Abstract
Fatty acyl-AMP ligase (FAAL) is a new member of a family of adenylate-forming enzymes that were recently discovered in Mycobacterium tuberculosis (Mtb). They are similar in sequence to fatty acyl-CoA ligases (FACLs). However, while FACLs perform a two-step catalytic reaction, AMP ligation followed by CoA ligation using ATP and CoA as cofactors, FAALs produce only the acyl adenylate and are unable to perform the second step. We report X-ray crystal structures of full length FAALs from E. coli (EcFAAL) and Legionella pneumophila (LpFAAL) bound to acyl adenylate, determined at resolution limits of 3.0 and 1.85 Å, respectively. The structures share a larger N-terminal domain and a smaller C-terminal domain, which together resemble the previously determined structures of FAAL and FACL proteins. Our two structures occur in quite different conformations. EcFAAL adopts the adenylate forming conformation typical of FACLs, whereas LpFAAL exhibits a unique intermediate conformation. Both EcFAAL and LpFAAL have insertion motifs that distinguish them from the FACLs. Structures of EcFAAL and LpFAAL reveal detailed interactions between this insertion motif and the interdomain hinge region and with the C-terminal domain. We suggest that the insertion motifs support sufficient interdomain motions to allow substrate binding and product release during acyl adenylate formation, whereas they preclude CoA binding thereby preventing CoA ligation.
Keywords: Fatty acyl-AMP ligase, Fatty acyl-CoA ligase, X-ray structure, AMP, CoA
Introduction
Adenylate forming enzymes are wide spread in nature. They play important roles in fatty acid metabolism in bacteria and eukaryotes. Fatty acid metabolism primarily involves two steps, adenylation and thioester-forming reactions catalyzed by a single enzyme, fatty acyl-CoA ligases (FACLs)1,2. Three-dimensional structures of many FACLs are available in the Protein Data Bank (PDB, www.pdb.org), both with and without bound cofactors2,3. FACLs occur as two domain structures, which display three functionally distinct conformations, including an adenylate forming conformation (e.g., PDB ID: 1T5D), a thioester-forming conformation (e.g., PDB ID: 2P2F, 1PG4, 1V26 and 3CW9), and so-called intermediate conformations (e.g., PDB ID: 3IPL)2,4. Reger et al. proposed that FACL uses interdomain movements to reconfigure the acyl CoA active site for the second catalytic step3.
Recently, an alternate mechanism of fatty acid activation has been described. This process occurs in Mycobacterium tuberculosis (Mtb) and involves a single step adenylation reaction catalyzed by Fatty Acyl AMP ligase (FAAL)5,6. To date, nine distinct FAAL encoding genes have been identified adjacent to genes encoding polyketide synthases (PKS) in Mtb6. Functional studies on FAALs from Mtb (MtFAAL) documented that the enzymes activate fatty acids as acyl-adenylates, but do not catalyze thioester-forming reactions5,6. It was therefore suggested that the acyl-adenylates serve as substrates for multifunctional PKSs to permit synthesis of complex lipids such as phthiocerol dimycocerosate (PDIM), sulfolipids, mycolic acids, and mycobactin6.
Studies on MtFadD32 demonstrated that it is a FAAL and catalyzes acyl-adenylate formation in the absence of an a acyl carrier protein (ACP), and in which the product is likely transferred to an ACP when this is present5,7. Since full-length MtFAAL has proven recalcitrant to crystallization, the crystal structure of the isolated N-terminal domain of MtFAAL was determined6. The overall fold of the N-termimal domain of MtFAAL is similar to that of FACL. It was proposed that the insertion motif present in all FAALs (as compared to FACLs) modulate interdomain movement and disrupt acyl-CoA formation6. MtFAAL does support thioester-forming reactions following removal of the insertion motif. To better understand fatty acid metabolism, FAALs from E. coli and Legionella pneumophila were targeted for structure determination. Herein, we report the first crystal structures of full length FAALs from E. coli and L. pneumophila with acyl adenylate bound in their active sites. With the benefit of these structures, we provide a phenomenological explanation for their restricted catalytic activity. We suggest that EcFAAL and LpFAAL can undergo some of the interdomain movements similar to those seen for the FACLs. However, the presence of the insertion motif restricts the FAALs to interdomain movements that support only the adenylation reaction5. Structure and sequence comparisons with FACLs reveal that the hinge region residues (this is often referred to as the A8 motif) are important for both interdomain movement and for catalytic activities. While both EcFAAL and LpFAAL appear capable of binding CoA in a fashion similar to that seen with the FACLs, we posit that the insertion motif prevents the conformational change required to enable the CoA thiol to approach and nucleophilically attack the acyl adenylate anhydride. Consequently, mutant forms of EcFAAL and LpFAAL lacking the insertion motif should support acyl-CoA formation as shown for MtFAAL. Although it is formally possible that binding of a protein or a co-factor imbues EcFAAL and/or LpFAAL with acyl CoA synthase activity, it is more likely that EcFAAL and LpFAAL transfer acyl adenylate onto an ACP or FACL protein. In this regard we note that both EcFAAL and LpFAAL genes are located adjacent to acyl carrier proteins.
Results and Discussion
Detection and characterization of acyl adenylate chain length
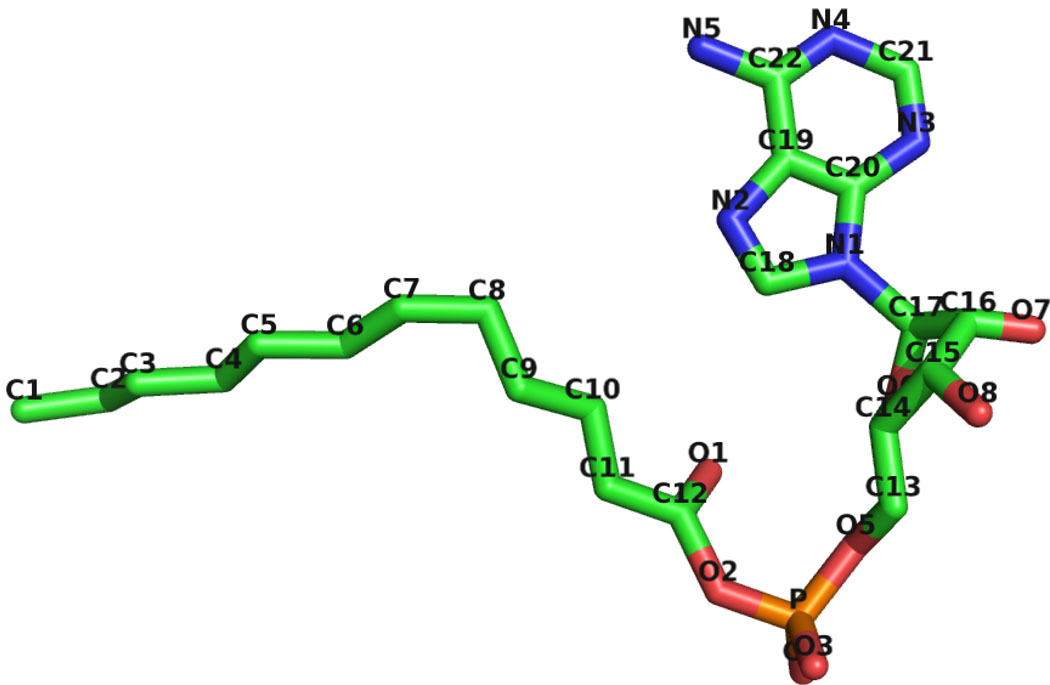
Although no substrate or product was added during crystallization of either EcFAAL or LpFAAL, continuous and significant residual electron density features were observed in the vicinity of the N-terminal domains of both structures. Based on the shape and size of these density features and their positions on the surface of the protein, this additional density was initially modeled as bound acyl adenylate. Subsequently, mass spectrometry was used to identify conclusively the bound ligands. Two additional molecular species with m/z 528 and m/z 556 predominated in purified EcFAAL protein preparations, whereas only molecular species corresponding to m/z 528 was detected in the corresponding LpFAAL protein preparation. In-source collision induced dissociation of these molecular species produced a common fragment ion at m/z 356, which matched the AMP standard. Therefore, the molecular species proved to be saturated C12 and C14 acyl adenylate, respectively. Thus, EcFAAL was co-purified from E. coli with a mixture of C12 and C14 acyl adenylates, while LpFAAL was co-purified with C12 adenylate (Figure 1).
