Abstract
The mitfa gene encodes a zebrafish ortholog of the microphthalmia-associated transcription factor (Mitf) which, like its counterparts in other species, is absolutely required for development of neural crest melanocytes. In order to evaluate mitfa’s role in different stages of melanocyte development, we have identified hypomorphic alleles of mitfa, including two alleles that are temperature-sensitive for melanocyte development. Molecular analysis revealed that the mitffh53ts results from a single base pair change producing an asparagine to tyrosine amino acid substitution in the DNA-binding domain, and the mitfavc7 ts allele is a mutation in a splice donor site that reduces the level of correctly-spliced transcripts. Splicing in the mitfavc7 allele does not itself appear to be temperature-dependent. A third, hypomorphic allele, mitfaz25 results in an isoleucine to phenylalanine substitution in the first helix domain of the protein. Temperature upshift experiments with mitfafh53ts show that mitfa is required at several stages of melanocyte differentiation, including for expression of the early melanoblast marker dct, again for progression from dct expression to differentiation, and again for maintenance of dendritic form following differentiation. mitfafh53ts mutants recover melanocytes within 2–3 days when downshifted at all stages of larval development. However, when melanocyte stem cells (MSCs) are ablated by early treatment with the erbB3 inhibitor AG1478, melanocyte recovery is lost by 48 hours. This result indicates first that the MSC is established at the restrictive temperature, and that melanoblasts die or lose the ability to recover after being held at the restrictive temperature for approximately one day.
Keywords: melanocyte, MITF, zebrafish, neural crest
INTRODUCTION
The microphthalmia-associated transcription factor (MITF), a protein of the basic helix-loop-helix leucine zipper family, has been described as the master regulator of vertebrate melanocyte development (Widlund and Fisher 2003; Steingrimsson, et al. 2004; Levy, et al. 2006). Mutations in the MITF gene cause one type of the deafness/pigmentation disorder Waardenburg syndrome in humans, and over 20 alleles at the mouse Mitf locus comprise a complex series of coat color, eye development, osteoclast and mast cell phenotypes. MITF mutations have been described in several other vertebrates, including rat (Opdecamp, et al. 1998), hamster (Hodgkinson, et al. 1998), quail (Mochii, et al. 1998), and zebrafish (Lister, et al. 1999). In zebrafish, null alleles of the MITF ortholog mitfa result in a complete absence of neural crest-derived melanocytes as well as earlier markers of melanoblast specification including dopachrome tautomerase (dct) and the receptor tyrosine kinase kit (Lister, et al. 1999), but spare the retinal pigment epithelium. Transient knockdown of mitfa with morpholino oligonucleotides delays but does not eliminate differentiated melanocytes (Mellgren and Johnson 2004), but the complete role of MITF/mitfa over the course of differentiation of the individual melanocyte, or the full life history of the zebrafish, or any other vertebrate, is not clearly understood.
The many transcriptional targets of MITF suggest its involvement in multiple aspects of melanocyte biology (reviewed in Cheli, et al. 2010). Not surprisingly the first identified were those involved directly in melanin synthesis (tyrosinase, dopachrome tautomerase, tyrp1). More recently, MITF has been linked to control of expression in genes involved in survival (bcl2), cell cycle control (cdk2, p16/INK4A, p21), and cell morphology (Dia1). The potential of MITF to promote a melanocyte differentiation program has been further demonstrated by induction of melanocyte development following overexpression in medaka ES cells (Bejar, et al. 2003), or ectopic development following expression in zebrafish (Lister, et al. 1999) and xenopus (Kumasaka, et al. 2005) embryos. Dysregulation of MITF may contribute to the initiation and progression of melanoma in a variety of ways (reviewed in Levy, et al. 2006).
Although the existence of subpopulations of zebrafish melanocytes with distinct genetic requirements has been recognized for some time (Johnson, et al. 1995), recent progress in this area has been rapid, as a result of the development of techniques to ablate melanocytes (and thereby induce their regeneration; (Yang, et al. 2004; Yang and Johnson 2006)) and identification of mutations specific to one population (Budi, et al. 2008). In zebrafish, the embryonic pattern is generated primarily from a direct-developing population of melanocyte precursors (Hultman and Johnson 2010), while the melanocytes that will begin to create the adult pattern at the onset of metamorphosis, as well as those that can be recruited to replace embryonic melanocytes lost due to ablation, arise from a self-renewing, stem cell-like population established during the first two days of development (Budi, et al. 2008; Hultman, et al. 2009).
Conditional mutations, in particular temperature–sensitive alleles, have been exploited in genetic model organisms for decades. Temperature-sensitive alleles in zebrafish have proven to be particularly useful for understanding mechanisms of regeneration (Johnson and Weston 1995; Poss, et al. 2002; Nechiporuk, et al. 2003) and pigmentation (Rawls and Johnson 2001; Parichy and Turner 2003; Parichy, et al. 2003; Rawls and Johnson 2003). Previous studies in zebrafish showed that mitfa is required for development of all embryonic and adult melanocytes (Lister, et al. 1999). However, because of the complete absence of melanoblasts and melanocytes in fish homozygous for mitfa null alleles we were previously unable to address stage-specific roles for mitfa in melanocyte differentiation. Here we describe new alleles of mitfa which are temperature-sensitive, and use these new conditional alleles of zebrafish mitfa to test the requirement for mitfa activity at different stages of development and different points of melanocyte differentiation. We find that mitfa activity is required at multiple stages of melanocyte differentiation, but does not appear to be required for establishment of the melanocyte stem cell population.
