT CGA ACC GTG ACC GGC AGC AAA ATG |
Underlined nucleotides are unpaired in the final substrate. Bold nucleotides are linked by phosphorothioate linkages.
DNA end-processing assay.
Assessment of duplex substrate degradation was performed as previously described23 with some modification. Reactions (25 µl) containing 25 µg of nuclear extract in reaction buffer (65.5 mM Tris-Cl pH 7.5, 10 mM MgSO4, 10 mM MnSO4, 91 nM EDTA, 9.1% glycerol, 1 mM ATP) and 45 pmoles of a DNA duplex were assembled on ice and then incubated at 30°C for various lengths of time, as described per experiment in the text. Reaction buffer was supplemented with Complete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics GmbH, Mannheim, Germany) used according to the manufacturer's instructions. Where indicated, extracts in reaction buffer were pre-treated for 5 min with Mirin (ChemBridge Corporation, San Diego, CA) at a concentration of 1 mM prior to addition of the duplex substrates and commencing the end-processing assay. End-processing reactions were stopped by adding 5 ul per reaction of a mixture of 2% SDS, 50 mM EDTA and 1 mg/ml Proteinase K followed by incubation at 37°C for 15 min.
Products from the DNA end-processing assay were separated on 12% acrylamide/7 M urea sequencing gels (Sequagel Sequencing System reagents, National Diagnostics, Atlanta, GA). Gels were imaged with a Typhoon 9410 Variable Mode Imager and product intensities determined using ImageQuant v5.2 software. Product intensities were corrected for background and then converted into percent intensities where percent intensity = (product intensity/total lane intensity) × 100.
Assessment of microhomology-mediated end joining in vitro.
To assess MMEJ in vitro, the repair of a linearized SupF22 plasmid in nuclear extracts was assayed as previously described.11
Assessment of microhomology-mediated end joining in vivo.
To examine MMEJ in vivo, the pMMEJ plasmid was constructed. This plasmid was derived from the pEGFP-C3 plasmid (Clontech, Palo Alto, CA) by PCR mutagenesis using QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) and employing the mutagenesis primer pair of the following sequences: 5′CAT GGT GAG CAA GGG CGA GTT ACG CTA GGG ATA ACA GGG TAA TAT AGG GGC GAG GAG CTG TTC ACC GGG GTG3′ and 5′CAC CCC GGT GAA CAG CTC CTC GCC CCT ATA TTA CCC TGT TAT CCC TAG CGT AAC TCG CCC TTG CTC ACC ATG3′. This resulted in an insertion of 35 bases at nucleotide position 628 of the pEGFP-C3 plasmid within the ORF of the wtEGFP gene. This insertion created an I-SceI megaendonuclease recognition site flanked by two 5 bp microhomologies (bold in the primer sequences). The sequence of the insert is underlined in the mutagenesis primer pair. The repair of a linearized pMMEJ plasmid by MMEJ results in reconstitution of the wtEGFP gene and consequently allows expression of EGFP. The pMMEJ plasmid was linearized with I-SceI (New England Biolabs, Ipswich, MA), treated with calf intestinal phosphatase, and then gel-purified using the Zymoclean gel DNA recovery kit (Zymo Research Corp., Orange, CA). Linear plasmid was transfected into cells and reconstitution of the wtEGFP gene via MMEJ was assessed by flow cytometry.
Cells were grown in six-well plates in DMEM supplemented with 10% FBS, allowed to reach a confluency of 60% and the medium was then replaced with DMEM. Linearized pMMEJ (2 µg) was transfected along with the mCherry transfection control plasmid (0.5 µg) (a generous gift from Dr. H. Fares at the Department of Molecular and Cellular Biology, University of Arizona, Tucson, AZ) using the Lipofectin (Sigma, St. Louis, MO) transfection reagent (6 µl) according to manufacturer instructions. Circular pMMEJ was used as a control where indicated. Cells were incubated overnight with the transfection medium at 37°C in 5% CO2, the medium was then replaced with DMEM containing 10% FBS and cells were incubated for another 48 hr. Assessment of EGFP and mCherry expression was conducted using the BD LSR II (BD Biosciences, Franklin Lakes, NJ) cytometer and data was analyzed with the FACSDiva v6.1.1 software. The pMMEJ was transfected in excess of the mCherry plasmid; consequently, cells expressing only EGFP were disregarded. The percentage of EGFP expression and consequently that of MMEJ, was calculated using the following formula: %EGFP expression = (number of cells expressing both EGFP and mCherry/number of cells expressing mCherry) × 100. Where indicated in the text, cells were treated with 25 mM Mirin overnight prior to transfection, and this concentration of the Mre11 nuclease inhibitor was maintained in the growth medium after transfection. Alternatively, Mre11 was knocked down, as described above, prior to transfection with the MMEJ assessment plasmids.