Figure 1.
Structure of C12 acyl adenylate from EcFAAL.
Enzyme assays
EcFAAL and LpFAAL were assayed for formation of octanoyl adenylate and octanoyl CoA. On incubating EcFAAL or LpFAAL with octanoic acid, ATP, and CoA, formation of octanoyl-CoA was not detected. However, both EcFAAL and LpFAAL were able to catalyze formation of octanoyl adenylate, with kcat and Km values of 0.07 ± 0.01 min−1/94 ± 19 µM and 0.022±0.001 min−1/811±111 µM, respectively. For reference, MtFAAL (FadD32)7 exhibits kcat values 0.015–0.15 min−1, depending on fatty acid chain length, and Km values of 3.2 µM for a C16 substrate to 2640 µM for a C12 substrate. Although MtFAAL accepts longer fatty acid substrates than those found in E. coli, it appears likely that the EcFAAL Km value for acyl substrates also varies as a function of length and that octanoic acid (Km = 94 µM) is not the optimal substrate. Also, MtFAAL can transfer acyl adenylate to an ACP and the lack of appropriate acceptor for acyl adenylate probably results in lower kcat and higher Km for EcFAAL and LpFAAL. The ability of EcFAAL to synthesize the acyl adenylate of octanoic acid is consistent with the presence of an acyl adenylate fortuitously co-purified and co-crystallized with the enzyme.
Sequence analysis
A sequence alignment of EcFAAL, LpFAAL, MtFAAL, and the FACL enzymes from Salmonella enterica (SeFACL), Thermus thermophilus (TtFACL), and Alcaligenes sp (AsFACL) is shown in Figure 2a. The substrate for AsFACL is 4-chlorobenzoate and it is a 4-chlorobenzoyl-CoA ligase, whereas TtFACL prefers myristate as the substrate. The sequence identity between MtFAAL, and EcFAAL and LpFAAL is 30% and 29%, respectively. Both EcFAAL and LpFAAL possess a ~27 residue insertion motif. However, the amino acid sequence of this insertion motif is not conserved among the three FAALs. Sequence alignment with the FACLs shows that the hinge region (A8 motif), the acyl adenylate binding sites, and the gate motif are highly conserved not only among FAALs but also with the FACLs (Figure 2a)2,8, suggesting that FAAL and FACL share the same mechanism of adenylate formation.
Figure 2.
a) ClustalW sequence alignment of FAALs from M. tuberculosis (MtFAAL), E. coli (EcFAAL), and L. pneumophila (LpFAAL) and FACL from Salmonella enterica (SeFACL) and Thermus thermophilus (TtFACL). Insertion motifs are highlighted in green, the hinge region in cyan and the gate motif in yellow. The highly conserved loop region in C-domain is highlighted in red. b) Arrangement of EcFAAL, LpFAAL and MtFAAL in their genome.
Unlike in M. tuberculosis, where nine MtFAAL genes are located adjacent to polyketide and non-ribosomal peptide sythetase (NRPS) genes5,6, EcFAAL and LpFAAL represent the only FAALs detectable in the genomes E. coli and L. pneumophila. The gene encoding EcFAAL is located adjacent to those encoding various CoA binding enzymes, acyl carrier protein and member associated proteins, while the gene for LpFAAL is located adjacent to genes for LpFACL, an acyl carrier protein, and (one or more) transporter proteins (Figure 2b). Since MtFAAL has acyl-ACP ligation function, EcFAAL and LpFAAL might also transfer acyl adenylate to an ACP for further metabolic elaboration.
