Abstract
Helicases use the energy of ATP hydrolysis to separate double-stranded nucleic acids to facilitate essential processes such as replication, recombination, transcription and repair. This article focuses on the human RECQ helicase gene and protein family. Loss of function of three different members has been shown to cause Bloom syndrome (BS), Werner syndrome (WS) and Rothmund–Thomson syndrome (RTS). This article outlines clinical and cellular features of these cancer predisposition syndromes, and discusses their pathogenesis in light of our understanding of RECQ helicase biochemical activities and in vivo functions. I also discuss the emerging role for RECQ helicases as predictors of disease risk and the response to therapy.
Keywords: RECQ helicases, Bloom syndrome, Werner syndrome, Rothmund–Thomson syndrome, DNA replication, DNA repair, Telomeres, Homologous recombination, Genetic instability, Cancer predisposition syndrome, Cancer chemotherapy, Premature aging syndrome
1. Prologue
Homer's Odyssey is a beguiling work, a bookend—literally—to Western literature together with the Illiad. The Odyssey is a literal and metaphorical travelogue, the forced decadal wanderings of Odysseus following the end of the Trojan war. Voyaging, wandering and sailing are recurrent themes in the Odyssey, and have served ever since as powerful metaphors for life's journey and all attempts to explore and to wrest meaning from the unknown. The muse in the following story is Nature; our goal is not Ithaca, home and rest, but an understanding of what experiments of Nature have revealed about our nature, the pathogenesis of disease and our fate.
2. Introduction
Helicase proteins are enzymes that use the energy of ATP hydrolysis to unwind double-stranded nucleic acids. DNA or RNA duplexes, together with DNA: RNA hybrid molecules all serve as physiologic substrates for these enzymes. Helicases can also perform a wider range of actions on nucleic acid or nucleoprotein templates to facilitate their metabolism. For example, helicases can act as DNA or RNA translocases, and as modulators of the structure and function of nucleoprotein filaments and complexes such as those involved in homologous recombination or replication fork restart [1,2]. The ubiquity of helicases reflects their important roles in virtually all aspects of nucleic acid metabolism.
This article focuses on the five members of the human RECQ helicase family, and their roles in DNA metabolism, human disease pathogenesis and the response to therapy. Mutations that lead to loss of function of three different human RECQ helicases cause different heritable cancer susceptibility syndromes. The observation that both Werner syndrome (WS) and Bloom syndrome (BS), two of the human RECQ helicase deficiency syndromes discussed below, were chromosomal instability and cancer predisposition syndromes provided early support for the idea that a mutator phenotype might provide particularly fertile soil for the emergence of cancer. We discuss and further elaborate on this idea below, and discuss how heritable or acquired loss of function of human RECQ helicase genes might promote genetic instability and cancer while, paradoxically, providing new opportunities to improve cancer therapy.
This review necessarily summarizes a large body of work from many different investigators. I have tried throughout to refer interested readers to recent reviews that cover some of the topics discussed here as well as other important areas that are not be discussed here. These reviews also provide fuller referencing of the primary literature on key points that could not be fully discussed and referenced here due to lack of space.
2.1. The RECQ helicase deficiency syndromes
The RECQ helicase deficiency syndromes were originally recognized and described on the basis of clinical findings and inheritance patterns. Bloom syndrome (BS), Werner syndrome (WS) and Rothmund–Thomson syndrome (RTS) are rare (≤1/50,000 live births), autosomal recessive Mendelian diseases that share an elevated risk of cancer together with additional, more variable features that include genetic instability and disease-specific developmental or acquired features. The clinical features of each syndrome are summarized below.
2.1.1. Bloom syndrome (BS)
Bloom syndrome (BS) was first identified in 1954 by David Bloom, who described three patients with congenital short stature and skin changes reminiscent of systemic lupus erythematosus [3,4]. Consistent features include marked intrauterine and post-natal growth retardation; congenital short stature; and a characteristic ‘butterfly’ rash across the bridge of the nose and cheeks that may extend to include the dorsum of the hands and forearms. This rash typically develops with sun exposure in the first years of life, then may become chronic with skin hyper- or hypopigmentation. Deficient cellular and humoral immunity is common, and may explain the elevated risk of otitis media and pneumonia. There is also an elevated risk of diabetes mellitus. BS patients have reduced fertility: males are typically infertile, whereas females are hypofertile but may give birth to normal offspring [5].
The most troubling aspect of BS is a high risk of cancer: BS patients are predisposed to a remarkably broad range of cancers, in contrast to almost all other genetically inherited cancer predispositions. There is an elevated risk of developing common adult epithelial tumors such as colon, breast and lung cancer; leukemias and lymphomas; sarcomas; and rare pediatric tumors such as Wilms' tumors [6]. Cancer is the most common cause of death in BS patients.
2.1.2. Werner syndrome (WS)
Werner syndrome (WS) alone among the human RECQ helicase deficiency syndromes has features strongly suggestive of premature aging. The key clinical findings, first reported by Otto Werner in 1904, include short stature; early graying and loss of hair; bilateral cataracts; and scleroderma-like skin changes [7–10]. The earliest and most consistent change observed is graying and loss of hair. This typically begins in the second decade of life with the scalp and eyebrows, and is progressive. Cataracts in WS are often bilateral; appear beginning in the second or third decade of life; and differ from the common ‘senile’ cataracts by involving the lens posterior cortex and subcapsular regions as opposed to lens nucleus. Vision is otherwise normal, and can often be restored by cataract removal. The short stature of WS patients results from a failure to undergo an adolescent or pubertal growth spurt. There is no suggestion that this acquired growth deficit is caused by an underlying endocrinopathy or other disease state. Subcutaneous connective tissue atrophy and dermal fibrosis together give skin a ‘tight, white and shiny’ or contracted appearance that over time contributes to a progressive sharpening of facial features, foot and ankle deformation with ulceration, and soft tissue calcification. WS patients are at increased risk to develop premature atherosclerosis, myocardial infarction and stroke; osteoporosis; and diabetes mellitus. The CNS is typically spared, and WS patients are not at elevated risk of Alzheimer or other types of dementia apart from those associated with vascular disease. Fertility is reduced in males and females [7–9,11].
Unlike BS, WS confers an elevated risk of only selected types of cancer [12–14]. The most frequently observed neoplasms in WS patients are soft tissue sarcomas, follicular thyroid carcinoma, meningioma, acral lentiginous malignant melanoma, malignant or pre-neoplastic hematologic disease (chiefly leukemias) and osteosarcoma. This spectrum is broader than the misleading characterization of the cancer predisposition in WS being limited to sarcomas or soft-tissue tumors. Cancer and premature cardiovascular disease are the leading causes of death in WS patients [15].
2.1.3. Rothmund–Thomson syndrome
Rothmund–Thomson syndrome (RTS) was first described by Rothmund in 1868 as a familial occurrence of unusual skin changes together with bilateral juvenile cataracts [16]. Subsequent cases were reported by Thomson in 1936, and in 1957 Taylor suggested that these reports were of patients with the same disease [16–18]. The characteristic skin changes of RTS typically appear within the first 3–6 months of life as a sun-sensitive rash with redness, swelling and blistering on the face. This rash spreads over the buttocks and extremities, while sparing the chest, back and abdomen. These skin lesions over time become variably pigmented with telangiectasias and areas of focal atrophy. Additional features include sparse or absent hair, eyelashes and eyebrows; congenital short stature in conjunction with frequent bone and tooth abnormalities; cataracts; and an elevated risk of cancer, most notably osteosarcoma [19].
The short stature of RTS patients is reminiscent, though not as severe, as that observed in BS: affected individuals are born small but proportionately developed and typically remain in the lower percentiles for height and weight throughout life. Bone and tooth abnormalities include dysplastic, malformed or absent bones, often involving the hand or thumbs; delayed bone formation or bone density loss; and malformed, missing or extra teeth. The cataracts originally noted by Rothmund have been found in only a minority of contemporary RTS patients [19,20]. Immunologic function appears to be intact, and fertility may be reduced although RTS females have given birth to normal offspring. Life expectancy in the absence of cancer appears to be normal [20].
Two additional, heritable human diseases have been associated with RTS, RAPADILINO syndrome and Baller–Gerold syndrome (BGS). RAPADILINO syndrome patients have joint dislocations and patellar hypoplasia or aplasia, but lack the characteristic poikiloderma seen in RTS patients. BGS patients have craniosynostosis with radial aplasia in addition to skin changes reminiscent of RTS [21,22]. Molecular analyses of clinically ascertained RTS patients have also identified RTS phenocopies, individuals whose clinical findings resemble RTS, though who lack RECQL4 mutations (see below; [20,23]). The recurrent themes of genetic heterogeneity, phenocopies, variable clinical expression of mutations in the same gene and ‘missing’ diseases are all discussed below: collectively these themes emphasize important work to be done to correlate clinical, pathologic and molecular findings in these diseases.