Acknowledgements
We thank the members of the Genomics Maintenance Group at the University of Arizona for helpful discussions and Eric G. Thompson, Hope Jones and Helen F. Smith for critical review of the manuscript. This work was supported by NIH grant R01-NS34782 to Kathleen Dixon and P01 CA92584 to John Tainer.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12363
References
- 1.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 2.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 3.Taylor AM, Byrd PJ. Molecular pathology of ataxia telangiectasia. J Clin Pathol. 2005;58:1009–1015. doi: 10.1136/jcp.2005.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6:931–942. doi: 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 5.DiBiase SJ, Zeng ZC, Chen R, Hyslop T, Curran WJ, Jr, Iliakis G. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res. 2000;60:1245–1253. [PubMed] [Google Scholar]
- 6.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 9.Wu W, Wang M, Wu W, Singh SK, Mussfeldt T, Iliakis G. Repair of radiation induced DNA double strand breaks by backup NHEJ is enhanced in G2. DNA Repair (Amst) 2008;7:329–338. doi: 10.1016/j.dnarep.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Carty MP, Oakley GG, Seidman MM, Medvedovic M, Dixon K. Expression of ATM in ataxia telangiectasia fibroblasts rescues defects in DNA double-strand break repair in nuclear extracts. Environ Mol Mutagen. 2001;37:128–140. doi: 10.1002/em.1020. [DOI] [PubMed] [Google Scholar]
- 12.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci USA. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 16.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 18.Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 19.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Ranganathan V, Weisman DS, Heine WF, Ciccone DN, O'Neill TB, et al. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 21.Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, et al. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 22.Kim JE, Minter-Dykhouse K, Chen J. Signaling networks controlled by the MRN complex and MDC1 during early DNA damage responses. Mol Carcinog. 2006;45:403–408. doi: 10.1002/mc.20221. [DOI] [PubMed] [Google Scholar]
- 23.Rahal EA, Henricksen LA, Li Y, Turchi JJ, Pawelczak KS, Dixon K. ATM mediates repression of DNA end-degradation in an ATP-dependent manner. DNA Repair (Amst) 2008;7:464–475. doi: 10.1016/j.dnarep.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 25.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 26.van den Bosch M, Bree RT, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan SS, Chang HL, Hou MF, Chan TF, Kao YH, Wu YC, et al. Neocarzinostatin induces Mre11 phosphorylation and focus formation through an ATM and NBS1-dependent mechanism. Toxicology. 2002;177:123–130. doi: 10.1016/s0300-483x(02)00220-2. [DOI] [PubMed] [Google Scholar]
- 28.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garner KM, Pletnev AA, Eastman A. Corrected structure of mirin, a small-molecule inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2009;5:129–130. doi: 10.1038/nchembio0309-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 32.Bentley J, Diggle CP, Harnden P, Knowles MA, Kiltie AE. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 2004;32:5249–5259. doi: 10.1093/nar/gkh842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin KH, Kang MK, Kim RH, Kameta A, Baluda MA, Park NH. Abnormal DNA end-joining activity in human head and neck cancer. Int J Mol Med. 2006;17:917–924. [PubMed] [Google Scholar]
- 34.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Windhofer F, Krause S, Hader C, Schulz WA, Florl AR. Distinctive differences in DNA double-strand break repair between normal urothelial and urothelial carcinoma cells. Mutat Res. 2008;638:56–65. doi: 10.1016/j.mrfmmm.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Yoshimoto M, Joshua AM, Chilton-Macneill S, Bayani J, Selvarajah S, Evans AJ, et al. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia. 2006;8:465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langland G, Elliott J, Li Y, Creaney J, Dixon K, Groden J. The BLM helicase is necessary for normal DNA double-strand break repair. Cancer Res. 2002;62:2766–2770. [PubMed] [Google Scholar]
- 39.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 40.Gottlich B, Reichenberger S, Feldmann E, Pfeiffer P. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur J Biochem. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- 41.Decottignies A. Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics. 2007;176:1403–1415. doi: 10.1534/genetics.107.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Current Opinion in Structural Biology. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 44.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 45.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, et al. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J Exp Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]