Overall Structure
The domain organization
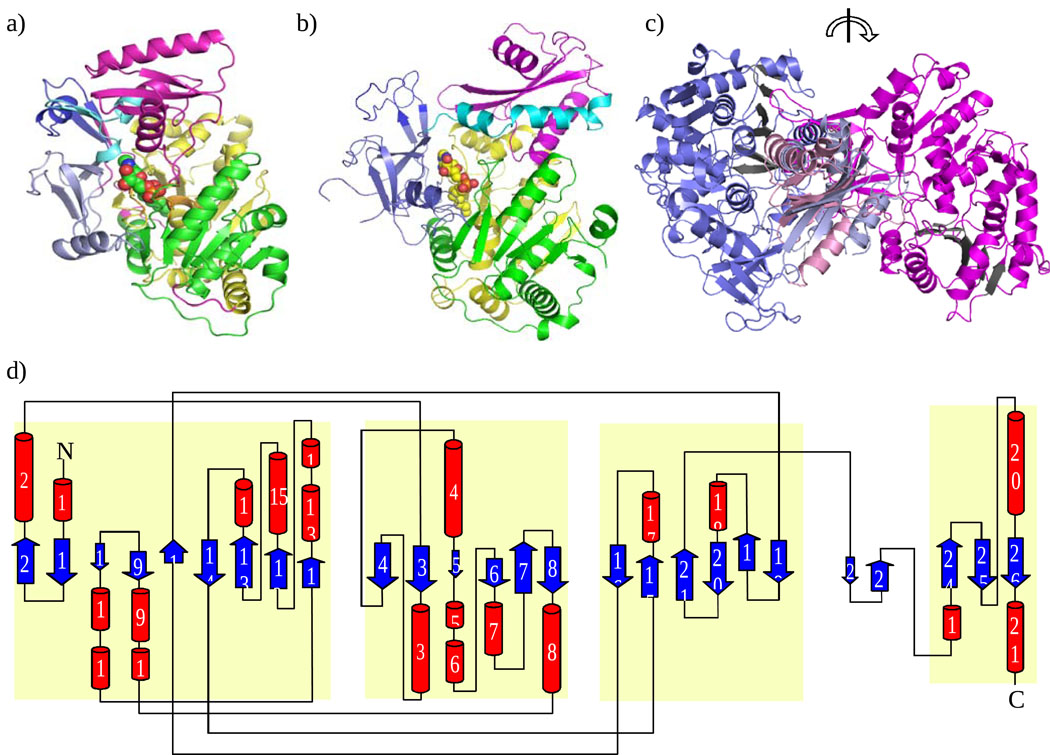
Crystal structures of EcFAAL (Figure 3a and 3d) and LpFAAL (Figure 3b) have been determined at 3 and 1.85 Å resolution, respectively. The crystal structure of LpFAAL cocrystallized with pyrophosphate (PPi) was also determined at 2.0 Å. The two LpFAAL structures are identical with some minor differences in the loop regions. PPi is bound to the gate region of the adenylation active site. Both FAALs exhibit a very similar fold typical of acyl CoA synthases. The folded polypeptide chains consist of two domains, a larger N-terminal domain (EcFAAL residues 1 – 457) and a smaller C-terminal domain (EcFAAL residues 478 – 568) connected by a flexible loop (EcFAAL residues 458 – 477, hereafter called the hinge region) that permits the C-terminal domain to adopt different orientations with respect to the N-terminal domain. The N-terminal domain can be conceptually divided into three subdomains. Subdomain 1 (green in Figure 3a) consists of an eight stranded β-sheet surrounded by ten α-helices. Subdomain 2 (yellow in Figure 3a) is made up of a six-stranded β-sheet flanked by six α-helices while subdomain 3 (violet in Figure 3a) is composed of a distorted four-stranded β-sheet and one α-helix. These three domains come together to form a pocket for substrate/product binding. The insertion motif (EcFAAL residues 356 – 382), a feature common to all FAALs, is shown in dark blue and occurs within the N-terminal domain. The N-terminal domain and the C-terminal domain (magenta) are connected by a hinge region (cyan in Figure 3a and Figure 2a). In FACL, the C-terminal domain undergoes a large conformational change to generate the active site that supports acyl CoA synthetase activity2,4. While both of the individual domains comprising EcFAAL and LpFAAL are structurally similar, their respective orientation differs between the two structures. The overall rmsd for the two proteins is ~1.6 Å for 567 Cα pairs9. Alignments of the individual domains show an rmsd of ~1.5 Å for the N-terminal domains (440 Cα pairs) and ~2.0 Å for the C-terminal domains (117 Cα pairs). The C-terminal domains of EcFAAL and LpFAAL share lower sequence similarity than do the N-terminal domains, which could explain the larger rmsd value. The N-terminal domain of MtFAAL is also very similar to that of LpFAAL (rmsd ~1.4 Å for 434 Cα pairs) and EcFAAL (rmsd ~1.5 Å for 430 Cα pairs). Structural comparison of FAAL and TtFACL shows structural similarity between both domains (N-terminal domains: EcFAAL versus TtFACL ~2.3 Å and LpFAAL versus TtFACL ~2.1 Å; C-terminal domains: EcFAAL versus TtFACL ~2.1 Å and LpFAAL versus TtFACL ~1.9 Å).
Figure 3.
Structure of FAAL from E. coli and L. pneumophila. (a) EcFAAL monomer showing location of acyl adenylate (sphere model) (b) Structure of LpFAAL monomer showing location of acyl adenylate (sphere model). N-terminal domain has three subdomains 1, 2, 3 colored in green, yellow, and pale violet, respectively. Insertion motif, hinge region, and C-terminal domain are colored in dark blue, light cyan and magenta, respectively. (c) EcFAAL (blue) and LpFAAL (magenta) overlaid at C-terminal domain. Sheet A of N-terminal domain is colored in black and is rotated about 180 degrees about the vertical axis. (d) Topology diagram of EcFAAL. N-terminal subdomain and C-terminal domain are highlighted in light yellow.
EcFAAL and LpFAAL are functional monomers
FACLs in general form functional dimers2,8,10. The FACL from T. thermophilus forms a domain-swapped dimer8. Our crystals of EcFAAL contain two nearly identical molecules in the asymmetric unit (rmsd ~0.4 Å, 568 Cα pairs). However, although EcFAAL does form a dimer in the crystalline state, it is not a functional dimer since the gel filtration profile shows only a monomeric peak (data not shown). The relatively small intermolecular surface area (~900 Å) buried between the two monomers strongly suggests that the crystalline dimer is not biologically relevant. LpFAAL is a monomer in our crystals and we presume that, like EcFAAL, this protein functions as a monomer in solution. The acyl adenylate adopts similar conformations and has similar enzyme-ligand contacts in both copies of EcFAAL. In the discussion below we consider only molecule A of EcFAAL.