2.2. RECQ genes, deleterious mutations and SNP variants
Cloning of the genes causally linked to BS, WS and RTS immediately identified all three as members of a human RECQ helicase gene family [24]. BLM was cloned in 1995 by making clever use of the mitotic recombination phenotype of BS cells [25], and led to naming of the family after the helicase domain shared with the E. coli RecQ protein. Positional cloning of the WRN gene followed in 1996, guided by prior linkage analyses [26], and confirmed earlier speculation that the gene responsible for WS might be a helicase [27]. The cloning of RECQL4 and RECQL5 were based on sequence homology [28,29]. The remaining family member, RECQL, was identified independently by two groups in 1994 as encoding a potent ATPase and helicase activity in human cell extracts [30,31]. RECQL and RECQL5 have not been linked to either heritable or acquired human disease states, although there is abundant evidence from biochemical, cellular and mouse modeling analyses that loss of function would likely lead to disease [32–34]. The five human RECQ helicase genes, their chromosomal locations, predicted protein products, conserved domains and encoded catalytic activities are shown in Fig. 1.
Fig. 1.
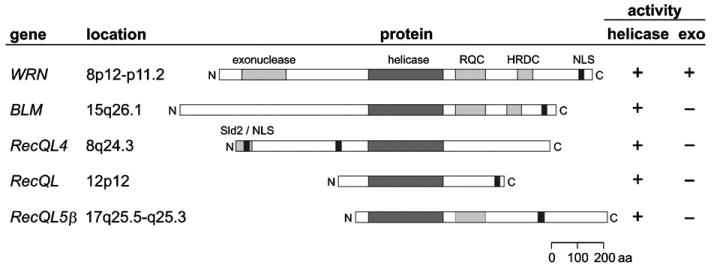
Human RECQ helicase gene and protein family. The five human RECQ helicase proteins are shown as boxes (center). Gene symbols and gene chromosomal locations are given to the left, and encoded catalytic activities to the right, of each protein diagram. All five proteins share a central, conserved RECQ helicase domain that encodes a 3′–5′ helicase activity. Three family members contain RECQ Consensus (RQC) domains, and two a Helicase and RNase D C-terminal (HRDC) domain. Nuclear localization signals (NLS) are depicted as short filled boxes. The 3′–5′ exonuclease domain is unique to WRN, whereas the Sld2 homology domain is found only in RECQL4.
Cloning of the human RECQ helicase genes provided a powerful stimulus for further research and allowed the identification of pathogenic mutations and sequence variants in all of the human RECQ helicase genes. A summary of clinically ascertained pathogenic mutations in BLM, WRN and RECQL4 is shown in Fig. 2 together with non-synonymous coding region SNP variants for each gene.
Fig. 2.
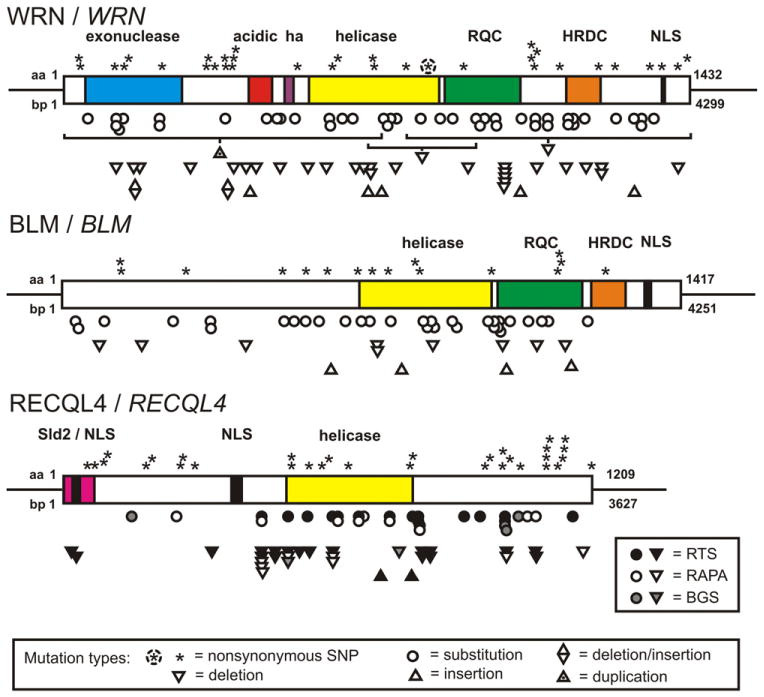
Disease-causing mutations in human RECQ helicase genes. WRN, BLM and RECQL4 open reading frames are depicted as boxes with domains indicated as in Fig. 1. Two additional acidic domains are shown for WRN: the acidic repeat domain (acidic) and a short hyperacidic (ha) stretch consisting of aspartic and glutamic acid residues preceding the helicase domain. Residue and bp coordinates are shown to the left of the beginning of each protein. Coding region non-synonymous SNP polymorphisms are shown above, and clinically ascertained mutations below, each RECQ protein. Mutations, not consequences, are shown; and only single examples of specific mutations. The WRN R834C SNP polymorphism is circled (see text), and large deletions and a duplication are indicated by a horizontal line linked to the appropriate type symbol. RECQL4 mutations identified in Rothmund–Thmonson, RAPADILINO or Baller–Gerold syndrome patients are further indicated by the symbol fill, with the key to the lower right.
BLM mutations in BS patients invariably lead to a loss of helicase function [35], though do not in all cases eliminate expression of the mutant protein. This suggests that BLM helicase activity is the major determinant of BLM-associated phenotypes in human somatic cells, regardless of whether the mutant protein is lost or not. WRN mutations, in contrast, lead to loss of the WRN protein and its two associated catalytic activities [36]. Recently reported missense mutations affecting the WRN helicase domain (Gly574→Arg) or located 20 residues upstream of the C-terminal NLS (Met1350→Arg) were identified in heterozygous WS patients in conjunction with other clearly deleterious mutations [37]. Both of these missense mutations have a high likelihood of affecting protein stability, as did a previously reported pair of homozygous missense mutations in the WRN exonuclease domain (Lys125→Asn/Lys135→Glu; [15]). These results are consistent with the autosomal recessive inheritance pattern of WS, and with prior work demonstrating loss of mutant WRN protein expression from WS patient cells [36].
Mutation analyses have also allowed a clear distinction to be made between WS linked to pathogenic mutations in WRN and a clinically overlapping disease referred to as ‘atypical Werner syndrome’ that is caused by splice-disrupting mutations in the lamin A/C gene LMNA [38]. One interesting—and as yet unanswered—question is whether there is a ‘missing’ disease associated with WRN missense mutations that selectively inactivate the WRN exonuclease or helicase activities. In prior work we demonstrated a requirement for both the WRN exonuclease and helicase activities to suppress WS cellular phenotypes [39]. These experiments predicted that WRN missense mutational diseases should display autosomal recessive inheritance, be recombinant-deficient, and may be clinically expressed as either cancer susceptibility or DNA damage sensitivity syndromes. These experiments also failed to identify a dominant negative effect of expression of single- or double-mutant WRN protein in control cells, a result that is again consistent with the autosomal recessive inheritance of WS [39].
A surprisingly large number of patients referred with potential WS lack either WRN or LMNA mutations. These patients represented 23% (41 of 176) individuals described in a recent update from the International Registry of Werner syndrome [37] (http://www.wernersyndrome.org). This group likely contains mistaken diagnoses; instances where WS was caused by WRN gene silencing, as opposed to mutation; individuals with mutations in proteins required for WRN function; or individuals with novel progeroid syndromes. Evidence for the existence of the latter group was first noted over a decade ago, in gene fusion experiments involving different WS patient-derived cell lines [40]. This patient cohort is thus a prime candidate for targeted exome sequencing to disentangle these possibilities.
As mentioned above, RECQL4 mutations have been linked to at least three different clinical syndromes: RTS, RAPADILINO and Baller–Gerold syndromes (BGS). RTS itself is genetically heterogeneous, with only a portion of clinically ascertained patients have mutations in RECQL4 [20,23]. BGS is similarly genetically heterogeneous: BGS patients have been identified that carry FGFR2 or TWIST, as opposed to RECQL4, mutations [20]. Two other features of clinically ascertained RECQL4 mutations are worth noting. First, there is an unusually high proportion of recurrent mutations that disrupt splicing. This is likely explained by the unusual genomic structure of the human RECQL4 gene, where 13 out of 20 exons are short (<100 bp long) and prone to stochastic or mutation-induced mis-splicing [41]. A second mechanistically intriguing observation is mutational sparing of the N-terminal portion of RECQL4. This RECQL4 region shares homology with the yeast replication protein Sld2, which suggests that the RECQL4 N-terminus and/or the Sld2 homology domain may encode an essential function (reviewed in [42]).