RESULTS
Identification of temperature-sensitive alleles of mitfa
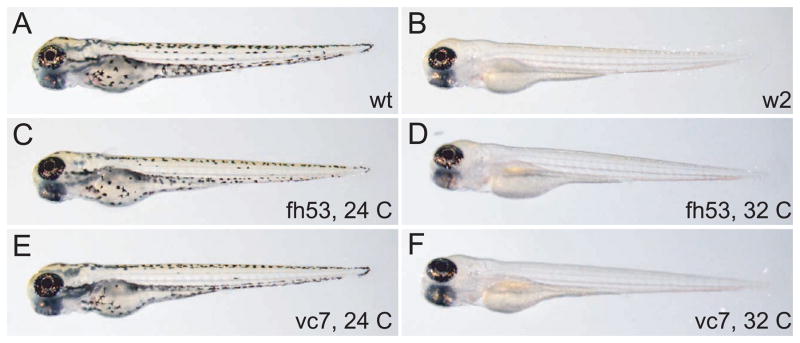
Three new ethlynitrosourea (ENU)-induced alleles of mitfa, mitfafh53, mitfavc7, and mitfaz25, were identified through non-complementation of existing mitfa alleles (B. Draper, T. Linbo, M. Jurynek, unpublished results). In contrast to the two previously described alleles of mitfa (mitfaw2 and mitfab692) in which neural crest-derived melanocytes are completely absent through all stages of development (Lister, et al. 1999; Lister, et al. 2001), fish homozygous for mitfafh53, mitfavc7, and mitfaz25, or in transheterozygous combination with the null alleles mitfaw2 and mitfab692, had variable numbers of melanocytes when reared under standard temperatures and conditions (28.5 C). (Figure 1, 2 and data not shown). Moreover, mitfafh53 and mitfavc7 developed no melanocytes at 32° C, but developed normal or nearly normal melanocyte patterns at 25° C (Table 1). Development at a variety of temperatures from 23–32 degrees suggest that the mitfavc7 allele is still somewhat compromised for function at the lowest permissive temperature attempted (23° C), while the mitfafh53 allele allows normal melanocyte development at 23° C, and nearly normal development at 24 and 25 degrees, significant deficits at 26° and 28.5° C, and complete ablation of melanocytes at 30° and 32°C. mitfafh53 showed similar temperature sensitivity when challenged to grow at different temperatures through mature stages (Figures 1, 2 and Table 1). In contrast, mitfaz25 embryos and adults developed similar numbers of melanocytes at all temperatures (not shown). These results indicate that mitfaz25 is a temperature-independent hypomorphic mutation, while mitfafh53 and to a lesser extent mitfavc7 are temperature-sensitive alleles of mitfa.
Figure 1.
Temperature sensitivity of two mitfa hypomorphic alleles: larval phenotypes. A) Wild-type, B) mitfaw2, C) mitfafh53 raised at 24°C, D) mitfafh53 raised at 32°C, E) mitfavc7 raised at 24°C, F) mitfavc7 raised at 32°C.
Figure 2.
Temperature sensitivity of the mitfa fh53 allele: adult phenotypes. A) Wild-type, B) mitfaw2, C) mitfafh53 raised at 29°C, D) mitfafh53 raised at 29°C to adulthood and then shifted to 25°C for one month, showing recovery of melanocytes.
Table 1.
Temperature dependence of mitfa alleles
| temp. | embryonic melanocytes(1) | adult melanocytes(2) | ||
|---|---|---|---|---|
| mitfavc7 | mitfafh53 | temp. range | mitfafh53 | |
| 23 | +* (n=7) | + | 25.0 – 26.9 | regular stripes (n=5) |
| 24 | +* (n=10) | +* (n=5) | 26.9 – 27.5 | stripes have fewer melanocytes (n=6) |
| 25 | +* (n=6) | +* (n=9) | ||
| 27 | +* | gaps (n=6) | 28.6 – 29.5 | very few melanocytes (< 50/side) (n=7) |
| 28.5 | 1–10 mels/emb. (n=9) | 10–50 mels/emb (n=6) | 31.0 – 31.8 | no melanocytes, solid xanthophore field (n=4) |
| 30 | none (n=8) | none (n=6) | ||
| 32 | none (n=4) | none (n=4) | ||
Embryos scored at ~ 80 hours post-fertilization, after shifting to indicated temperatures at ~ 10 hpf.
Fish from heterozygous intercrosses were were reared at restrictive temperatures for 3 days, mutant embryos selected and returned to 25 degrees for a further 10 days. They were then shifted to aquaria with individual heaters for 5 weeks, and scored for qualitative defects. Temperature in each tank was then recorded daily, and temperature range over the course of experiment shown
Full pattern development slower at these temperatures
Identification of distinct mutations
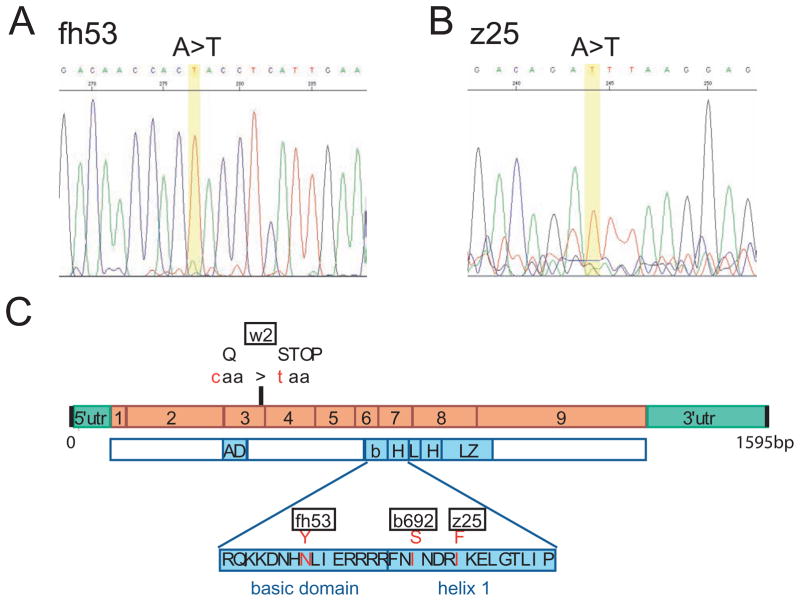
To further explore the hypomorphic or temperature-sensitive nature of the new mitfa alleles we sought to identify their molecular lesions. We first performed RT-PCR with gene specific primers to the 5′ and 3′ untranslated regions on mRNA from homozygous embryos (see Material and Methods). mitfafh53 and mitfaz25 gave single PCR products (data not shown), which were sequenced directly to search for coding sequence changes. This analysis showed that mitfafh53 is an A to T substitution that changes an asparagine to a tyrosine at amino acid 205 (N205Y) in the basic domain known to be the DNA binding portion of the molecule (Fig. 3A). This asparagine is absolutely conserved in all mitf/tfe proteins as well as in a large fraction of basic-helix-loop-helix proteins generally, including USF and all of the Myc/Mad/Max family proteins (Atchley and Fitch 1997). Although an MITF crystal structure has not been published, the structures of the Max and USF homodimers suggest that this asparagine makes contacts with the phosphate backbone (Ferre-D’Amare, et al. 1993; Ferre-D’Amare, et al. 1994; Hallsson, et al. 2007); changing this position to a tyrosine may therefore destabilize DNA binding by the mutant protein at higher temperatures. Mutation of this same codon in human MITF has been identified in a pedigree of a family with Tietz syndrome, although the substitution is for a lysine instead of a tyrosine residue (Smith, et al. 2000).
Figure 3.