The C-terminal domains occur in different relative orientations
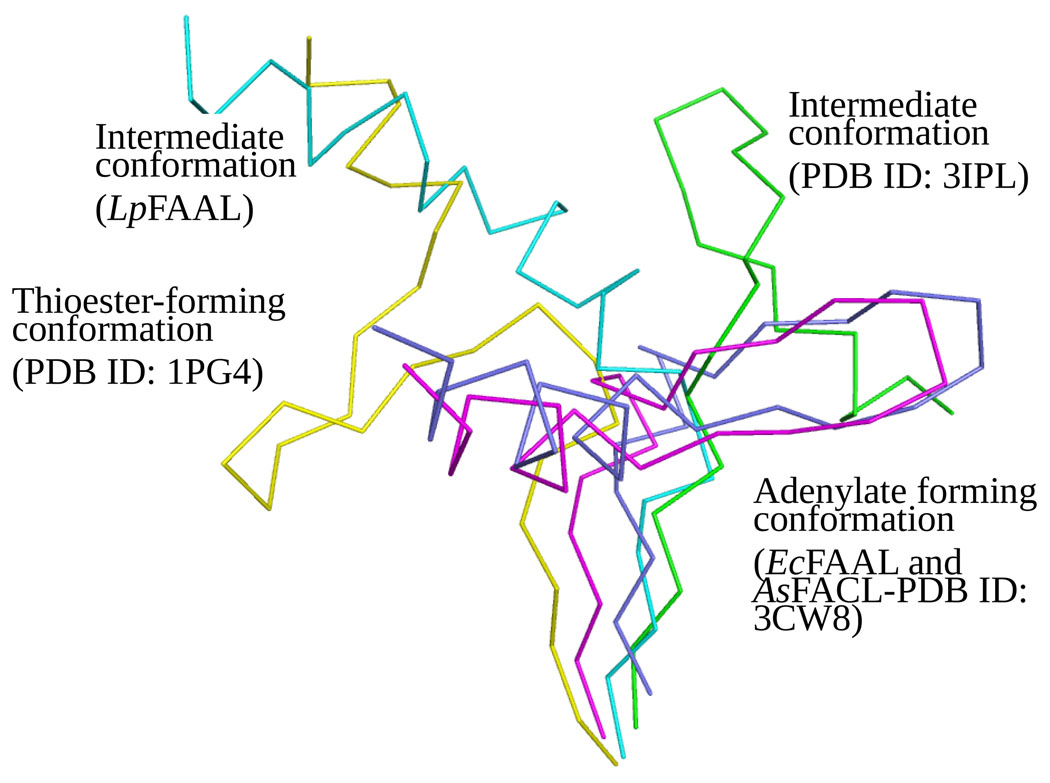
The N and C-terminal domains of FAAL and FACL proteins are connected by a highly flexible hinge region that allows the domains to adopt different relative orientations (Figure 3c). Structures have been determined of the Alcaligenes sp FACL in both the adenylate forming conformation and the thioester forming conformation, which differ by a 140° rotation of one domain with respect to the other3,4,10. The hinge regions are highly conserved among the FAALs (Figure 2a) and, despite some differences with the FACLs (Figure 2a), the hinge regions in both protein families share a R-X-K/D-D-X-(I/L)-X3-GX4-(P/T/S)-X-(D/E)-(I/L)-E signature motif (Figure 2a). This similarity suggests that EcFAAL and LpFAAL, and other FAAL family members, could undergo reorientation of the two domains with respect to one another during catalysis, but perhaps not to the same extent as seen for the FACLs, a hypothesis that is supported by Hingeprot analysis11,12 (data not shown). Our structures provide experimental support for this proposal (Figure 4). Both EcFAAL and LpFAAL were co-crystallized with acyl adenylate, the binding pocket for which is located immediately below the interface of the two domains and is formed by the three β-sheets of the N-terminal domain. However, while EcFAAL adopts a relative domain orientation very similar to that seen in the adenylate forming conformation of Alcaligenes sp FACL3,4,10, the relative orientation of the two domains in LpFAAL (180° rotation of the C-terminal domain with respect to N-terminal domain) differs from all structures reported previously (Figure 4). The distinct conformations revealed by our structures of EcFAAL and LpFAAL result from differences between the hinge regions of the two proteins (a hairpin structure in EcFAAL and an unusual loop-helix-loop structure in LpFAAL) in addition to minor differences in amino acid sequence. In neither case do the FAAL proteins adopt the CoA thioester bound conformation observed for Alcaligenes sp FACL. Removal of the insertion motif in MtFAAL yielded modest CoA ligase activity in this enzyme5, suggesting that the presence of insertion motifs is responsible for the absence CoA ligase activity in FAALs. Below we describe the structure of the insertion motif in our structures of the two full-length FAAL enzymes.
Figure 4.
Comparison of hinge regions when the N-terminal domains (not shown) of FAAL and FACL structures are overlaid. Adenylate forming conformation from EcFAAL and AsFACL (3CW8) are colored in magenta and blue, respectively. Intermediate conformations from LpFAAL and FACL (3IPL) are colored in cyan and green, respectively. CoA binding conformation from FACL (1PG4) colored in yellow. LpFAAL N-terminal domain is colored in grey.
Insertion motif and its effect on C-terminal domain movement
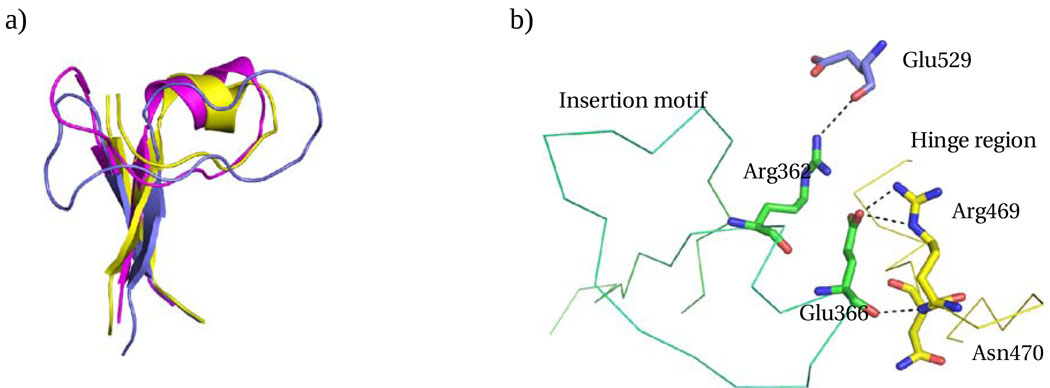
All FAALs contain an insertion motif which differentiates them from the FACLs. For EcFAAL, the insertion motif includes residues 355 – 381, while in LpFAAL it is formed by residues 343 – 368 (Figures 2a and 3a). These insertion motifs adopt a similar local structure in both enzymes, including a common two stranded β-sheet and a long loop connecting the two strands, with a short α helix present in the center of the EcFAAL loop (Figure 5a). The two stranded β-sheet of the insertion motif makes extensive interactions with the N-terminal domain in the three FAAL structures known to date. Electrostatic surface analysis demonstrates that all three insertion motifs form a negatively charged pocket facing the C-terminal domain (Supplementary material S1). In EcFAAL, Asp363 and Glu366 occur in the negatively charged pocket, and Arg469 from the hinge region forms a salt bridge with Glu366 (Fig. 5b). In addition, hinge residues Asn470 (N) and Asp475 (OD2) form hydrogen bonds with Glu366 and Ser382, respectively, while the guanidino NH2 group of Arg362 hydrogen bonds with the backbone carbonyl oxygen of Glu529. The loop region of the insertion motif also makes hydrophobic contacts with the N-terminal domain. We propose that the observed interactions between the insertion motif and the remainder of the protein prevent large excursions of the C-terminal domain. Our analysis suggests that interactions between the insertion motif and the hinge region effectively precludes formation of the CoA binding conformation, and, hence, catalysis of the CoA ligation reaction by the FAALs.
Figure 5.
Insertion motif. a) Insertion motifs of EcFAAL (magenta), LpFAAL (blue) and MtFAAL (yellow) are superposed. b) The C-alpha trace of EcFAAL insertion motif (green) and hinge region (yellow) is shown with the critical interactions between insertion motif and the C-terminal domain or hinge region highlighted.
Active site of FAAL and bound acyl adenylate
Although no substrate or product was added during crystallization, both our EcFAAL and LpFAAL structures contain a bound acyl adenylate, which allowed us to identify active site residues and characterize their interactions with the product. Three loops form the entrance to the active site. These loops are highlighted in yellow in Figure 2a. In addition, residues 545–553 from the C-terminal domain may also form part of the opening to the active site. The acyl adenylate reaction product occupies a pocket created by all three subdomains of the N-terminal domain. The C-terminal domain movement facilitated by the highly flexible hinge region exposes the interface between the two domains. Although the FAALs do not possess acyl CoA ligation activity, FAALs retain the capacity for interdomain movement and hinge regions similar to FACLs, suggesting that domain movement and hinge region residues are important for acyl adenylate formation and, potentially, also for product dissociation.