All of the human RECQ genes have polymorphic variants of unknown significance in addition to the clearly pathogenic mutations summarized by molecular type and location in Fig. 2. One WRN polymorphic variant, R834C, has been shown thus far to affect WRN helicase activity [43], and has the potential to disrupt function if homozygous or found in combination with a second, clearly deleterious allele.
2.3. RECQ helicase biochemical activities and DNA metabolic function
The presence of a conserved RECQ helicase domain in each of the human RECQ helicase proteins provided both a common name for the protein family and predicted all of the family members would unwind duplex DNA in an ATP hydrolysis-dependent manner by translocating along one strand in the 3′ to 5′ direction (reviewed in [2,44]). This prediction was subsequently confirmed for all five of the human RECQ helicases using short oligonucleotide substrates in vitro (Fig. 3). RECQL4 was initially thought to lack 3′ to 5′ helicase activity, though helicase activity was eventually documented when it was realized that RECQL4 possesses an unusually potent, competing strand annealing activity [45]. In vivo counterparts of the substrates used to define RECQ helicase catalytic activities are likely to be key intermediates in several important DNA metabolic processes (see below; Fig. 4).
Fig. 3.
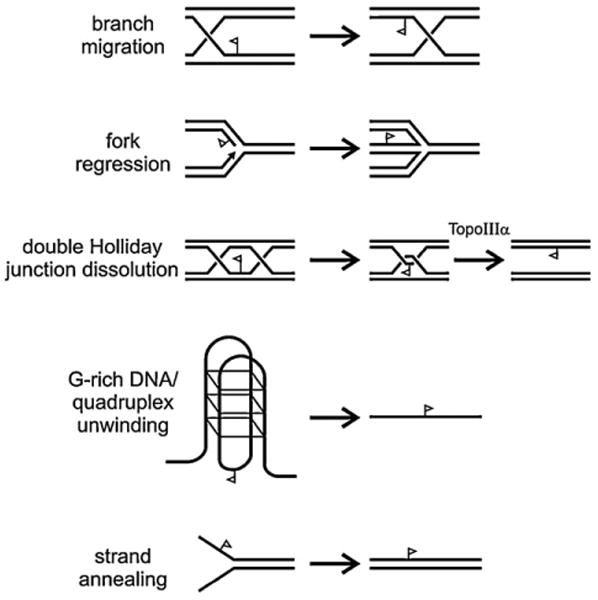
DNA metabolic activities of human RECQ helicases. Activities of human RECQ helicases on model DNA substrates is depicted, and can be thought of as different combinations of unwinding, translocation/displacement, strand annealing and, in the case of WRN exonucleolytic degradation activities. Different human RECQ helicases encode different combinations of the activities shown (see text for detail). The flag symbol on DNA strands provides a reference point to aid visualization of the consequences of RECQ-mediated biochemical activities.
Fig. 4.
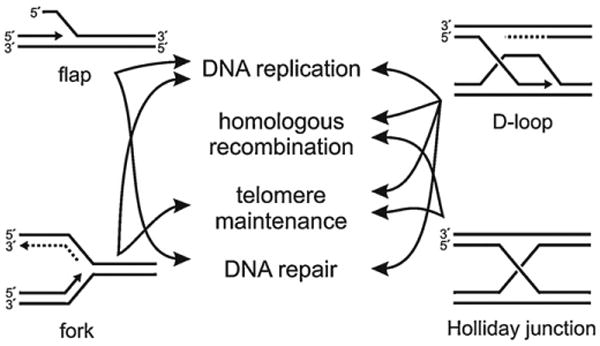
Inferred RECQ helicase substrates and their roles in DNA metabolism. The common model substrates depicted in Fig. 3 and used to define RECQ helicase biochemical activities have direct counterparts in cellular DNA metabolism. RECQ helicases are able to unwind and release DNA flaps (upper left); promote replication fork progression, regression or remodeling (lower left); release an invading 3′ DNA tail in a D-loop (upper right); and branch migrate and, in the case of BLM in conjunction with topoisomerase IIIα, resolve separate DNA duplexes joined in a Holiday junction (lower right).
BLM and WRN, but not RECQ1 or RECQL4, can unwind non-B-form DNAs such as G-quadruplex (G4) DNA in addition to B-form DNAs [2,24,47]. Likely genomic targets for RECQ activity to unwind G4 DNA include G-rich telomeres, ribosomal RNA genes and some G-rich simple repeat sequences that need to be unwound to facilitate replication, recombination, repair or transcription [48]. BLM and WRN alone can branch migrate (i.e. translocate) 3- and 4-stranded DNA junctions such as Holliday junctions (HJs) and displacement loops (D-loops) along double-stranded DNA. HJs are strand exchange intermediates formed during homology-dependent recombination (HR), whereas D-loops form when a duplex DNA is invaded by another homologous, single-stranded 3′ DNA end (Fig. 4).
RECQ1, WRN, and BLM can dissociate HJs into double-stranded DNA products, and WRN and BLM can in similar fashion dissociate D-loops [47]. RECQ5, in contrast, cannot efficiently dissociate D-loops [49]. BLM and WRN can also convert three-way junctions into a HJ by fork regression [2,47]. WRN can unwind flap structures (Fig. 3, top left that have 5′ ends and may form during Okazaki fragment processing during lagging strand DNA replication. WRN can also unwind DNA:RNA duplexes, as well as substrates in which one strand has mixed RNA–DNA content similar to that observed in Okazaki fragments [50]. Okazaki fragments are short DNA:RNA hybrid fragments that have RNA on their 5′ ends that are formed during lagging strand replication. All five RECQs unwind three-way DNA junctions resembling replication forks [2,24,47].
WRN alone among the human RECQ helicases contains a 3′ to 5′ exonuclease activity in addition to the canonical 3′ to 5′ helicase activity [51,52]. WRN exonuclease is active on many of the same substrates that can be unwound by WRN helicase activity, suggesting that the two activities might be coordinated either within a single polypeptide, or by interaction with other WRN molecules or proteins [53]. WRN exonuclease can degrade recessed 3′ ends in double-stranded DNA, and can initiate DNA degradation from a nick or a gap in dsDNA. These types of activities might be particularly useful during DNA replication, where both nicked and gapped substrates are plentiful; in post-replication repair; and in other repair pathways (Fig. 4).
There are now several well-described examples of the activities of RECQ helicases modulating, or being modulated by, interactions with other DNA metabolic and DNA damage response signaling proteins. Some of these interactions are common to all five of the human RECQ helicases, e.g., interactions with RPA, whereas others are specific to individual RECQ proteins and thus provide mechanistic and functional clues. For example, BLM interacts with topoi-somerase IIIα and two RMI (RECQ-Mediated Genome Instability) proteins to form a resolution complex. BLM is also part of the BRAFT supercomplex that includes Fanconi anemia-associated proteins [24,54]. BLM interacts with RAD51 [55], a crucial component of the homologous recombination (HR) machinery [56].
WRN-interacting proteins include XRRC4/Ligase IV [57] involved in DNA double strand break (DSB) repair; base excision repair (BER) proteins including DNA polymerase β [58]; and uniquely with topoisomerase I [59] and the HR proteins RAD54B and RAD52 [60,61]. WRN and BLM also been shown to interact with one another [47]. RECQ5, BLM and WRN all interact with the MRE11/RAD50/NBS1 (MRN) complex that detects and processes DSBs [47]. RECQL4 alone among the RECQ helicases forms a cytoplasmic complex with UBR1 and UBR2, and interacts with replication initiation components including MCM10, MCM2-7, GINS and CDC45 in the nucleus. These results reflect the important role for RECQL4 in replication initiation and, to a lesser extent, replication fork progressing [42,62,63].
Several general themes emerge from the above observations. First, RECQ helicases typically work in concert with other proteins to facilitate specific DNA transactions, and are rarely essential for any given process. These functional ‘collaborations’ may reflect direct physical interaction or functional interactions that are difficult to demonstrate using conventional protein–protein association assays. The best studied of the functional partnerships are between RECQ proteins and exo- and endonucleases, or different topoisomerases. Second, RECQ function mediated by physical and/or functional interactions can promote or inhibit specific DNA metabolic processes. One good example is HR, that can be promoted by WRN and BLM, or alternatively antagonized by the actions of BLM and RECQL5 [64,65]. This and other observations suggest that RECQ protein activity may dynamically modulate DNA metabolic processes to favor specific outcomes under different circumstances. A third general observation is that RECQs are partially redundant for at least some specific functions. This conclusion is supported by recent work from my lab that analyzed the functional consequences of depleting WRN, BLM or both proteins from human cells on cell proliferation and cell survival after DNA damage [66]. Finally, it is important to remember that our suppositions about what RECQ helicase proteins do often reflect conclusions drawn initially—or solely—from in vitro biochemical data. A clear challenge now is to reinterpret these data in the context of cellular biochemistry and function.