Identification of mutations. A), B) Electropherogram of sequence obtained from mitfafh53 (A) and mitfaz25 (B) alleles indicate single base substitutions that change amino acids. C) Location of fh53 and z25 mutations in the mitfa cDNA sequence. Previously characterized alleles w2 and b692 are also shown. Orange boxes indicate coding portions of exons.
Our analysis of the mitfaz25 mutant sequence found that it results in an A-to-T substitution that changes an isoleucine to a phenylalanine (I219F)(Fig. 3B). Interestingly, this is the same amino acid change described for the Mitfmi-enu122 allele in mouse (Steingrimsson, et al. 1998). Crystal structure analysis of homologous bHLH proteins show that this position is an an internal hydrophobic region of the protein that promotes dimerization (Ferre-D’Amare, et al. 1993; Ferre-D’Amare, et al. 1994; Hallsson, et al. 2007). Lack of temperature-dependence of this mutation suggests that the phenotype isn’t simply a matter of thermodestabilization.
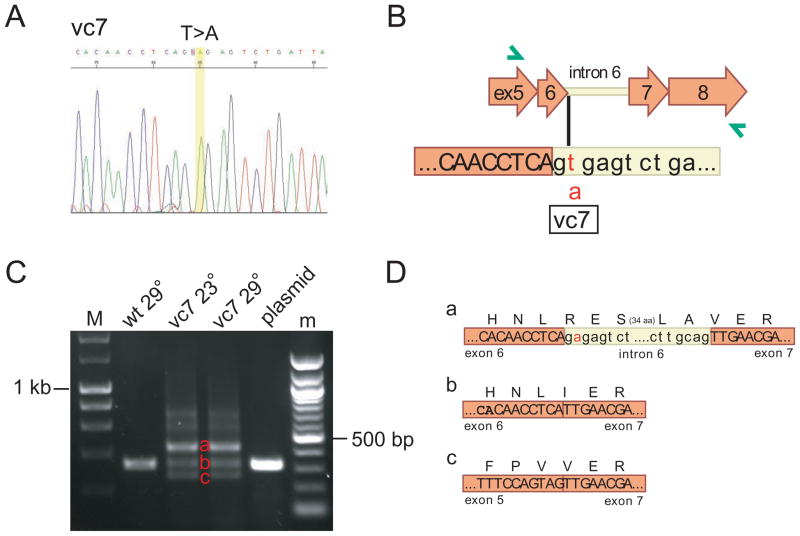
In contrast, RT-PCR from the mitfavc7 mutant resulted in multiple PCR products. To understand this, we cloned each of the products, then sequenced them. The sequence from the predominant RT-PCR product contained a T-to-A substitution in the intron 6 splice donor site (Figure 4), and retention of intron 6. The translation product of this mRNA is expected to extend through intron 6 and continue into exon 7 while preserving the correct reading frame, thus introducing 39 additional amino acids in the middle of the basic region. A minor RT-PCR product of the correct size showed proper splicing, suggesting that this splice donor mutation does not completely abrogate splicing, while a second minor product showed skipping of exon 6, which also preserves the reading frame but truncates the basic region. Semiquantitative RT-PCR shows that the proportion of intron-retaining message to properly-spliced message is identical in RNA harvested from animals reared at the permissive or restrictive temperatures (Fig. 4C), indicating that the temperature sensitivity of the mitfavc7 mutation is not a result of temperature-sensitive splicing. Instead it raises the possibility that the threshold requirement for mitfa is less at lower temperatures. Thus, the small amount of normal protein produced from the minor, correctly spliced mRNA species may be sufficient at 23°C, but insufficient at 32°C. Alternatively, one or both of the translation products resulting from the misspliced messages may have temperature-dependent function.
Figure 4.
The vc7 allele affects spicing of mitfa. A) Electropherogram of sequence obtained from mitfavc7 allele. B) Location of vc7 mutation in genomic sequence. C) Semi-quantitative RT/PCR shows that missplicing is not temperature-sensitive. vc7 results in transcripts that (a) include intron 6 and (c) skip exon 6, in addition to correctly-spliced transcript (b). M, 200 basepair ladder; m, 100 basepair ladder. D) Reading frame is preserved in aberrantly-spliced transcripts a and c.
mitfa is required at multiple steps in melanocyte development
We and others have previously shown that Mitf/mitfa is required for specification of melanocyte fate: neural crest cells in mitfa null mutants fail to express melanoblast markers such as dopachrome tautomerase (dct) (Opdecamp, et al. 1997; Lister, et al. 1999). This early and absolute requirement for mitfa for specification of the melanocyte lineage has prevented a more thorough understanding of subsequent roles for mitfa in melanocyte development and differentiation. We reasoned that we could use our ability to remove or add back mitfa function with temperature upshifts or downshifts at different stages to determine if mitfa is required once, to specify the lineage, or required at one or more successive stages in the differentiation and physiology of the melanocyte.