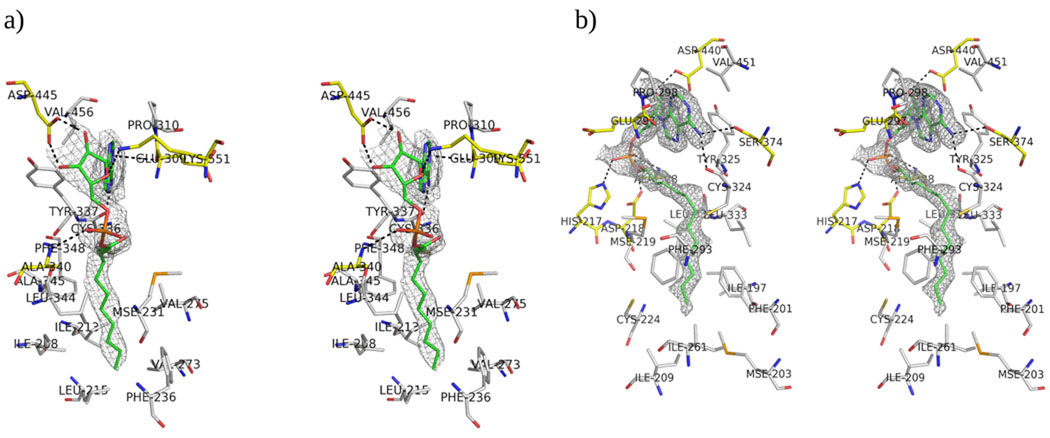
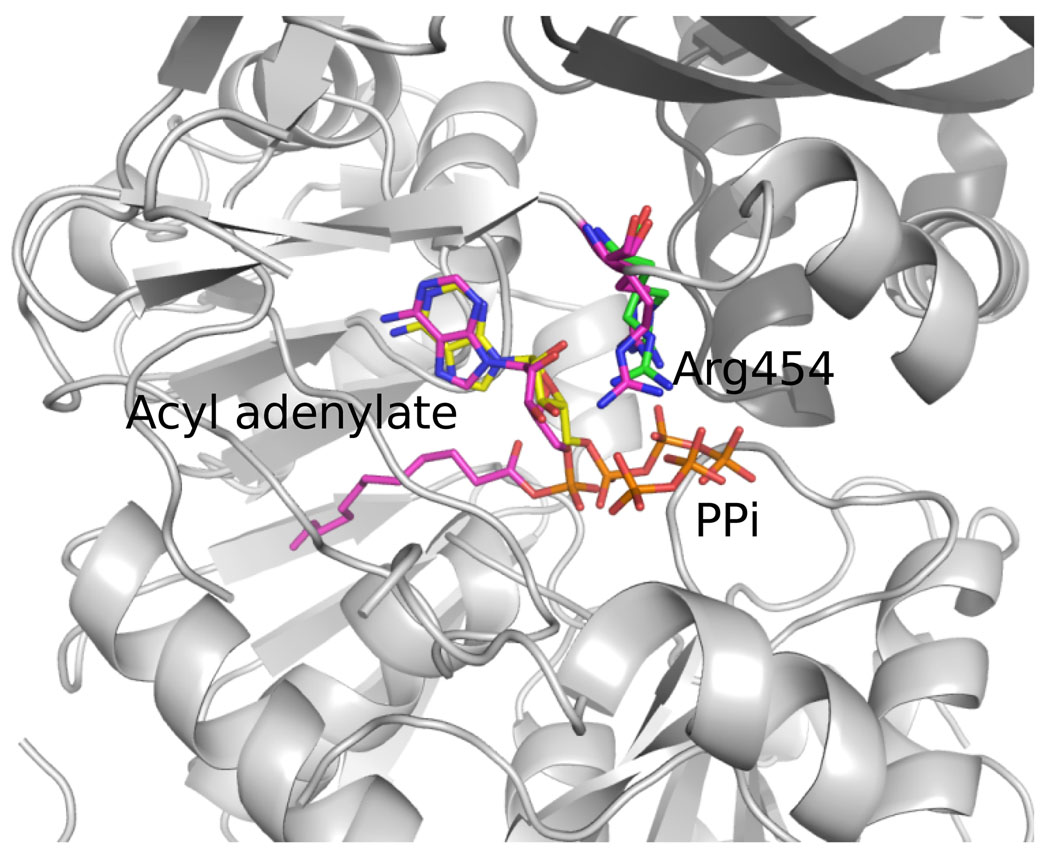
The acyl adenylate forms extensive hydrogen bonds with active site residues, either directly or indirectly via water mediated contacts. In both of our structures, the AMP moieties occur in very similar orientations and interact with highly conserved residues (Supplementary material S2). Both adenyl rings are located in a hydrophobic pocket formed by the hydrophobic residues of Pro310/298, Tyr337/325 and Val456/451 in EcFAAL/LpFAAL, respectively (Figure 6). It also makes hydrogen bonds with main chain and side chain atoms of Cys336/324 and -/Ser374 in EcFAAL/LpFAAL (Figure 6). A water molecule mediates the interaction between O5 of the AMP moiety and the carbonyl oxygen of Ser549, while the two ribose hydroxyl oxygens are hydrogen bonded to OD1 and OD2 of Asp445. The AMP moiety also forms extensive van der Waals interactions with active site residues. In LpFAAL, more water mediated indirect interactions are observed between the AMP moiety and the active site residues, which simply may be the result of higher resolution data. Water molecule 37 mediates the interaction between N3 of the AMP moiety and peptide nitrogen of Val301, while the AMP N6 also forms hydrogen bonds with the Ser374 OG as well as Cys324. Similar to EcFAAL, the two ribose hydroxyl oxygens form hydrogen bonds with OD1 and OD2 of Asp440 in LpFAAL. This Asp is conserved in FAALs and FACLs and is important for AMP binding since alanine mutation of this residue drastically lowers fatty acyl-CoA formation in FACL11. The phosphate oxygens form hydrogen bonds to the amide NH of Ala340/Ala328 in EcFAAL/LpFAAL. Both the adenyl binding hydrophobic pocket and the adenylate-interacting residues are highly conserved within and between the FAAL and FACL families, indicating that they may share a similar adenylation mechanism. The Asp sidechain that interacts with the hydroxyl groups of the ribose moiety is conserved in most FAALs and FACLs (Figure 2a) and appears important for the ATP binding. In addition, the ribose oxygen (O) hydrogen bonds to Lys551 in EcFAAL which is part of a highly conserved loop region (highlighted in red in Figure 2a) in the C-terminal domain of both the FAALs and FACLs. This interaction does not occur in LpFAAL, because of the alternative C-terminal conformation adopted by this enzyme, which results in solvent exposure of the equivalent Lys553. The equivalent lysine residue is also exposed to solvent in the thioester forming conformation in AsFACL (3CW9). In our structure of LpFAAL bound to pyrophosphate and acyl adenylate, the acyl adenylate molecule and binding site residues adopt the same conformations seen in LpFAAL with acyl adenylate, with the exception of the hinge region residue Arg454 that adopts two alternate conformations (Figure 7). A similar double conformation was observed for Arg461, the equivalent residue in the human FACL (PDB ID: 3C5E) (HsFACL). An overlay of the HsFACL and LpFACL N-termimal domains shows that the LpFACL pyrophosphate binding site is very close to the β and γ phosphates of ATP in HsFACL (Figure 7). In addition, Arg461 of HsFACL and Arg454 of LpFAAL form hydrogen bonds with the β-phosphate oxygen of ATP and pyrophosphate, respectively. Arg454 of LpFAAL and Arg461 of HsFACL also hydrogen bond to Asp440 of LpFAAL and Asp446 of HsFACL. This arginine is conserved in all FAALs and FACLs, and site directed mutation of the equivalent Arg453 in EcFACL yielded significant loss of enzyme activity11, indicating that this residue is critical for catalytic function.