2.4. RECQ helicase roles in cellular nucleic acid metabolism
The in vivo biochemical characterization of RECQ helicases proteins provided immediate clues to in vivo function. The following is a brief overview of functional roles for the human RECQ helicases in specific aspects of nucleic acid metabolism.
2.4.1. Homologous recombination/other DNA repair pathways
Homologous recombination (HR) in somatic cells plays an important role in DNA double strand break repair, immunoglobulin class switching and the successful completion of DNA replication [56]. WS cells were first found to have a recombination resolution defect over a decade ago [67,68]. This defect is in the post-synaptic stage of recombination, after the generation of recombinant DNA molecules, and results in the loss of viable recombinant daughter cells containing conversion-type or non-crossover products. An HR resolution defect provides a plausible explanation for the chromosomal rearrangements and deletion mutator phenotype observed in WS cells: failed resolution products can still give rise to viable, albeit mutant, daughter cells following the breakage and end-joining of unresolved HR products.
BS cells were suspected to have a recombination defect on the basis of elevated sister chromatid exchanges, and evidence for a mutator phenotype driven by aberrant HR [4]. Consistent with these findings is the ability of BLM (and other RECQ helicases such as WRN) to unwind and/or branch migrate 3- and 4-way junctions such as D-loops or Holliday junctions (see above; Figs. 3 and 4). More recent data indicate that BLM participates both in early and late stages of HR to promote or antagonize recombination [69,70]. BLM can stimulate recombination by working with exonuclease 1/EXO1 to resect one strand of DNA duplex in the 5′ to 3′ direction to generate single-stranded DNA with a 3′ end for formation of a RAD51 filament [71,72]. RAD51 filaments can invade homologous DNA to form a D-loop recombination intermediate (Fig. 4). BLM (but not WRN or QL) stimulates this strand exchange activity of active, ATP-bound RAD51 filaments, while disrupting inactive, ADP-bound RAD51 filaments [69,73]. Portions of this biochemistry, e.g., the interaction with EXO1, are also shared by WRN [74].
BLM has the unique property among the human RECQ helicases of being able to dissociate HJ intermediates while suppressing the generation of crossover products. This junction dissolution activity requires interaction with topoisomerase IIIα with RMI1 and RMI2 to form a functional ‘dissolvasome’ that branch-migrates and collapses HJs into a hemi-catenane, prior to removing the remaining linked single strands by making use of topoisomerase IIIα's strand pass activity (Fig. 3). This dissolution reaction topologically resolves recombination products, and suppresses the generation of crossover products [75–77]. A failure to resolve double HJ intermediates to form non-crossover products could explain the high levels of SCE's (sister chromatid exchanges), as well as the frequent appearance of UFBs (ultra-fine bridges) in BS cells (see below).
One surprise in light of the recent, rapid progress to understand the biochemical basis of recombination resolution has been the absence of new information on WRN: WRN was not found associated with any of the recently identified resolution proteins or protein complexes (e.g., GEN1, MUS81–EME1, and the SLX4 complex; [78,79]), and no new endonucleolytic activity has been found associated with or encoded by WRN that would allow direct resolution by strand cleavage. These results support initial suggestions that WRN resolves recombination products by strand unwinding or end degradation, as opposed to strand cleavage [67,68]. Consistent with this model, loss of either the WRN exonuclease or helicase activities confers an HR defect with preferential loss of the predominant conversion-type event in human cells, while still allowing the generation of cross-over or ‘popout’ type recombinants at reduced frequency [67,68].
RECQ helicases may be involved in DNA repair pathways in addition to HR. WRN has been implicated in nonhomologous end-joining (NHEJ) repair and in base excision repair (BER). These links are supported by WRN physical interaction with the MRN complex [80] and the KU70/80 protein [81], and with the APE1 endonuclease [82] and DNA polymerase β [58], respectively. Functional interactions have also been observed between WRN and NHEJ or BER factors [47]. Despite these observations, WRN-deficient cells are not appreciably radiosensitive, or highly sensitive to base damage as would be predicted if either appreciable DSB repair or BER-deficient. Moreover, WS patients do not display the immunodeficiency and variable radiation sensitivity observed in NHEJ-deficient patients, or the organ- or tissue-specific defects that can be predicted from the phenotypes of BER-deficient mice [83]. These observations suggest that roles for WRN in NHEJ or BER are likely to be subsidiary to the primary physiologic roles played by WRN in DNA replication, telomere maintenance and HR repair.
2.4.2. DNA replication
BLM and WRN interact with several proteins that play key roles in replication, e.g. FEN1 or flap endonuclease 1, DNA polymerase δ and proliferating cell nuclear antigen (PCNA). Both BS and WS cells show replication defects that may reflect in part the loss of these interactions [47,84]. Recent analyses have demonstrated that RECQ helicase deficiencies can affect unperturbed replication, and that RECQ function is critical during replication stress when fork progression is impeded by DNA damage or a shortage of dNTPs [66,84].
In contrast to BLM and WRN, RECQL4 and RECQL play important roles in replication initiation as opposed to fork progression, stabilization or restart. RECQL4, and to a lesser extent RECQL, bind directly to replication origins [62]. RECQL4 is found associated with the replicative MCM2-7 helicase proteins and other components of replication initiation complex via an interaction with MCM10 [63]. RECQL4 depletion reduces origin firing efficiency, and suppresses cell proliferation [62]. These observations point to an important role of RECQL4, and to a lesser extent RECQL, in origin unwinding and replication initiation. Cells deficient in BLM or RECQL show slight (10–20%) reductions in replication fork progression rates [62,85]. This may reflect transient fork pausing on the leading strand, as the fork encounters DNA secondary or tertiary DNA structures or bound proteins; or on the lagging strand if Okazaki fragment maturation is slowed.
The basal requirement for WRN and BLM, and perhaps RECQL during an unperturbed S-phase is likely to become more pronounced when replication forks encounter DNA template lesions, or are slowed or stalled by a shortage of dNTPs due to hydroxyurea (HU) treatment. Stalled replication forks need to be maintained in an active state if they are to resume replication, or protected from breakage if they are to be passively replicated from adjacent active origins. Both WRN and BLM are required for the efficient resumption of replication in hydroxyurea-treated cells, where forks have been stalled by a dNTP shortage [86,87]. WRN and BLM may ensure replication restart by fork remodeling to protect nascent DNA strands, even if replicative DNA polymerase(s) have been lost from the fork. WRN is required for optimal fork progression after the resumption of HU-mediated replication arrest [87]. This may reflect a role for WRN in DNA polymerase proofreading, perhaps via its 3′ to 5′ exonuclease activity, as fork elongation after stalling may lead to elevated levels of nucleotide misincorporation.
2.4.3. DNA damage checkpoint signaling
DNA damage and replication stress are detected by signal transduction networks that arrest the cell cycle, initiate repair or recovery, or trigger cell death [88,89]. For example the ATM and ATR kinases, key components of the response to DNA damage and replication stress, phosphorylate BLM and WRN in a DNA damage/replication stress-inducible manner [90,91]. Other post-translational modifications likely also occur, e.g., ubiquitylation and SUMO addition (see, e.g., [92]. RECQ helicases may, in turn, modify checkpoint activity via protein interactions, or by generating or disrupting DNA substrates that modulate checkpoint signaling activity [93]. Impaired checkpoint signaling, or adaptation in the face of persistent, abnormal DNA metabolism may be an additional source of genomic instability in RECQ deficient cells.
2.4.4. Telomere metabolism
Telomeres, specialized structures that cap the ends of eukaryotic chromosomes, serve two important roles: they distinguish chromosome ends from DNA double strand breaks, and they facilitate the replication of chromosome ends. Telomeric DNA consists of tandem repeats of a short G-rich DNA sequence unit (TTAGGG in humans) that, together with the multiprotein shelterin complex, form a specialized telomeric D-loop (T-loop) structure [94,95]. Telomeres protect chromosome ends from being recognised and processed as double-strand breaks, but their very nature—a complex, highly structured G-rich nucleoprotein assembly—they impede normal replication and repair.
WRN and BLM may facilitate telomere replication by disrupting telomeric T-loops and G4 DNA [95,96]. The helicase activity of WRN is required for the efficient replication of the G-rich lagging strand of telomeric DNA [95,97,98]. During replication this G-rich strand may remain single-stranded, and thus prone to the formation of secondary structures. BLM appears to be partially redundant with WRN for telomere maintenance [47,96]. Telomere end replication of the lagging strand also requires telomerase that is able to add new, templated telomere repeats directly to chromosome ends. The interplay between telomere length and RECQ helicase activity has been best demonstrated in mice that are later generation (>G3) telomerase-deficient and lacking Wrn and/or Blm [99,100]. The corresponding story in human cells, however, appears more complicated (reviewed in [95]).