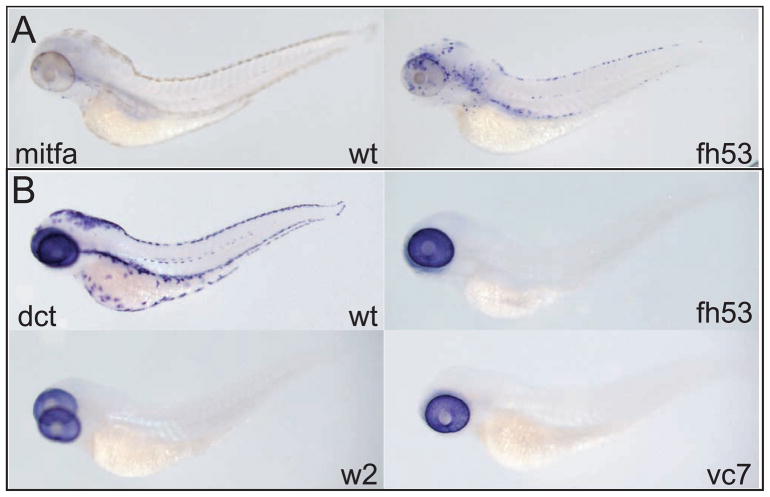
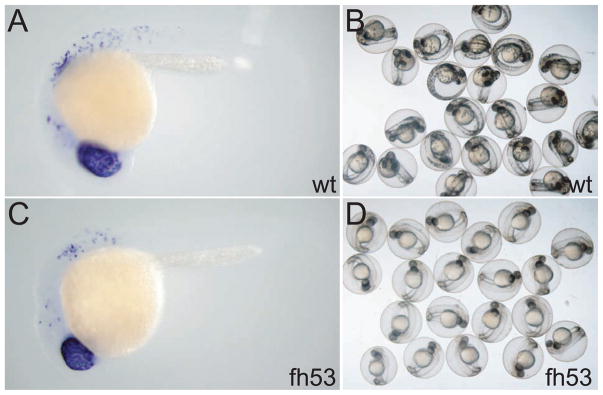
We first asked whether melanocyte lineage specification is similarly defective in the mitf-ts alleles as we had shown for the null mutations. We reared mutants for these alleles at the restrictive temperature for 70 hours at 31.5°C (equivalent of 82 hours standard development (Kimmel, et al. 1995)), fixed and probed them for expression of mitfa and dct. As a missense mutation, the mitfafh53 transcript should be as stable as the wild-type message. At this stage, all of the direct-developing embryonic melanocytes have differentiated in wild-type larvae and strong expression of mitfa is restricted to a small number of cells in the head (Figure 5A). (Weak mitfa expression is detectable in differentiated melanocytes after 2 to 3 days of color development; data not shown.) In contrast, in mitfafh53 homozygous larvae at this stage, strongly mitfa-expressing cells are still abundant, and although a large number around the ear appear not to have undergone extensive migration, many of these cells are found in positions characteristic of the dorsal, lateral, and ventral stripes. Despite abundant mitfa+ cells in the temperature-sensitive mutants at the restrictive temperature (Fig. 5A), no dct expressing cells could be detected outside the pigmented retinal epithelium (Fig. 5B). Similarly, we find no expression of a variety of melanocyte lineage reporters such as a kita enhancer trap (Distel, et al. 2009), the Gpnmb enhancer reporter (Loftus, et al. 2009), the fTyrp1 promoter reporter (Zou, et al. 2006), and TYR- and DCT- promoter and enhancer reporters (A. McCallion and S. Johnson, unpublished) (not shown). These results suggest that similar to the null mutants, the mitfa-ts mutants block melanocyte specification at early (pre-kita or pre-dct expressing) stages. The identity/fate of the mitfa+ cells is not known (but see below).
Figure 5.
mitfa is required at multiple steps in melanocyte development. A) mitfa expression persists at restrictive temperature. Wild-type and mitfafh53 larvae are shown. B) No dct is expressed in neural crest cells when held at restrictive temperature, but retinal expression of dct is not affected. Wild-type, mitfaw2, mitfafh53, and mitfavc7 larvae are shown. Embryos were treated with 0.2 mM PTU to suppress melanin synthesis. All larvae are at the equivalent of 82 hours at standard temperature.
In order to ask whether mitfa has subsequent requirements in differentiation we took embryos reared to the equivalent of stage 22 hpf at the permissive temperature and shifted them to the restrictive temperature. Embryos examined for dct expression at the time of the upshift showed abundant dct expression, indicating that melanoblasts had been specified and were beginning to differentiate at the permissive temperatures (Fig. 6). However, no melanin was produced in mitfafh53 embryos (n=58) that were upshifted but allowed to develop for an additional day or longer, with the exception of the RPE, which has no requirement for mitfa. These results show that mitfa is required both for initial specification of melanoblasts, but additionally after specification has been demonstrated, there remain additional roles for mitfa in melanocyte differentiation, including melanin production.
Figure 6.
Embryos raised at permissive temperature to dct+ stage, when shifted show no melanized cells. A) Wild-type embryo showing dct expression in retinal pigment epithelium (RPE) and neural crest melanoblasts, B) wild-type embryos 20 hours after upshift (stage equivalent to 45 hours at standard temperature) showing differentiated melanocytes, C) mitfafh53 embryo showing dct expression at time of temperature upshift, D) mitfafh53 embryos 20 hours after upshift do not display any melanized cells save for RPE. In A and C, embryos were treated with 0.2 mM PTU to suppress melanin synthesis.
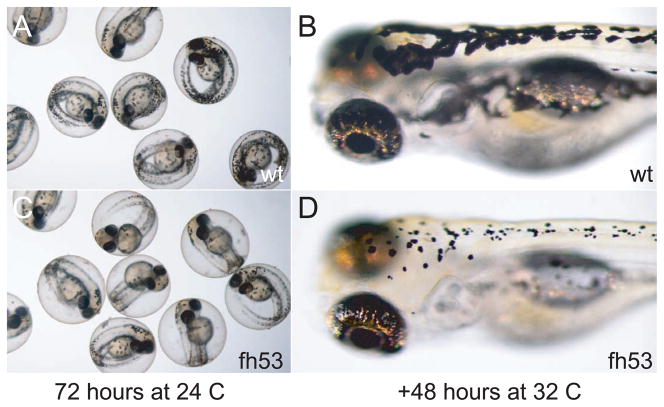
We were also interested in whether mitfa has roles in maintaining the normal physiology of the melanocyte after differentiation. When mitfafh53 larvae carrying the fTyrp>GFPj900 transgene for differentiated melanocytes were shifted to the restrictive temperature following melanization and expression of fTyrp>GFP, GFP fluorescence was extinguished within 24 hours (data not shown). Possible explanations for this result include that mitfa is continuously required for expression of the fTyrp>GFP transgene, or that the melanocytes are dying, or both. One possibility is that mitfa is required for survival of melanocytes, which might be mediated through the requirement for kit (or kita) function. Use of a temperature-sensitive allele of kit has shown that kit is required at multiple stages for different melanocyte functions, including promoting survival after 48 hours (Rawls and Johnson 2003). Since kit expression is dependent on mitfa function, one prediction is that melanocytes shifted to the restrictive temperature at 48 hours would also die. To address this, we took mitfafh53 embryos reared at the permissive temperature through 72 hours, at which stage they have the full complement of differentiated and melanized melanocytes, and shifted them to restrictive temperature. When larvae were examined 48 hours later, their melanocytes had lost dendricity and taken on a small, rounded appearance (Fig. 7). Although this is characteristic of dying melanocytes (Parichy, et al. 1999), we were unable to detect markers of cell death, and did not observe extrusion of cell carcasses from the epidermis characteristic of melanocyte death in kit mutants, even after several days. One possible explanation for the absence of overt melanocyte death and extrusion was that the mitfafh53 mutation has residual activity at the restrictive temperature. This seems unlikely, since halving the dosage of mitfafh53 by generating transheterozygotes with a null mitfa mutation (mitfafh53/mitfaw2) also showed little or no extrusion. A second possible explanation, that lack of overt death and extrusion is due to a deficit in macrophages, seems unlikely since mitfa mutants stain normally for macrophages (data not shown) with the marker neutral red (Herbomel and Levraud, 2005). Whether there is residual mitfa activity or not, these results show that mitfa is still required for melanocyte physiology even after the transcript has become only weakly detectable.