Figure 6.
Stereo view of acyl adenylate and FAAL residues. a) Stereo view of interactions between acyl adenylate and EcFAAL residues. b) Stereo view of interactions between acyl adenylate and LpFAAL residues. The residues that form hydrogen bonds with acyl adenylate are colored yellow and the residues that form the hydrophobic pockets are colored gray. Both a and b show the Sigma weighted 2Fo-Fc electron density contoured at 1σ level around the acyl adenylates.
Figure 7.
Overlay of LpFAAL with acyl adenylate/PPi bound and HsFACL with ATP bound. The N- (grey) and C-(darker grey) terminal domain of LpFAAL are shown as cartoons. Acyl adenlylate, PPi and Arg454 are colored in magenta. HsFACL were only presented with ATP and Arg461, colored in yellow and green, respectively.
The acyl moiety of acyl adenylate
The acyl moieties of EcFAAL and LpFAAL were found in a predominantly hydrophobic pocket in which no water molecules were observed in our electron density maps. The acyl moieties are well-ordered in both structures. In LpFAAL, C3, C7, C11 and the carbonyl oxygen (Figure 6) of the acyl group form van der Waals interactions with Phe201, SG of Cys326, CD1 of Leu332, and Ala296. Similarly, in EcFAAL, the C2, C8 and carbonyl oxygen of the acyl group form van der Waal interactions with Phe236, SG of Cys336, and CD1 of Leu344. Although mass spectrometry suggested that both C12 and C14 acyl adenylates were bound to the enzyme, the observed electron density is only consistent with a C12 acyl group, indicating that one or both of the two terminal atoms in the C14 acyl adenylate are disordered in our crystal structure or that crystals were only formed from the proteins bound to C12 acyl adenylate. Although an acyl adenylate, ATP or substrate is not present in the crystal structure of MtFAAL, the N-terminal domain and the active site residues in this enzyme have the same conformation as those in EcFAAL and LpFAAL. Finally, a study of the FACL from T. thermophilus demonstrates that a fatty acid (myristic acid) can diffuse into the enzyme active site after the protein has been co-crystallized in the presence of AMP-PNP to form myristoyl-AMP, suggesting that binding of ATP and/or the fatty acid substrate do not occur at the interface between the C- and N-terminal domains and may not require domain movement. In our structure obtained from pre-formed crystals of LpFAAL soaked with ATP, we observed only the pyrophosphate moiety suggesting that the ATP was hydrolyzed by the enzyme.
CoA binding cavity – FAAL vs FACL
In the structure of SeFACL bound to CoA (PDB Code: 1PG4), the hinge region residue Arg526 interacts with the phosphate oxygen of the adenosine-5’-propylphosphate (used in the crystal structure) while two amide nitrogens in the mercaptoethylamine and pantothenic acid CoA motifs also hydrogen bond to the peptide backbone oxygen atoms of Gly524 and Ser523 (hinge region residues), respectively. Similar interactions were observed in the structure of AsFACL bound to 4-chlorophenacyl-CoA (PDB Code: 3CW9)3. Thus, the two amide nitrogens hydrogen bond to the peptide oxygen atoms of Gly409 (equivalent to SeFACL Gly524) and Gly408 (equivalent to SeFACL Ser523), while the adenyl nitrogens N1A and N6A are hydrogen bonded to OG and O of Ser407, which resides in a loop. Such interactions are hallmarks of adenine recognition13. In addition, the O7A and O8A oxygen atoms of the sugar phosphate group are hydrogen bonded to the NH1 and NH2 groups of Arg475, while the O1A α-phosphate oxygen is hydrogen bonded to the NH1 and NH2 groups of Arg87 from the N-terminal domain. Finally, Trp440 forms a stacking interaction with the adenine ring. These interactions position CoA for catalysis. In the structure without CoA, such interactions are absent, because the residues engaging these interactions are more than 20Å away and a rotation of the C-terminal domain by 140° would be required to bring them close to the CoA thiol group3. Arg87, in the N-terminal domain, takes a different rotamer position because of the absence of CoA and its phosphate group. Interestingly, the C-terminal domain of EcFAAL assumes an orientation similar to that seen in AsFACL. Loop 403–413 of AsFAAL, containing Ser407 (interacting with adenyl nitrogens), corresponds to loop 462–472 of EcFAAL. Arg469 in this loop makes a strong salt bridge with Glu366 of the insertion motif and appears to prevent this loop from binding CoA. Similarly, residues 456–478 containing Arg475 (which contacts the phosphate oxygen atoms) corresponds to 518–535 of EcFAAL. Glu529 makes a strong hydrogen bond with Arg362 of the insertion motif, again hindering its movement. In summary a cavity is present between the C- and N-terminal domains to accommodate CoA, but the productive orientation of the C-terminal domain required to make the necessary contacts is not possible because of the insertion motif (Supplementary material S7). This forms the structural basis for the observation that removal of the insertion motif results in CoA ligase activity in FAAL enzymes5.