Although telomerase activity is absent from most human somatic cells, it can be readily detected in many cancer cells where it may serve as an important facilitator of neoplastic growth [101]. Telomerase-negative cancer cells take advantage of a second process, recombination-mediated ‘alternative lengthening of telomeres’ (ALT), that may also be modulated or facilitated by RECQ helicase proteins. In support of this idea, BLM and WRN have been found to colocalize with POT1, TRF1 and TRF2 in telomerase-deficient, ALT-positive immortalized cells, and BLM depletion from ALT cells leads to rapid telomere shortening [95,102].
2.5. Origins of cellular phenotypes
The RECQ helicase DNA metabolic functions outlined above provide mechanistic insight into the origins of common cellular phenotypes that have been defined for RECQ deficient cells. These include proliferative defects, DNA damage sensitivity and genetic or genomic instability.
2.5.1. Proliferative defects
These were first described for primary fibroblasts isolated from WS patients [103], and subsequently identified in BS cells as well. Slow growth in culture could reflect a prolonged cell cycle time, reduced growth fraction or increased cell death, or increased cellular senescence. The proliferative defect in WS cells reflects a combination of cell cycle abnormalities leading to longer cell cycle times, especially in response to DNA damage; a reduced growth fraction; and high levels of cellular senescence especially in long term cultures of primary cells [87,104]. Similar defects have been observed in BLM-deficient cells, and in comparative analyses the depletion of BLM has a stronger growth-suppressive effect than comparable levels of depletion of WRN. Of note, co-depletion of WRN and BLM in hese experiments did not suppress proliferation beyond that observed in cells depleted of BLM alone [66]. Human cells depleted of RECQ1 and RECQL4 also display reduced proliferation in culture [32,62].
2.5.2. DNA damage sensitivity
Patient-derived BS and WS cells as well as normal cells depleted of BLM or WRN are sensitive to many DNA-damaging or replication-blocking agents (chemicals, radiation, reactive oxygen species). Some of these agents also selectively kill cells defective in RECQL, RECQL4 and RECQL5. These DNA damage sensitivity profiles provide additional mechanistic insight into functional roles of RECQ helicases in human cells. For example, the selective killing of WS cells by topoisomerase I inhibitors such as camptothecin or by DNA cross-linking agents such as mitomycin C or cis-Pt provides additional evidence that WRN is involved in replication and recombination pathways (see [66] for additional discussion).
2.5.3. Genomic instability
Genetic instability is a hallmark of both BS and WS, and this was first identified as chromosomal instability. Primary BS cells have elevated frequencies of DNA exchanges between different chromosomes, as well as breaks, gaps, and fusions in addition to sister chromatid exchanges (SCEs) [4]. Both spontaneous and DNA damage-induced SCEs are elevated in BS cells. In contrast, SCE frequencies are normal in WS cells, as predicted from our current understanding of functional roles of WRN in HR (see above). Regular and ultrafine (UFB) anaphase bridges are also increased in BS cells. Anaphase bridges form when segregating chromosomes remain partially linked as mitosis progresses, whereas UFBs are thin threads tethering otherwise segregated chromosomes [105,106]. WS cells accumulate clonal chromosome deletions and translocations, a characteristic cytogenetic phenotype that was originally termed ‘variegated translocation mosaicism’ [107,108]. These cytogenetic abnormalities in WS likely reflect the capture of stable karyotypic abnormalities that result from defective recombination, replication or telomere maintenance as described above.
The evidence for genomic instability in RTS and associated syndromes is still fragmentary, but there are interesting hints of a coherent story: RECQL4 is located on chromosome 8q, and trisomy 8 or 8q isochromosomes have been identified in several RTS patients. Chromosome 8 abnormalities have been identified in osteosarcomas, as has upregulation of RECQL4 mutations, and elevated levels of glycophorin-A variant red cells have been identified in the blood of one RTS patient [109,110]. It will be important in pursuing these clues to use cell lines and patient material that has been mutation-typed at the RECQL4 locus to avoid the confusions of genetic heterogeneity and phenocopies described above.
2.6. Mechanistic origins of RECQ helicase deficiency syndromes
The cellular defects discussed above provide a way to link DNA metabolic defects resulting from RECQ deficiencies to pathogenesis of the RECQ helicase deficiency syndromes as illustrated in the general model outlined in Fig. 5.
Fig. 5.
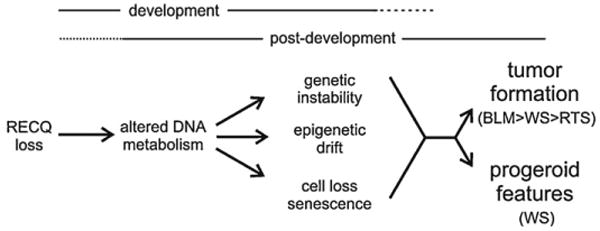
Pathogenesis of human RECQ helicase deficiency syndromes. A model is depicted that summarizes cellular and organismal consequences of loss of RECQ helicase function during and after development. Heritable loss of RECQ function leads to altered DNA metabolism in most or all cell lineages during and after development. Altered or aberrant DNA metabolism, in turn, leads to genetic instability, epigenetic ‘drift’ and cell loss or senescence that over time may compromise tissue structure and function while promoting the emergence of cells with a proliferative advantage to form specific neoplasms (upper right). Tumor generation is strongest in BS. Loss of WRN function also strongly promotes cellular senescence that contributes to global progeroid changes and may provide a non-specific tumor suppressive mechanism that limits tumor formation to a few susceptible cell lineages such as osteoblasts.
The human RECQ helicases appear to be ubiquitously expressed during and after development in most or all cell lineages. Thus an inherited loss of function of a specific RECQ helicase will disrupt DNA metabolism during and after development. A key consequence of disrupted DNA metabolism is continued genetic instability and mutagenesis. Additional, less direct consequences include the potential for continued epigenetic ‘drift’ due to persistent cycles of DNA damage and repair due to ‘low fidelity’ DNA metabolism, and progressive loss of otherwise viable and useful cells due to replication defects, senescence or apoptosis. These cellular consequences of loss of function constitute ‘intermediate phenotypes’ that may limit the number or quality of cells needed to complete developmental tasks or to maintain tissue structure and function after development.
Two examples illustrate how this model could explain developmental features of BS and RTS, or the development of progeroid features in WS patients. BS and RTS patients are typically born small though are proportionately developed. This finding is particularly striking in BS, where affected individuals are often born, and typically remain, at or below the 5th percentile for height and weight throughout life [111]. Cell loss due to DNA replication defects in BS and RTS could lead to proportionate dwarfing by providing too few cells to complete otherwise normal developmental processes. This quantitative deficit, if further compounded by ongoing genetic instability and cellular dysfunction, provides a partial explanation for the elevated risk of developmental abnormalities in both BS and RTS patients.
Persistently abnormal DNA metabolism may have similar long term consequences. These include the seeding of all cell lineages with mutant cells during development; persistent DNA damage signaling; epigenetic ‘churning’ and drift due to higher levels DNA turnover and associated DNA- or chromatin protein-associated epigenetic marks; and the suppression of global regulatory pathways such as the growth hormone (GH)/insulin-like growth factor 1 (IGF-1) signaling pathway that regulates metabolism and longevity in many organisms [112,113]. The combination of genetic instability, epigenetic drift and cell loss in continuously or conditionally replicating cell lineages would provide a fertile environment for the emergence of pre-neoplastic cells with proliferative advantage. Some cell lineages may be particularly susceptible to this combination of events, e.g., the osteoblast lineage that gives rise to osteosarcoma in BS, WS and RTS and RAPADILINO patients. A systematic comparison of genetic and epigenetic alterations in osteosarcomas arising in WS, BLM and RTS/RAPADILINO patients would be of considerable interest, as it might identify mutations in each syndrome that reflected underlying genetic instability in addition to common mutations that promoted osteosarcoma.
The conceptual model in Fig. 5 also provides an explanation for the striking, prematurely aged appearance of many WS patients. The proliferative defects, genetic instability and DNA damage sensitivity of WRN-deficient cells would lead to the progressive accumulation of high levels of senescent cells in many cell lineages, though this alone would not be of sufficient magnitude during development to produce a high risk of developmental defects. Loss of WRN function substantially increases the probability of generating senescent cells, as cellular senescence is one important outcome of DNA damage sensitivity and disrupted DNA metabolism [104,114]. The progressive accumulation of senescent cells has been documented in aging primates [115], and one testable prediction is that senescent cells are present—and likely substantially elevated—in many tissues in WS patients. Cell loss and elevated levels of cellular senescence could compromise tissue or organ structure and function with time, and may further compromise function by negative trophic effects on surrounding normal cells [116]. These senescent cell-associated ‘trans’ effects represent an important area for further exploration in both WS and normal aging, as they may represent an avenue for therapeutic intervention or disease prevention.