Figure 7.
Loss of dendricity in melanocytes at restrictive temperature. Wild-type and mitfafh53 embryos were raised at permissive temperature for 72 hours, photographed (A,C) then shifted to 32°C for 48 hours (B,D).
mitfa is not required in the stem cell, but is required for maintenance of direct-developing precursors
Melanocytes in embryonic and larval zebrafish originate primarily from direct-developing embryonic (ontogenetic) precursors, however upon their ablation new melanocytes can be regenerated from a stem cell population which may be the same as that responsible for addition of melanocytes to the forming adult pigment pattern at metamorphosis (Hultman, et al. 2009). To determine if there are differential requirements for mitfa in these populations we performed temperature shift experiments. Melanocytes in mitfafh53 homozygotes were observed to recover when animals were raised at restrictive temperature and downshifted after as many as 9 days, and indeed even in fish shifted to permissive temperature as adults (Figures 2, 8). These results suggest that embryonic melanoblasts may be arrested (albeit prior to becoming dct-positive) in the absence of mitfa activity, but capable of differentiating after mitfa is restored, and/or that mitfa activity is not required for the establishment or survival of the melanocyte stem cell.
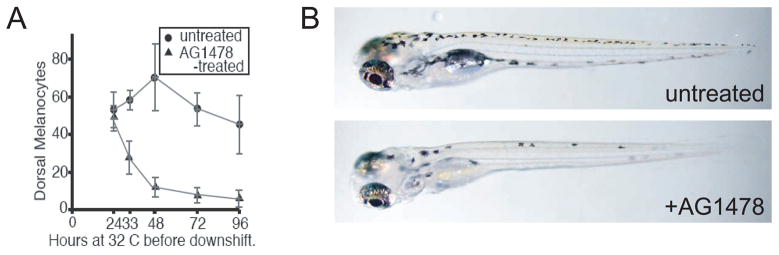
Figure 8.
mitfa is not required in the stem cell, but is required for survival of direct-developing melanocyte progenitors. A) graph showing dorsal melanocyte number in mitfafh53 homozygous larvae held at 32°C for the indicated times and then shifted to permissive temperature, and melanocytes counted three days later. B) Homozygous mitfa fh53 larvae held at 32°C in the presence of DMSO (top) or AG1478 (bottom) then downshifted to permissive temperature.
Although the melanocyte stem cell has not yet been isolated in zebrafish, it is known that its establishment is dependent upon the activity of the erbb3b gene during a narrow window of embryonic development (Budi, et al. 2008; Hultman, et al. 2009). The stem cell can be eliminated by treatment with the drug AG1478 between 9 and 48 hours post fertilization (Budi, et al. 2008; Hultman, et al. 2009). To further test the relationship between the melanocyte stem cell and mitfa activity, we placed mitfa fh53 homozygous embryos at restrictive temperature and treated them with AG1478, then downshifted to permissive temperature at various times to assess recovery of melanocytes (Figure 8). While AG1478-treated embryos downshifted at 24 hours showed comparable recovery to those treated with DMSO, embryos downshifted at later times showed progressively reduced capacity to recover (Figure 8). This sensitivity suggests that the majority of the melanocytes that develop in larvae downshifted after 48 h at the restrictive temperature do indeed derive from the erbb3b-dependent (stem cell) population. Moreover, because in untreated embryos these cells recover even when larvae are held at restrictive temperature during the critical period for MSC establishment, it argues that mitfa is not itself required for that establishment. We investigated the possibility that residual mitfa activity from the ts allele was responsible for promoting establishment of the MSC, by combining mitfafh53 (or mitfavc7) with the null mitfa allele, mitfaw2. These animals should have one-half the residual mitfa activity as the homozygotes for the ts alleles. In both cases, the transheterozygotes recovered melanocytes similar to the homozygotes, tending to argue that there is little or no residual mitfa activity in the ts mutations at the restrictive temperature (not shown).
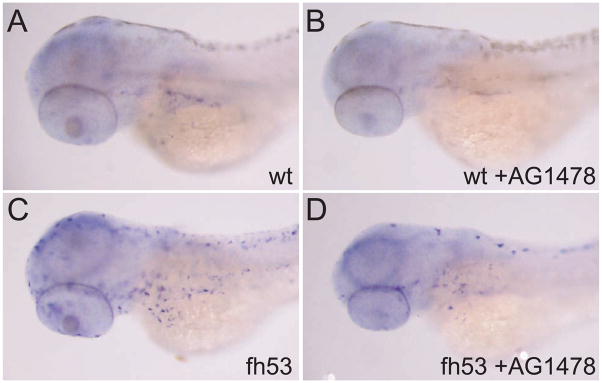
Finally, we examined the expression of mitfa by in situ hybridization under these conditions in wild-type and fh53 homozygous embryos and larvae. Embryos were raised at restrictive temperature in the presence or absence of AG1478, and then fixed at the equivalent of 65 hours’ development at standard temperature. As noted earlier, mitfa expression in wild-type larvae has greatly subsided by this stage; in AG1478-treated larvae, expression is reduced to an even greater degree (Figure 9). Likewise, the abundant expression of mitfa mRNA observed in fh53 larvae at restrictive temperature is also greatly diminished by AG1478 treatment. Together with the loss of melanocyte recovery, these results raise the possibility that direct developing mitfa+ melanoblasts, in the absence of mitfa function, eventually die. Presumably, the MSC derived melanoblasts share a similar requirement on mitfa for their survival. Whether any of the mitfa-expressing population represents MSC’s or rather committed progenitors recruited from the MSC awaits further investigation.
Figure 9.
in situ hybridization for mitfa in wild-type (A,B) and mitfafh53 (C,D) larvae held at restrictive temperature with or without AG1478 treatment from 9–48 hours development. AG1478 treatment reduces the number of mitfa-expressing cells in both wild-type and mutant embryos. Larvae are at the equivalent of the 65 hour stage at standard temperature. Embryos were treated with 0.2 mM PTU to suppress melanin synthesis.
DISCUSSION
An understanding of the central role of Mitf in vertebrate pigmentation has been greatly aided by the large number of mouse Mitf alleles available for study (Hallsson, et al. 2000; Steingrimsson, et al. 2004). In this study we describe characterization of three new alleles of the zebrafish Mitf ortholog mitfa. We demonstrate that two of these alleles display temperature-dependent phenotypes, and exploit this conditionality to test the requirement for mitfa activity at distinct points of melanocyte differentiation and stages of ontogeny. Interestingly, although the fh53 and vc7 alleles display similar temperature dependence, the nature of the two mutations is quite distinct. mitfafh53 results from an amino acid substitution at a conserved position in the basic domain, which is responsible for DNA binding. In contrast, the mitfavc7 mutation is in a splice site, and leads to similar levels of mis-splicing both at permissive and restrictive temperatures. Whether the temperature sensitivity is a result of overall lower levels of Mitfa protein, or due to antimorphic effects from translation products of one or more of the mis-spliced transcripts, remains to be resolved.