CoA ligase activity of FAAL is hindered by the insertion motif
The CoA binding pocket is located in the interface of the FACL two domains (PDB Code: 1PG4). According to Engel et al.14, there is no common binding mode for CoA ligands. Three FACL co-crystal structures with CoA are available (PDB Codes: 1PG4, 3EQ6, and 3CW9)3. These three structures adopt the same thioester-forming conformation. The adenylate motifs of CoA bind to a surperficial hydrophobic pocket formed by non-polar residues from the N- and C-terminal domain, while the mercaptoethylamine and pantothenic acid group bind in a hydrophobic tunnel that runs from the surface of the protein toward the buried active site. The positively charged residues lining the opening of this binding pocket interact with the phosphate oxygen atoms of CoA. As a result, the adenylate moieties of the three CoAs adopt very different conformations and interact with different residues from both the N- and C-terminal domains. Residues in the adenylate binding site and the hydrophobic pocket are not conserved. In contrast, mercaptoethylamine and pantothenic acid moieties assume similar conformations and make similar interactions with the active site residues. Comparison of the AsFACL and EcFAAL structures suggests that a cavity capable of accommodating CoA does exist in the FAALs, but the necessary interactions of the C-terminal domain with CoA are possible only if the C-terminal domain can reorient itself. This reorientation is hindered by contacts with the insertion motif (Supplementary material S3–6). As discussed earlier, MtFAAL can perform CoA ligation if its insertion motif is deleted. In vivo FAALs, therefore, could transfer acyl-adenylate to a suitable acceptor like ACP. Alternatively, the acyl adenylate products of FAALs may be substrates of other as yet uncharacterized enzymes5.
MATERIALS AND METHODS
Cloning, Expression, and Purification of FAALs
FAAL genes were amplified by PCR from E. coli (Genbank AAN82156, residues 5–576 from ATCC 700926D) and L. pneumophila (Genbank AAU28294, residues 2–579 from ATCC 33152D) genomic DNA using primers:
| E. coli | 5'-TCTAATAAAATCTTTACGCATTCCC-3' |
| 5'-CTGCCAGGGATTCCTGCACATTAAGA -3' | |
| L. pneumophila: | 5'-AAAAAAGAATATTTGCAGTGCCAGT-3' |
| 5'-CCTCAATTTTATTGAGTTGCCAGGTA-3' |
Both of the resulting PCR products were cloned into the C-terminal His6-tagged pSGX3 vector and the encoded proteins were over-expressed in E. coli BL21(DE3)-Codon+RIL. Protein expression and purification protocols and results are described in detail in PepcDB (www.pepcdb.pdb.org); the TargetID’s are “NYSGXRC-11191g” and “NYSGXRC-11191l”, respectively.
Acyl Adenylate Chain Length Determination
The two protein samples were dialyzed against a buffer containing 20 mM HEPES and 150 mM NaCl. Mass spectra were acquired on thermo TSQ Quantum Access (Thermo Fisher) triple quadrupole mass spectrometer (TSQ). All samples (1 mg/ml) were diluted in 50% (acetonitrile/water) and infused at 5µl/min. Full scan negative ion mass spectra were carried out. For MS analysis the scan ranged from m/z 300 to 600 in 0.5 sec. The parent ions were selected and isolated for further Masspec analysis.
Enzyme Kinetic Studies
FAAL PPi release assay
EcFAAL and LpFAAL activity was monitored by coupling the release of pyrophosphate (PPi) to the oxidation of NADH (Sigma)15 so that the reaction can be monitored at 340 nm (ε340 6,300 M−1cm−1). Assays were performed at 30°C in 20 mM Tris-HCl buffer pH 7.5 containing 1mM MgCl2. Reactions were initiated by adding FAAL (final concentration 82 nM) and initial velocities were obtained at various concentrations of octanoic acid (12, 60, 120, 240, 480 µM) at a fixed ATP concentration (240 µM). Initial velocities were fit to the Michaelis-Menten equation using GraFit 4.0 (Erithacus).
Reaction of octanoic acid, ATP and CoA with FACL
FACL activity (50 nM) was determined by monitoring the formation of octanoyl-CoA from octanoic acid (120 µM), ATP (240 µM) and CoA (240 µM). Reactions were performed at 37°C in 20 mM Tris-HCl buffer pH 7.5 containing 1mM MgCl2, and product formation was monitored by HPLC following a 1 hr incubation using a Waters XTerra MS C18 analytical column (3.5µm particle diameter, 4.6 mm i.d., 100 mm length) with 20 mM NH4OAc in water as buffer A and acetonitrile as B buffer. Chromatography was performed using the following elution profile: 0–20 min 0% acetonitrile, 20–40min 0–100% acetonitrile, 40–50min 100-0% acetonitrile.
Crystallization and structure determination
EcFAAL and LpFAAL were each concentrated to ~10 mg/ml and subjected to sitting-drop, vapor-diffusion by mixing 1 µl of protein solution and 1 µl of reservoir from PEG/Ion screen of Hampton Research. EcFAAL formed rod-shaped crystals overnight from 0.2 M NaF and 20% w/v polyethylene glycol 3350. LpFAAL formed diamond shaped crystals overnight from 25% PEG 3350 and 0.2 M NaCl. LpFAAL was also cocrystallized in the presence of 10 mM ATP from 25% PEG 3350 and 0.2 M NaCl. All crystals were cryoprotected with mother liquor + 20% glycerol (v/v) and flash-frozen by direct immersion in liquid nitrogen.
For EcFAAL and LpFAAL, X-ray diffraction data at the selenium absorption edge (λ=0.9795 Å) were recorded at beamlines X25 and X12C, respectively, at the National Synchrotron Light Source (NSLS). All data were processed with HKL200016. EcFAAL crystallized in the orthorhombic space group P212121 with two molecules/asymmetric unit and diffracted to ~3.0 Å resolution. The structure of EcFAAL was determined by molecular replacement using Mtb FAAL (PDB Code: 3E53) as a partial search model. LpFAAL crystallized in the monoclinic space group P21 with one molecule in the asymmetric unit, and diffracted to 1.85 Å resolution. The structure of LpFAAL was determined by AUTOSOL17, followed by autobuilding in ARP/wARP18,19. Atomic models for both structures were subsequently manually adjusted using COOT20. The structure of LpFAAL cocrystallized with ATP (LpFAAL-ppi) was determined by molecular replacement using PHASER21 with the LpFAAL structure as the search model. All three structures were refined to convergence with PHENIX17. Data collection and refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| EcFAAL | LpFAAL | LpFAAL-PPi | |
|---|---|---|---|
| PDB ID | 3GQW | 3KXW | 3LNV |
| Data collection statistics | |||
| Wavelength (Å) | 0.9795 | 0.9795 | 0.9795 |
| Resolution (Å) | 50-3.00 | 50-1.85 | 50-2.0 |
| Outershell Resolution (Å) | 2.95-3.00 | 1.79-1.85 | 1.95-2.0 |
| Space group | P212121 | P21 | P212121 |
| Cell dimensions (Å) | 91.47,118.34,137.97 | 48.59,112.7,67.73 | 70.19,78.24,108.39 |
| (°) | 90.0, 90.0, 90. | 90.0, 104.84, 90.0 | 90.0, 90.0, 90.0 |
| Number of molecules/asymmetric unit | 2 | 1 | 1 |
| Redundancy (overall/outermost shell) | 7.1 (6.5) | 2.5 (2.3) | 13.0 (12.8) |
| I/σ(I) (overall/outermost shell) | 10.7 (2.7) | 10.9 (4.0) | 20.8 (3.7) |
| Rmerge (overall/outermost shell) | 0.198 (0.648) | 0.075 (0.213) | 0.141 (0.73) |
| Completeness (%) (ovrall/outmost shell) | 99.7 (98.9) | 89.8 (97.2) | 100.0 (99.9) |
| No. of Reflections | 30715 | 54,877 | 41,113 |
| Refinement statistics | |||
| Resolution range (Å) | 50-3.00 | 31.43-1.85 | 50-2.00 |
| No. of reflections | 30439 | 54,241 | 41038 |
| Completeness (work+test)(%) | 99.22 | 90.56 | 99.97 |
| R factor (%) | 19.27 | 18.94 | 19.34 |
| R free (%) | 25.03 | 22.89 | 24.43 |
| No. of protein atoms | 8573 | 4571 | 4525 |
| No. of water molecules | 87 | 611 | 388 |
| No of ligand atoms | 72 | 36 | 45 |
| Mean B value (overall, Å2) | 32.54 | 19.56 | 22.82 |
| rmsd bonds (Å) | 0.07 | 0.014 | 0.01 |
| rmsd angles (degrees) | 1.008 | 1.42 | 1.27 |
| Ramachandran plot analysis: | |||
| Most favored region (additionally allowed) (%) | 93.90 (5.00) | 95.85 (2.89) | 95.46 (3.27) |
| Disallowed region (%) | 1.09 | 2.89 | 1.27 |
Accession Numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with accession numbers 3GQW, 3KXW and 3LNV.