The dominant role of cellular senescence in WS might have one modest silver lining: senescence is an effective, non-specific mechanism to suppress the emergence of tumors, even in the face of persistent genetic instability [117]. It should be possible to gain additional insight into these different aspects of disease pathogenesis by examining ‘intermediate’ phenotypes in patients with RECQ helicase deficiency syndromes, and by making judicious use of mouse models of several of the human RECQ deficiency syndromes.
2.7. RECQ helicase roles in sporadic cancer
Heritable loss of RECQ function is associated with an elevated risk of cancer in BS and WS patients, and the subset of RTS and RAPADILINO patients who carry RECQL4 mutations. Less clear is whether heterozygous carriers of known pathogenic RECQ mutations are at elevated risk of cancer, or of enhanced toxicity following cancer therapy with DNA damaging agents. WRN heterozygotes carrying known pathogenic mutations have in vivo genetic instability [118], and cell lines from these individuals show intermediate sensitivity to killing by DNA damaging chemotherapeutic agents that selectively kill WRN-deficient cells [119]. Persistent genetic instability associated with inherited or somatic mutations and haploinsufficiency for WRN—and perhaps for other human RECQ genes—might be sufficient to initiate a vicious cycle with frequent loss of remaining intact RECQ alleles and DNA metabolic and cellular defects. Resulting fully RECQ-deficient cells would drive the pathogenetic sequence outlined in Fig. 5, and the associated organismal endpoints of tumorigenesis and tissue dysfunction and hypofunction. These arguments suggest that heterozygote phenotypes associated with partial loss of RECQ function may be of considerable practical significance. Moreover, they may be common as current estimates of the frequency of individuals carrying single known deleterious WRN mutations, of 1 in 250 individuals in the U.S. [120], predicts that there may be ∼1.5 × 106 individuals at risk of this type of deleterious positive feedback loop that could lead to an elevated risk of cancer, therapy-related toxicity or other diseases.
Few somatic mutations in RECQ genes have been reported in human tumors, or identified by large scale cancer genome sequencing. Loss or silencing of expression of RECQ genes, in contrast, may be frequent in common adult epithelial cancers such as breast and colorectal cancer. Promoter region or gene methylation have been suggested as one mechanistic explanation for the loss of RECQ expression [121,122]. However, the relationship between methylation and loss of expression does not appear to be consistent enough, in our and others' hands, to use methylation alone as an expression marker. Thus new reagents or assays will be needed to reliably determine whether loss of RECQ expression is common in sporadic human cancer.
RECQ expression loss in tumors is practically important to identify, as it would provide a potentially useful therapeutic biomarker of immediate utility in conjunction with the large body of data on the drug sensitivity of RECQ-deficient human cells [66]. The idea of targeting RECQ helicase proteins directly to treat cancer has also been suggested, as the helicase and exonuclease catalytic activities of the human RECQ helicase proteins provide ready targets for the identification of new drugs or small molecule inhibitors [123]. One advantage of this approach is that the consequences of RECQ helicase inhibition are readily predictable (see above). Direct targeting of tumors with RECQ helicase inhibitors might provide a clear tumor-specific therapeutic advantage if combined with conventional chemotherapy, or with pre-existing genetic instability or higher levels of replication stress in tumors [124,125]. The targeting of survival pathways specific to one—or common to several—RECQ helicases could provide a second approach to improve the therapy of patients with tumor-specific RECQ defects. These pathways could be identified by the use of RNAi or drug/small molecule screens to identify pathways or proteins that are synthetically lethal with RECQ deficiencies [126–129].
2.8. Epilogue: RECQ futures
Loss of function of three different members of the human RECQ helicase family, BLM, WRN and RECQL4, lead to distinct diseases with developmental and acquired features including a markedly elevated risk of specific cancers. These ‘experiments of nature’ have, after a decade of work, begun to provide useful insight into the role of the human RECQ helicases in cellular DNA metabolism, the origins and consequences of DNA metabolic defects following loss of function, and how acquired loss of RECQ function may provide new opportunities to improve cancer therapy.
One important ‘take-home’ from the past decade's work on RECQ helicase deficiency syndromes is that genetic instability requires context-dependent information to predict phenotypic effects. The RECQ helicase deficiency syndromes illustrate how different DNA metabolic defects may lead to a mutator phenotype, and how genetic instability may promote developmental defects, the early emergence of tumors, or progeroid features in different contexts. Tumors represent a practically important focus for further developing context-dependent analyses of mutator phenotypes: many of the needed data are in hand or are being rapidly acquired, and cancer therapy represents an important practical opportunity to determine how mutator phenotypes can be identified, rapidly characterized and turned to therapeutic advantage to aid individual patients. The progress summarized here provides hope that our voyage, though far from over, has prospect for a satisfying conclusion.
Web links.
Bloom syndrome/BLM
Entrez gene record with reference sequence (RefSeq) gene and protein links: http://www.ncbi.nlm.nih.gov/gene/641.
On-line Mendelian inheritance in man (OMIM) record: http://www.ncbi.nlm.nih.gov/omim/210900.
GeneClinics gene review: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=bloom
Werner syndrome/WRN
Entrez gene record with reference sequence (RefSeq) gene and protein links: http://www.ncbi.nlm.nih.gov/gene/7486.
On-line Mendelian Inheritance in Man (OMIM) record: http://www.ncbi.nlm.nih.gov/omim/277700.
GeneClinics gene review: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=werner.
Werner syndrome locus-specific mutation database: http://www.pathology.washington.edu/research/werner/database/
International registry of Werner syndrome: http://www.pathology.washington.edu/research/werner/index.html. http://www.pathology.washington.edu/research/werner/registry/registry.html.
Rothmund–Thomson syndrome/RTS
Entrez Gene record with reference sequence (RefSeq) gene and protein links: http://www.ncbi.nlm.nih.gov/gene/9401.
On-line Mendelian inheritance in man (OMIM) record: http://www.ncbi.nlm.nih.gov/omim/268400.
GeneClinics gene review: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=rts.
RAPADILINO syndrome
On-line Mendelian inheritance in man (OMIM) record: http://www.ncbi.nlm.nih.gov/omim/266280.
Baller–Gerold syndrome
On-line Mendelian inheritance in man (OMIM) record: http://www.ncbi.nlm.nih.gov/omim/218600.
GeneClinics gene review: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=bgs.
RECQL
Entrez gene record with reference sequence (RefSeq) gene and protein links: http://www.ncbi.nlm.nih.gov/gene/5965.
RECQL5
Entrez Gene record with reference sequence (RefSeq) gene and protein links: http://www.ncbi.nlm.nih.gov/gene/9400.
Acknowledgments
Work in the author's laboratory has been supported by grants from the NIA, NCI and the Nippon Boehringer Ingelheim Virtual Research Institute of Aging. The author is grateful to lab members who have contributed many ideas and the hard work that made this review possible. This article is dedicated to three pioneers I have had the good fortune to know as mentors and colleagues: George Martin and Arno Motulsky, who first brought Werner syndrome to my—and many others'—attention; and Larry Loeb, a pioneer in recognizing the critical role of mutator phenotypes in cancer risk, pathogenesis and the response to therapy.
Footnotes
Conflicts of Interest
R.J.M., Jr. has no relevant conflicts of interest to declare.