Our results support previous studies of null alleles that mitf activity is required for specification of melanocytes (Lister, et al. 1999), as under restrictive conditions (high temperature) we never observe expression of the specification marker dct. Additionally, we find that mitfa is required for later steps, as when the temperature is raised shortly after the onset of neural crest dct expression (i.e. specification) melanized cells are still not observed. Melanocytes that have undergone differentiation still require mitf function; although we did not observe melanocyte cell death at restrictive temperature, differentiated melanocytes lost dendricity and took on the appearance of dying cells (Parichy, et al. 1999). Similarly, melanoma cells depleted of Mitf by siRNA arrest in G1 and round up but do not die (Carreira, et al. 2006). It may be that even at restrictive temperature these conditional zebrafish alleles retain sufficient residual mitfa function to keep the melanocyte alive.
The fate of those cells which initiate mitfa expression but lack mitfa function is a key question. The finding that recovery of melanocytes following restoration of mitfa function is rapidly lost when MSCs were first ablated leads us to favor the model that these cells die. An alternative possibility is that these cells, failing in their efforts to specify along the melanocyte lineage, take on other pigment cell fates. Some support for this model is provided by previous reports of co-expression of mitfa with markers of other pigment lineages in wild-type embryos (Parichy, et al. 2000; Curran, et al. 2010) and of excess iridophores in mitfa null mutants (Lister et al, 1999). Similar increases in iridophores are also observed in the ts alleles at the restrictive temperature (data not shown). However, the number of excess iridophores (~ 20) can only partially account for the more than 400 direct developing melanocytes that fail to differentiate in mitfa mutants. Similarly, we fail to observe an excess of xanthophores in mitfa mutants.
In addition to addressing different stages in development of the melanocyte, we were able to use the conditional alleles to investigate the requirement of mitfa at different stages of the life history and in different populations of melanocyte precursors. Previous work has shown that the embryonic zebrafish melanocyte pattern is derived primarily from a population of direct-developing precursors, and that filling in (Hultman and Johnson 2010) or regenerating (Hultman, et al. 2009) the final pattern, as well as generating the metamorphic pattern (Budi, et al. 2008), is dependent on a stem cell population that is dependent on activity of the erbb3b gene between 9 and 48 hours post-fertilization. By combining temperature-shift experiments with drug treatments that block erbb3b activity, we find that mitfa activity is not required for establishment of the melanocyte stem cell: in mutant animals raised initially at restrictive temperature, shifts to permissive temperature even as late as adulthood result in recovery of melanocytes. This recovery arises largely from the erbb3b-dependent stem cell population as it is sensitive to early treatment with the antagonist AG1478. Similarly, the number of mitfa-expressing cells in mitfafh53 embryos at restrictive temperature is greatly reduced by AG1478. This could argue that these mitfa expressing cells in mitfafh53 embryos at restrictive temperature represent the MSC itself or their committed daughters at the first mitfa-requiring stage. That wild-type embryos have few cells expressing mitfa at this stage tends to argue that mitfa is not expressed in the stem cell, but rather in the committed daughter, at the time that mitfa is required to initiate melanocyte differentiation. We note that that in the absence of biochemical assays for Mitf activity, we have not completely excluded the unlikely possibility that there remains sufficient function at the restrictive temperature to rescue the MSC. Nevertheless, our finding is consistent with work in mouse, where Mitf does not appear to be a consistent marker of the MSC (Osawa, et al. 2005). Stable, long-term lineage tracing methods (such as via Cre/lox, or transposon marking) that have been available in other systems and are beginning to be established in zebrafish (Boniface, et al. 2009; Hans, et al. 2009; Hesselson, et al. 2009; Tu and Johnson 2010) are likely to aid in the resolution of this and other questions.
These results show that mitfa is required both for initial specification of melanoblasts, but additionally after specification has been demonstrated by dct expression, there remain additional roles for mitfa in melanocyte differentiation. These roles may include expression of structural genes for melanosome biogenesis, such as pmel17, mlana, or gpnmb, or other enzymes that contribute to melanin synthesis, such as tyr or tyrp1 (Cheli, et al. 2010). Expression of each of these genes is dependent on the transcription promoting activity of Mitf in mammalian cells. Whether these genes are expressed simultaneously with dct in the zebrafish melanoblast, or sufficiently late to account for the failure of mitfafh53 mutants to melanize after upshift to the restrictive temperature is not clear. These upshift experiments may provide an additional tool to understand the possible roles for temporal differences in control of gene expression by mitfa.
MATERIALS AND METHODS
Fishkeeping and husbandry
Adult fish of the wild-type AB strain, and mitfa alleles w2 (ZDB-ALT-990423-22), fh53, and vc7 were maintained on a 14 hour/10 hour light/dark cycle. Embryos were obtained from natural matings and staged according to Kimmel et al. (Kimmel, et al. 1995). In some cases, melanin synthesis was first suppressed by prior incubation with 0.2mM pheylthiourea (PTU, Sigma, P7629). All experiments were performed in accordance with Institutional Animal Care and Use Committee (IACUC) protocols. For all experiments, conditions, or datapoints, we analyzed at least 10 embryos, unless otherwise stated.
Identification of lesions
First strand cDNA was generated from total RNA isolated from homozygous mitfafh53 and mitfavc7 or transheterozygous (mitfafh53/mitfaw2 or mitfavc7/mitfaw2) embryos, and used as a template for PCR to amplify the mitfa open reading frame in two overlapping sections with the following primers: 5′forward: GGC CAA GAC GAC TGG TCA GTT CTT GCA C, 5′ reverse: TCT CTC TTT TGC CAG GGC TCT GAC TTC TGC, 5′ reverse2: ACG GAT CAT TTG ACT TGG GAA TTA AAG, 3′forward: GCA GAA GTC AGA GCC CTG GC, 3′reverse: GGT TCA TGA AAT TTA GTT GGC ATT GC. PCR products were sequenced directly, or subcloned by TOPO TA cloning (Invitrogen) and then sequenced. Sequence changes identified in cDNA were confirmed in genomic DNA.
Temperature shift experiments
For most experiments, mitfats embryos were generated from homozygous intercrosses. For the experiments shown in Table 1, mitfafh53 embryos were generated from heterozygous intercrosses. At 24–27 degrees, mitfafh53 mutant embryos were identified by ectopic iridophores, while at higher temperatures mutant embryos were clearly discerned by melanocyte phenotypes.