Conclusion
EcFAAL can adopt the adenylate forming conformation common to FACLs, while LpFAAL adopts a unique intermediate conformation not previously described in the literature. Detailed inspection of the interactions formed between the acyl adenylate and the enzyme active site provide insights into the FAAL catalyzed adenylation reaction. Interactions observed between the insertion motif and the C-terminal domain and the hinge region prevent CoA from adopting a bound conformation that allows CoA ligation to occur, thereby explaining the absence of acyl CoA activity among the FAALs. This gives experimental support to the earlier report5. Although the FAAL enzymes do not possess CoA ligase activity, the overall structure of the FAALs and FACLs are very similar. Analyses of near neighbors of the genes encoding EcFAAL and LpFAAL revealed in both cases the presence of at least one gene that appears to encode an acyl CoA synthetase. We think it likely, therefore, that the enzymatic products of these two FAAL proteins are “handed off” to these or other ACPs.
Supplementary Material
Acknowledgments
Research was supported by a U54 award from the National Institute of General Medical Sciences to the NYSGXRC (GM074945, PI: S.K.B.) under DOE Prime Contract No. DEAC02-98CH10886 with Brookhaven National Laboratory, and by NIH grant AI044639 (PI: P.J.T.). We thank Drs. Howard Robinson and Anand Saxena for providing data collection facilities (X29 and X12C) at the National Synchrotron Light Source.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bar-Tana J, Rose G. Studies on medium-chain fatty acyl-coenzyme a synthetase. Enzyme fraction I: mechanism of reaction and allosteric properties. Biochem. J. 1968;109:275–282. doi: 10.1042/bj1090275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gulick AM. Conformational Dynamics in the Acyl-CoA Synthetases, Adenylation Domains of Non-ribosomal Peptide Synthetases, and Firefly Luciferas. ACS Chem. Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reger AS, Wu R, Dunaway-Mariano D, Gulick AM. Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase4-chlorobenzoate:CoA ligase. Biochemistry. 2008;47:8016–8025. doi: 10.1021/bi800696y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu R, Reger AS, Lu X, Gulick AM, Dunaway-Mariano D. The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase. Biochemistry. 2009;48:4115–4125. doi: 10.1021/bi9002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arora P, Goyal A, Natarajan VT, Rajakumara E, Verma P, Gupta R, Yousuf M, Trivedi OA, Mohanty D, Tyagi A, Sankaranarayanan R, Gokhale RS. Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat. Chem. Biol. 2009;5:166–173. doi: 10.1038/nchembio.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trivedi OA, Arora P, Sridharan V, Tickoo R, Mohanty D, Gokhale RS. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 7.Léger M, Gavalda S, Guillet V, van der Rest B, Slama N, Montrozier H, Mourey L, Quémard A, Daffé M, Marrakchi H. The Dual Function of the Mycobacterium tuberculosis FadD32 Required for Mycolic Acid Biosynthesis. Chem. Biol. 2009;16:510–519. doi: 10.1016/j.chembiol.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Hisanaga Y, Ago H, Nakagawa N, Hamada K, Ida K, Yamamoto M, Hori T, Arii Y, Sugahara M, Kuramitsu S, Yokoyama S, Miyano M. Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer. J. Biol. Chem. 2004;278:31717–31726. doi: 10.1074/jbc.M400100200. [DOI] [PubMed] [Google Scholar]
- 9.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 10.Gulick AM, Lu X, Dunaway-Mariano D. Crystal Structure of 4-Chlorobenzoate:CoA Ligase/Synthetase in the Unliganded and Aryl Substrate-Bound States. Biochemistry. 2004;43:8670–8679. doi: 10.1021/bi049384m. [DOI] [PubMed] [Google Scholar]
- 11.Black PN, Zhang Q, Weimar JD, DiRusso CC. Mutational Analysis of a Fatty Acyl-Coenzyme A Synthetase Signature Motif Identifies Seven Amino Acid Residues That Modulate Fatty Acid Substrate Specificity. J. Biol. Chem. 1997;272:4896–4903. doi: 10.1074/jbc.272.8.4896. [DOI] [PubMed] [Google Scholar]
- 12.Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: Automated Prediction of Hinges in Protein Structures. Proteins. 2008;70:1219–1227. doi: 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]
- 13.Denessiouk KA, Rantanen VV, Johnson MS. Adenine recognition: A motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins. Proteins. 2001;44:282–291. doi: 10.1002/prot.1093. [DOI] [PubMed] [Google Scholar]
- 14.Engel C, Wierenga R. The diverse world of coenzyme A binding protein. Curr. Opin. Struct. Biol. 1996;6:790–797. doi: 10.1016/s0959-440x(96)80009-1. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien EW. A continuous sepctrophotometric assay for argininosuccinate synthetase based on pyrophosphate formation. Anal. Biochem. 1976;76:426–430. doi: 10.1016/0003-2697(76)90337-7. [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collection in oscillation mode. In: Carter CW, Sweet R, editors. Methods Enzymol. Vol. 276. 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 17.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 18.Hattne J, Lamzin VS. Patter recognition-based detection of planar objects in 3D electron density maps. Acta Crystallogr. 2008;D64:834–842. doi: 10.1107/S0907444908014327. [DOI] [PubMed] [Google Scholar]
- 19.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protocols. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.