References
- 1.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76(1):23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Vindigni A, Marino F, Gileadi O. Probing the structural basis of RecQ helicase function. Biophys Chem. 2010;149:67–77. doi: 10.1016/j.bpc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Bloom D. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs. Am J Dis Child. 1954;88:754–8. [PubMed] [Google Scholar]
- 4.German J. Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 5.German J. Bloom's syndrome VIII Review of clinical and genetic aspects. In: Goodman RM, Motulsky AG, editors. Genetic diseases among Askenazi jews. New York: Raven Press; 1979. pp. 121–39. [Google Scholar]
- 6.German J. Bloom's syndrome: XX. The first 100 cancers. Cytogenet Cell Genet. 1997;93:100–6. doi: 10.1016/s0165-4608(96)00336-6. [DOI] [PubMed] [Google Scholar]
- 7.Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner's syndrome: a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine. 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Goto M. Hierarchical deterioration of body systems in Werner's syndrome: implications for normal ageing. Mech Ageing Dev. 1997;98:239–54. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- 9.Monnat RJ., Jr . Werner syndrome as a model of human aging. In: Conn PM, editor. Handbook of models for human aging. Amsterdam: Elsevier Academic Press; 2006. pp. 961–76. [Google Scholar]
- 10.Werner O. On cataract in conjunction with scleroderma [Hoehn H, Trans.] In: Salk D, Fujiwara Y, Martin GM, editors. Werner's syndrome and human aging. New York: Plenum Press; 1985. pp. 1–14. [Google Scholar]
- 11.Tollefsbol TO, Cohen HJ. Werner's sydrome: an underdiagnosed disorder resembling premature aging. Age. 1984;7:75–88. [Google Scholar]
- 12.Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–46. [PubMed] [Google Scholar]
- 13.Monnat RJ., Jr . Cancer pathogenesis in the human RecQ helicase deficiency syndromes. In: Goto M, Miller RW, editors. From premature gray hair to helicase – Werner syndrome: implications for aging and cancer. Tokyo, Japan: Japan Scientific Societies Press; 2001. pp. 83–94. [Google Scholar]
- 14.Monnat RJ., Jr . Werner syndrome. In: Fletcher C, Unni K, Mertens F, editors. Monograph on Pathology and Genetics of Tumours of Soft Tissue and Bone. WHO/IARC; Lyon: 2002. pp. 273–4. [Google Scholar]
- 15.Huang S, Lee L, Hanson NB, et al. The spectrum of WRN mutations in Werner syndrome patients. Human Mutat. 2006;27:558–67. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothmund A. Ueber cataracten in verbindung mit einer eigentümlichen hautdegenera-tion. Arch Klin Exp Ophtal. 1868;14(1):159–82. [Google Scholar]
- 17.Thomson MS. Poikiloderma congenitale. Br J Dermatol. 1936;48:221–34. [Google Scholar]
- 18.Taylor WB. Rothmund's syndrome–Thomson's syndrome: congenital poikiloderma with and without juvenile cataracts. A review of the literature, report of a case, and discussion of the relationship of the two syndromes. AMA Arch Dermatol. 1957;75:236–44. doi: 10.1001/archderm.1957.01550140080013. [DOI] [PubMed] [Google Scholar]
- 19.Wang LL, Levy ML, Lewis RA, et al. Clinical manifestations in a cohort of 41 Rothmund–Thomson syndrome patients. Am J Hum Genet. 2001;102:11–7. doi: 10.1002/1096-8628(20010722)102:1<11::aid-ajmg1413>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 20.Siitonen HA, Sotkasiira J, Biervliet M, et al. The mutation spectrum in RECQL4 diseases. Eur J Hum Genet. 2009;17:151–8. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siitonen HA, Kopra O, Kaariainen H, et al. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12(21):2837–44. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 22.Van Maldergem L, Siitonen HA, Jalkh N, et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller–Gerold syndrome caused by mutations in the RECQL4 gene. J Med Genet. 2006;43:148–52. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang LL, Gannavarapu A, Kozinetz CA, et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund–Thomson syndrome. J Natl Cancer Inst. 2003;95:669–74. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- 24.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9(9):644–54. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 25.Ellis NA, Groden J, Ye TZ, et al. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–66. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 26.Yu CE, Oshima J, Fu YH, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–62. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 27.Monnat RJ., Jr Werner syndrome: molecular genetics and mechanistic hypotheses. Exp Gerontol. 1992;27:447–53. doi: 10.1016/0531-5565(92)90080-j. [DOI] [PubMed] [Google Scholar]
- 28.Kitao S, Shimamoto A, Goto M, et al. Mutations in RECQ4L cause a subset of cases of Rothmund–Thomson syndrome. Nat Genet. 1999;22:82–4. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 29.Kitao S, Ohsugi I, Ichikawa K, et al. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–52. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 30.Puranam KL, Blackshear PJ. Cloning and characterization of RecQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–45. [PubMed] [Google Scholar]
- 31.Seki M, Miyazawa H, Tada S, et al. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli RecQ helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 1994;22:4566–73. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma S, Stumpo DJ, Balajee AS, et al. RECQL, a Member of the RecQ Family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol. 2007;27(5):1784–94. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aygün O, Svejstrup JQ. RECQL5 helicase: connections to DNA recombination and RNA polymerase II transcription. DNA Repair. 2010;9(3):345–53. doi: 10.1016/j.dnarep.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Wu Y, Brosh J. Distinct roles of RECQ1 in the maintenance of genomic stability. DNA Repair. 2010;9(3):315–24. doi: 10.1016/j.dnarep.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. Syndrome-causing mutations of the BLM gene in persons in the Bloom syndrome registry. Human Mutat. 2007;28:743–53. doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- 36.Moser MJ, Kamath-Loeb AS, Jacob JE, et al. WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 2000;28:648–54. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedrich K, Lee L, Leistritz D, et al. WRN mutations in Werner syndrome patients: genomic rearrangements, unusual intronic mutations and ethnic-specific alterations. Human Genet. 2010;128(1):103–11. doi: 10.1007/s00439-010-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L, Lee L, Kudlow BA, et al. LMNA mutations in atypical Werner's syndrome. Lancet. 2003;362(9382):440–5. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- 39.Swanson C, Saintigny Y, Emond MJ, Monnat RJ., Jr The Werner syndrome protein has separable recombination and viability functions. DNA Repair. 2004;3:475–82. doi: 10.1016/j.dnarep.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Prince PR, Ogburn CE, Moser MJ, et al. Cell fusion corrects the 4-nitroquinoline 1-oxide sensitivity of Werner syndrome fibroblast cell lines. Human Genet. 1999;105:132–8. doi: 10.1007/s004399900078. [DOI] [PubMed] [Google Scholar]
- 41.Wang LL, Worley K, Gannavarapu A, et al. Intron-size constraint as a mutational mechanism in Rothmund-Thomson syndrome. Am J Hum Genet. 2002;71(1):165–7. doi: 10.1086/341234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y. Rothmund–Thomson syndrome helicase. RECQL4: on the crossroad between DNA replication and repair. DNA Repair. 2010;9:325–30. doi: 10.1016/j.dnarep.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Kamath-Loeb AS, Welcsh P, Waite M, Adman ET, Loeb LA. The enzymatic activities of the Werner syndrome protein are disabled by the amino acid polymorphism R834C. J Biol Chem. 2004;279(53):55499–505. doi: 10.1074/jbc.M407128200. [DOI] [PubMed] [Google Scholar]
- 44.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund–Thomson syndrome protein, RECQ4. EMBO J. 2009;28(5):568–77. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair. 2010;9(3):331–44. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13(12):1055–9. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 49.Ghosh A, Rossi ML, Aulds J, Croteau D, Bohr VA. Telomeric D-loops containing 8-oxo-2′-deoxyguanosine are preferred substrates for Werner and Bloom syndrome helicases and are bound by POT1. J Biol Chem. 2009;284(45):31074–84. doi: 10.1074/jbc.M109.027532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakraborty P, Grosse F. WRN helicase unwinds Okazaki fragment-like hybrids in a reaction stimulated by the human DHX9 helicase. Nucl Acids Res. 2010;38(14):4722–30. doi: 10.1093/nar/gkq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang S, Li B, Gray MD, et al. The premature aging syndrome protein, WRN, is a 3′ to 5′ exonuclease. Nat Genet. 1998;20:114–6. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen JC, Gray MD, Oshima J, et al. Werner syndrome protein I: DNA helicase and DNA exonuclease reside on the same polypeptide. J Biol Chem. 1998;273:34139–44. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 53.Kamath-Loeb AS, Shen JC, Loeb LA, Fry M. Werner syndrome protein II: characterization of the integral 3′→5′ DNA exonuclease. J Biol Chem. 1998;273:34145–50. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- 54.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–48. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 55.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276(22):19375–81. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 56.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11(3):196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kusumoto R, Dawut L, Marchetti C, et al. Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing. Biochemistry. 2008;47(28):7548–56. doi: 10.1021/bi702325t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrigan JA, Wilson DM, Prasad R, et al. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase β. Nucl Acids Res. 2006;34(2):745–54. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–9. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- 60.Baynton K, Otterlei M, Bjørås M, et al. WRN interacts physically and functionally with the recombination mediator protein RAD52. J Biol Chem. 2003;278:36476–86. doi: 10.1074/jbc.M303885200. [DOI] [PubMed] [Google Scholar]