For adult experiments, fish from heterozygous intercrosses were were reared at restrictive temperatures for 3 days, mutant embryos selected and returned to 25 degrees for a further 10 days. They were then shifted to aquaria with individual heaters for 5 weeks, and scored for qualitative defects. Temperature in each tank was then recorded daily, and temperature range over the course of experiment is shown.
Drug treatments
AG1478/tyrphostin (LC Laboratories cat. no. T-7310) was prepared at a concentration of 6 millimolar in DMSO, and diluted further with DMSO before use to yield a final concentration of 3 micromolar in 0.5% DMSO in system water or embryo medium. Drug was added at 9 hours post fertilization and washed out at 48 hours post-fertilization unless otherwise noted. 10 fish were examined per timepoint for each treatment, and the experiment was replicated with similar results to those presented.
In situ hybridization
In situ hybridization was carried out as previously described (Thisse and Thisse 2008). Riboprobes for mitfa (ZDB-GENE-990910-11) and dct (ZDB-GENE-000508-1) have been described previously (Lister, et al. 1999; Kelsh, et al. 2000). For photography, samples were equilibrated in 50% glycerol/PBS and imaged with an Olympus SZ12 stereo dissecting scope and DP70 digital camera. Images were adjusted and color balanced in Photoshop (Adobe).
Acknowledgments
Supported by American Cancer Society grant IRG-99-225-04 and a Conquer Cancer Now award from the Concern Foundation to J.A.L. and NIH RO1-GM56988 to S.L.J. Thanks to Bruce Draper, Tor Linbo, Mick Jurynek for sharing the new mitfa alleles and Alex Scott for help in counting melanocytes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atchley W, Fitch W. A Natural Classification of the Basic Helix-Loop-Helix Class of Transcription Factors. Proc Natl Acad Sci U S A. 1997;94:5172–516. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar J, Hong Y, Schartl M. Mitf Expression is Sufficient to Direct Differentiation of Medaka Blastula Derived Stem Cells to Melanocytes. Development. 2003;130:6545–6553. doi: 10.1242/dev.00872. [DOI] [PubMed] [Google Scholar]
- Boniface EJ, Lu J, Victoroff T, Zhu M, Chen W. FlEx-Based Transgenic Reporter Lines for Visualization of Cre and Flp Activity in Live Zebrafish. Genesis. 2009;47:484–491. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budi EH, Patterson LB, Parichy DM. Embryonic Requirements for ErbB Signaling in Neural Crest Development and Adult Pigment Pattern Formation. Development. 2008;135:2603–2614. doi: 10.1242/dev.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf Regulation of Dia1 Controls Melanoma Proliferation and Invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-Year Quest for Microphthalmia-Associated Transcription Factor Target Genes. Pigment Cell Melanoma Res. 2010;23:27–40. doi: 10.1111/j.1755-148X.2009.00653.x. [DOI] [PubMed] [Google Scholar]
- Curran K, Lister JA, Kunkel GR, Prendergast A, Parichy DM, Raible DW. Interplay between Foxd3 and Mitf Regulates Cell Fate Plasticity in the Zebrafish Neural Crest. Dev Biol. 2010;344:107–118. doi: 10.1016/j.ydbio.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Koster RW. Optimized Gal4 Genetics for Permanent Gene Expression Mapping in Zebrafish. Proc Natl Acad Sci U S A. 2009;106:13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D’Amare AR, Pognonec P, Roeder RG, Burley SK. Structure and Function of the b/HLH/Z Domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D’Amare AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its Cognate DNA through a Dimeric b/HLH/Z Domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Haflidadottir BS, Schepsky A, Arnheiter H, Steingrimsson E. Evolutionary Sequence Comparison of the Mitf Gene Reveals Novel Conserved Domains. Pigment Cell Res. 2007;20:185–200. doi: 10.1111/j.1600-0749.2007.00373.x. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Favor J, Hodgkinson C, Glaser T, Lamoreux ML, Magnusdottir R, Gunnarsson GJ, Sweet HO, Copeland NG, Jenkins NA, Steingrimsson E. Genomic, Transcriptional and Mutational Analysis of the Mouse Microphthalmia Locus. Genetics. 2000;155:291–300. doi: 10.1093/genetics/155.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-Controlled Site-Specific Recombination in Zebrafish. PLoS ONE. 2009;4:e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P, Levraud JP. Imaging early macrophage differentiation, migration, and behaviors in live zebrafish embryos. Methods Mol Med. 2005;105:199–214. doi: 10.1385/1-59259-826-9:199. [DOI] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct Populations of Quiescent and Proliferative Pancreatic Beta-Cells Identified by HOTcre Mediated Labeling. Proc Natl Acad Sci U S A. 2009;106:14896–14901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Nakayama A, Li H, Swenson LB, Opdecamp K, Asher JH, Jr, Arnheiter H, Glaser T. Mutation at the Anophthalmic White Locus in Syrian Hamsters: Haploinsufficiency in the Mitf Gene Mimics Human Waardenburg Syndrome Type 2. Hum Mol Genet. 1998;7:703–708. doi: 10.1093/hmg/7.4.703. [DOI] [PubMed] [Google Scholar]
- Hultman KA, Johnson SL. Differential Contribution of Direct-Developing and Stem Cell-Derived Melanocytes to the Zebrafish Larval Pigment Pattern. Dev Biol. 2010;337:425–431. doi: 10.1016/j.ydbio.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman KA, Budi EH, Teasley DC, Gottlieb AY, Parichy DM, Johnson SL. Defects in ErbB-Dependent Establishment of Adult Melanocyte Stem Cells Reveal Independent Origins for Embryonic and Regeneration Melanocytes. PLoS Genet. 2009;5:e1000544. doi: 10.1371/journal.pgen.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Africa D, Walker C, Weston JA. Genetic Control of Adult Pigment Stripe Development in Zebrafish. Dev Biol. 1995;167:27–33. doi: 10.1006/dbio.1995.1004. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Weston JA. Temperature-Sensitive Mutations that Cause Stage-Specific Defects in Zebrafish Fin Regeneration. Genetics. 1995;141:1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh R, Schmid B, Eisen J. Genetic Analysis of Melanophore Development in Zebrafish Embryos. Dev Biol. 2000;225:277–93. doi: 10.1006/dbio.2000.9840. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of Embryonic Development of the Zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kumasaka M, Sato S, Yajima I, Goding CR, Yamamoto H. Regulation of Melanoblast and Retinal Pigment Epithelium Development by Xenopus Laevis Mitf. Dev Dyn. 2005;234:523–534. doi: 10.1002/dvdy.20505. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: Master Regulator of Melanocyte Development and Melanoma Oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Lister JA, Close J, Raible DW. Duplicate Mitf Genes in Zebrafish: Complementary Expression and Conservation of Melanogenic Potential. Dev Biol. 2001;237:333–344. doi: 10.1006/dbio.2001.0379. [DOI] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. Nacre Encodes a Zebrafish Microphthalmia-Related Protein that Regulates Neural-Crest-Derived Pigment Cell Fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Loftus SK, Antonellis A, Matera I, Renaud G, Baxter LL, Reid D, Wolfsberg TG, Chen Y, Wang C, Prasad MK, Bessling SL, McCallion AS, Green ED, Bennett DC, Pavan WJ NISC Comparative Sequencing Program. Gpnmb is a Melanoblast-Expressed, MITF-Dependent Gene. Pigment Cell Melanoma Res. 2009;22:99–110. doi: 10.1111/j.1755-148X.2008.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellgren EM, Johnson SL. A Requirement for Kit in Embryonic Zebrafish Melanocyte Differentiation is Revealed by Melanoblast Delay. Dev Genes Evol. 2004;214:493–502. doi: 10.1007/s00427-004-0428-y. [DOI] [PubMed] [Google Scholar]
- Mochii M, Ono T, Matsubara Y, Eguchi G. Spontaneous Transdifferentiation of Quail Pigmented Epithelial Cell is Accompanied by a Mutation in the Mitf Gene. Dev Biol. 1998;196:145–159. doi: 10.1006/dbio.1998.8864. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Poss K, Johnson S, Keating M. Positional Cloning of a Temperature-Sensitive Mutant Emmental Reveals a Role for sly1 during Cell Proliferation in Zebrafish Fin Regeneration. Dev Biol. 2003;258:291–306. doi: 10.1016/s0012-1606(03)00129-5. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Vanvooren P, Riviere M, Arnheiter H, Motta R, Szpirer J, Szpirer C. The Rat Microphthalmia-Associated Transcription Factor Gene (Mitf) Maps at 4q34-q41 and is Mutated in the Mib Rats. Mamm Genome. 1998;9:617–621. doi: 10.1007/s003359900832. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte Development in Vivo and in Neural Crest Cell Cultures: Crucial Dependence on the Mitf Basic-Helix-Loop-Helix-Zipper Transcription Factor. Development. 1997;124:2377–2386. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Osawa M, Egawa G, Mak SS, Moriyama M, Freter R, Yonetani S, Beermann F, Nishikawa S. Molecular Characterization of Melanocyte Stem Cells in their Niche. Development. 2005;132:5589–5599. doi: 10.1242/dev.02161. [DOI] [PubMed] [Google Scholar]
- Parichy D, Turner J. Temporal and Cellular Requirements for Fms Signaling during Zebrafish Adult Pigment Pattern Development. Development. 2003;130:817–833. doi: 10.1242/dev.00307. [DOI] [PubMed] [Google Scholar]
- Parichy D, Turner J, Parker N. Essential Role for Puma in Development of Postembryonic Neural Crest-Derived Cell Lineages in Zebrafish. Dev Biol. 2003;256:221–241. doi: 10.1016/s0012-1606(03)00016-2. [DOI] [PubMed] [Google Scholar]
- Parichy D, Ransom D, Paw B, Zon L, Johnson S. An Orthologue of the Kit-Related Gene Fms is Required for Development of Neural Crest-Derived Xanthophores and a Subpopulation of Adult Melanocytes in the Zebrafish, Danio Rerio. Development. 2000;127:3031–344. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish Sparse Corresponds to an Orthologue of c-Kit and is Required for the Morphogenesis of a Subpopulation of Melanocytes, but is Not Essential for Hematopoiesis Or Primordial Germ Cell Development. Development. 1999;126:3425–3436. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 Defines a Proximal Blastemal Proliferative Compartment Essential for Zebrafish Fin Regeneration. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- Rawls J, Johnson S. Temporal and Molecular Separation of the Kit Receptor Tyrosine Kinase’s Roles in Zebrafish Melanocyte Migration and Survival. Dev Biol. 2003;262:152–161. doi: 10.1016/s0012-1606(03)00386-5. [DOI] [PubMed] [Google Scholar]
- Rawls J, Johnson S. Requirements for the Kit Receptor Tyrosine Kinase during Regeneration of Zebrafish Fin Melanocytes. Development. 2001;128:1943–1949. doi: 10.1242/dev.128.11.1943. [DOI] [PubMed] [Google Scholar]
- Smith SD, Kelley PM, Kenyon JB, Hoover D. Tietz Syndrome (hypopigmentation/deafness) Caused by Mutation of MITF. J Med Genet. 2000;37:446–448. doi: 10.1136/jmg.37.6.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the Microphthalmia Transcription Factor Network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Favor J, Ferre-D’Amare AF, Copeland NG, Jenkins NA. Mitfmi-enu122 is a Missense Mutation in the HLH Dimerization Domain. Mamm Genome. 1998;9:250–252. doi: 10.1007/s003359900736. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-Resolution in Situ Hybridization to Whole-Mount Zebrafish Embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Tu S, Johnson SL. Clonal Analyses Reveal Roles of Organ Founding Stem Cells, Melanocyte Stem Cells and Melanoblasts in Establishment, Growth and Regeneration of the Adult Zebrafish Fin. Development. 2010;137:3931–3939. doi: 10.1242/dev.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund HR, Fisher DE. Microphthalamia-Associated Transcription Factor: A Critical Regulator of Pigment Cell Development and Survival. Oncogene. 2003;22:3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- Yang CT, Johnson SL. Small Molecule-Induced Ablation and Subsequent Regeneration of Larval Zebrafish Melanocytes. Development. 2006;133:3563–3573. doi: 10.1242/dev.02533. [DOI] [PubMed] [Google Scholar]
- Yang CT, Sengelmann RD, Johnson SL. Larval Melanocyte Regeneration Following Laser Ablation in Zebrafish. J Invest Dermatol. 2004;123:924–929. doi: 10.1111/j.0022-202X.2004.23475.x. [DOI] [PubMed] [Google Scholar]
- Zou J, Beermann F, Wang J, Kawakami K, Wei X. The Fugu tyrp1 Promoter Directs Specific GFP Expression in Zebrafish: Tools to Study the RPE and the Neural Crest-Derived Melanophores. Pigment Cell Res. 2006;19:615–627. doi: 10.1111/j.1600-0749.2006.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]