- 61.Otterlei M, Bruheim P, Ahn B, et al. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci. 2006;119(24):5137–46. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- 62.Thangavel S, Mendoza-Maldonado R, Tissino E, et al. The human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol. 2009;30(6):1382–96. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009;28(19):3005–14. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu Y, Raynard S, Sehorn MG, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21(23):3073–84. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwendener S, Raynard S, Paliwal S, et al. Physical Interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem. 2010;285(21):15739–45. doi: 10.1074/jbc.M110.110478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The Human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 2010;70(16):6548–55. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prince PR, Emond MJ, Monnat RJ., Jr Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 2001;15:933–8. doi: 10.1101/gad.877001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in Werner syndrome. Mol Cell Biol. 2002;22(20):6971–8. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bugreev DV, Mazina OM, Mazin AV. Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J Biol Chem. 2009;284(39):26349–59. doi: 10.1074/jbc.M109.029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chu WK, Hanada K, Kanaar R, Hickson ID. BLM has early and late functions in homologous recombination repair in mouse embryonic stem cells. Oncogene. 2010;29(33):4705–14. doi: 10.1038/onc.2010.214. [DOI] [PubMed] [Google Scholar]
- 71.Nimonkar AV, Özsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A. 2008;105(44):16906–11. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22(20):2767–72. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu L. Wrestling off RAD51: a novel role for RecQ helicases. BioEssays. 2008;30(4):291–5. doi: 10.1002/bies.20735. [DOI] [PubMed] [Google Scholar]
- 74.Aggarwal M, Sommers JA, Morris C, Brosh J. Delineation of WRN helicase function with EXO1 in the replicational stress response. DNA Repair. 2010;9(7):765–76. doi: 10.1016/j.dnarep.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–4. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 76.Plank JL, Wu J, Hsieh Ts. Topoisomerase IIIα and Bloom helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci U S A. 2006;103(30):11118–23. doi: 10.1073/pnas.0604873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu L, Bachrati CZ, Ou J, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103(11):4068–73. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Svendsen JM, Harper JW. GEN1/Yen1 and the SLX4 complex: solutions to the problem of Holliday junction resolution. Genes Dev. 2010;24(6):521–36. doi: 10.1101/gad.1903510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–39. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng WH, von Kobbe C, Opresko PL, et al. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J Biol Chem. 2004;279(20):21169–76. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- 81.Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly (ADP-ribose) polymerase-1. J Biol Chem. 2004;279(14):13659–67. doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- 82.Ahn B, Harrigan JA, Indig FE, Wilson DM, Bohr VA. Regulation of WRN helicase activity in human base excision repair. J Biol Chem. 2004;279(51):53465–74. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- 83.Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage (Version 6) DNA Repair. 2004;3(12):1617–38. doi: 10.1016/j.dnarep.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 84.Sidorova JM. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair. 2008;7:1776–86. doi: 10.1016/j.dnarep.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rao VA, Conti C, Guirouilh-Barbat J, et al. Endogenous g-H2AX-ATM-Chk2 checkpoint activation in Bloom's Syndrome helicase-deficient cells is related to DNA replication arrested forks. Mol Cancer Res. 2007;5(7):713–24. doi: 10.1158/1541-7786.MCR-07-0028. [DOI] [PubMed] [Google Scholar]
- 86.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14(7):677–9. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 87.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 89.Warmerdam DO, Kanaar R. Dealing with DNA damage: relationships between checkpoint and repair pathways. Mut Res/Rev Mut Res. 2010;704(1–3):2–11. doi: 10.1016/j.mrrev.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 90.Pichierri P, Rosselli F, Franchitto A. Werner's syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during S-phase of the cell cycle. Oncogene. 2003;22:1491–500. doi: 10.1038/sj.onc.1206169. [DOI] [PubMed] [Google Scholar]
- 91.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's Syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24(3):1279–91. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ouyang KJ, Woo LL, Zhu J, et al. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 2009;7(12):e1000252. doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng WH, Muftic D, Muftuoglu M, et al. WRN Is required for ATM activation and the S-phase checkpoint in response to interstrand cross-link-induced DNA double-strand breaks. Mol Biol Cell. 2008;19(9):3923–33. doi: 10.1091/mbc.E07-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Griffith JD, Comeau L, Rosenfield S, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–14. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 95.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genomic instability. Nat Rev Mol Cell Biol. 2010;11:171–81. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Opresko PL. Telomere ResQue and preservation—roles for the Werner syndrome protein and other RecQ helicases. Mech Ageing Dev. 2008;129(1–2):79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 97.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306(5703):1951–3. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 98.Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez H, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc Natl Acad Sci U S A. 2007;104(7):2205–10. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang S, Multani AS, Cabrera NG, et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36(8):877–82. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 100.Du X, Shen J, Kugan N, et al. Telomere shortening exposes functions for the mouse Werner and Bloom Syndrome genes. Mol Cell Biol. 2004;24(19):8437–46. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31(1):9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bhattacharyya S, Sandy A, Groden J. Unwinding protein complexes in ALTernative telomere maintenance. J Cell Biochem. 2010;109(1):7–15. doi: 10.1002/jcb.22388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martin GM, Sprague CA, Epstein CJ. Replicative life-span of cultivated human cells. Effects of donor's age, tissue, and genotype. Lab Invest. 1970;23:86–92. [PubMed] [Google Scholar]
- 104.Dhillon KK, Sidorova J, Saintigny Y, et al. Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6(1):53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- 105.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26(14):3397–409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lahkim Bennani-Belhaj K, Rouzeau S, Buhagiar-Labarchede G, et al. The Bloom syndrome protein limits the lethality associated with RAD51 deficiency. Mol Cancer Res. 2010;8(3):385–94. doi: 10.1158/1541-7786.MCR-09-0534. [DOI] [PubMed] [Google Scholar]
- 107.Hoehn H, Bryant EM, Au K, et al. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenet Cell Genet. 1975;15:282–98. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- 108.Salk D, Au K, Hoehn H, Martin GM. Cytogenetic aspects of Werner syndrome. Adv Exp Med Biol. 1985;190:541–6. doi: 10.1007/978-1-4684-7853-2_27. [DOI] [PubMed] [Google Scholar]
- 109.Grant S, Wenger S, Latimer J, Thull D, Burke L. Analysis of genomic instability using multiple assays in a patient with Rothmund–Thomson syndrome. Clin Genet. 2000;58(3):209–15. doi: 10.1034/j.1399-0004.2000.580308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maire G, Yoshimoto M, Chilton-MacNeill S, et al. Recurrent RECQL4 imbalance and increased gene expression levels are associated with structural chromosomal instability in sporadic osteosarcoma. Neoplasia. 2009;11:260–8. doi: 10.1593/neo.81384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Keller C, Keller KR, Shew SB, Plon SE. Growth deficiency and malnutrition in Bloom syndrome. J Pediatr. 1999;134(4):472–9. doi: 10.1016/s0022-3476(99)70206-4. [DOI] [PubMed] [Google Scholar]
- 112.Monnat RJ., Jr From broken to old: DNA damage. IGF1 endocrine suppression and aging. DNA Repair. 2007;6(9):1386–90. doi: 10.1016/j.dnarep.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garinis GA, van der Horst GTJ, Vijg J, Hoeijmakers HJ. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10(11):1241–7. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8(7):512–22. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 115.Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech Ageing Dev. 2007;128(1):36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16(5):238–46. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10(1):51–7. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moser MJ, Bigbee WL, Grant SG, et al. Genetic instability and hematologic disease risk in Werner syndrome patients and heterozygotes. Cancer Res. 2000;60:2492–6. [PubMed] [Google Scholar]
- 119.Ogburn CE, Oshima J, Poot M, et al. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum Genet. 1997;101:121–5. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- 120.Schellenberg GD, Miki T, Yu CE, Nakura J. Werner syndrome. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic & Molecular Basis of Inherited Disease. New York: McGraw-Hill; 2001. pp. 785–97. [Google Scholar]
- 121.Agrelo R, Cheng WH, Setien F, et al. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci U S A. 2006;103(23):8822–7. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kawasaki T, Ohnishi M, Suemoto Y, et al. WRN promoter methylation possibly connects mucinous differentiation, microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2007;21:150–8. doi: 10.1038/modpathol.3800996. [DOI] [PubMed] [Google Scholar]
- 123.Aggarwal M, Brosh RM., Jr Hitting the bull's eye: novel directed cancer therapy through helicase-targeted synthetic lethality. J Cell Biochem. 2009;106:758–63. doi: 10.1002/jcb.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gupta R, Brosh RM., Jr Helicases as prospective targets for anti-cancer therapy. Anticancer Agents Med Chem. 2008;8:390–401. doi: 10.2174/187152008784220339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Helleday T. Amplifying tumour-specific replication lesions by DNA repair inhibitors—a new era in targeted cancer therapy. Eur J Cancer. 2008;44(7):921–7. doi: 10.1016/j.ejca.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 126.Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5(9):689–98. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 127.Martin SA, Lord CJ, Ashworth A. DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev. 2008;18(1):80–6. doi: 10.1016/j.gde.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 128.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 129.Martin SA, Hewish M, Lord CJ, Ashworth A. Genomic instability and the selection of treatments for cancer. J Pathol. 2010;220(2):281–9. doi: 10.1002/path.2631. [DOI] [PubMed] [Google Scholar]


