Abstract
Objective
The role of post-transcription regulation in preeclampsia is largely unknown. We investigated preeclampsia related placental microRNA (miRNA) expression using microarray and confirmatory qRT-PCR experiments.
Study design
Placental expressions of characterized and novel miRNAs (1,295 probes) were measured in samples collected from 20 preeclampsia cases and 20 controls. Differential expression was evaluated using Students T-test and fold change analyses. In pathway analysis, we examined functions/functional relationships of targets of differentially expressed miRNAs.
Results
Eight miRNAs were differentially expressed (1 up- and 7 down-regulated) among preeclampsia cases compared with controls. These included previously identified candidates (miR-210, miR-1 and a miRNA in the 14q32.31 cluster region) and others that are novel (miR- 584 and miR-34c-5p). These miRNAs target genes that participate in organ/system development (cardiovascular and reproductive system), immunologic dysfunction, cell adhesion, cell cycle and signaling.
Conclusion
Expression of microRNAs that target genes in diverse pathophysiological processes is altered in the setting of preeclampsia.
Keywords: expression, microRNA, placenta, preeclampsia
Introduction
Preeclampsia, a vascular disorder characterized by hypertension and proteinuria during pregnancy, is a consequence of diverse pathophysiological processes involving impaired implantation, endothelial dysfunction and systemic inflammation.1–3 The placenta, a source and site of action of mediators of these processes, plays a central role in preeclampsia pathogenesis.1–4 Placenta-based investigations can potentially enhance our understanding of disease initiation and progression. While substantial evidence supports differential placental gene expression in preeclampsia,5–8 the role of related placental post-transcription regulation that may contribute to pathway/network perturbations is largely unknown.
MicroRNAs (miRNAs), small non-coding RNAs, are highly conserved post-transcription regulators of gene expression.9–10 Through inhibition of protein translation or promotion of mRNA degradation, miRNAs have roles in physiological and pathological processes such as cell differentiation, proliferation/growth, apoptosis, angiogenesis, inflammation, redox signaling and other endothelial cell functions.9–12 Since these processes are disrupted in preeclampsia, miRNAs can potentially play significant roles in preeclampsia pathogenesis. Few investigators have studied placental miRNA expression in relation to preeclampsia and even fewer describe microarray based global placental miRNA profiling in preeclampsia.13–16 In order to address this paucity of data, we investigated global placental miRNA expression using microarray and confirmatory qRT-PCR methodologies among 20 preeclampsia cases and 20 normotensive control subjects. In pathway analysis, we also examined functions and functional relationships of targets of differentially expressed miRNAs.
Materials and Methods
Study population and data collection
Study participants were selected from participants of the Omega study (a cohort study) and the Placenta MicroArray study (a case control study). Study populations and data collection procedures, described before, were briefly as follows.8 Omega study participants were women who initiated prenatal care before 16 weeks gestation and attended prenatal care clinics affiliated with Swedish Medical Center, Seattle, Washington. The Placenta MicroArray study participants comprised of women who delivered at Swedish Medical Center. Preeclampsia was diagnosed when both pregnancy-induced hypertension (PIH) and proteinuria were present according to ACOG 2000 guidelines.17 PIH was defined as a sustained (≥2 measures 6 hours apart) blood pressure elevation (>140/90 mmHg) after 20 weeks of gestation. Proteinuria was defined as a sustained (≥2 measures 4 hours apart) presence of elevated protein in the urine (>30 mg/dL or >1+ on a urine dipstick). Controls were selected from those women who had normotensive pregnancies uncomplicated by proteinuria. Women who had history of chronic hypertension and/or pre-gestational diabetes as well as current non-singleton pregnancies were excluded. Cases (N=20) and controls (N=20) were frequency matched for parity, maternal race/ethnicity and labor status. Medical records were used to obtain information on risk factors, pregnancy history and perinatal outcome. The Institutional Review Board of the Swedish Medical Center approved study protocols. All participants provided written informed consent.
Placental sample collection
Placental tissue were collected immediately after delivery, weighed, double bagged and transported in coolers to the placenta-processing lab where it was processed within 15 minutes post-delivery. The chorionic plate and overlying membranes were removed and tissue biopsies (~0.5 cm3 each) were obtained from 16 sites (8 maternal and 8 fetal sides) using a systematic sampling technique to achieve uniformity and adequate sampling.8 Briefly, the placenta was laid flat with the fetal side facing up and mapped into four quadrants. Two samples were obtained from each quadrant; one medial (about 2 centimeters from the center) and one lateral (about 2 centimeters from the margin). The placenta was then turned over and eight corresponding samples were taken from the maternal side. For this analysis, biopsy samples taken from the maternal side consisting primarily of the villous tissue, utero-placental arteries and some decidua basalis were evaluated. Biopsy samples were placed in cryotubes containing RNAlater (Qiagen Inc, Valencia, CA), at 10µl per 1 mg of tissue and stored at −80°C.
RNA extraction and quality control
Total RNA was extracted from samples using a modification of the acid guanidinium thiocyanate-phenol chloroform extraction method.18 Briefly, up to 30 mg of tissue was homogenized in 1,000 µL of RNASTAT60 (Tel-Test, Friendswood, TX), followed by addition of 250 µL of chloroform and vortexing for 1 min. The RNA was precipitated with 2X volumes of isopropanol overnight at −20 °C before being pelleted with a 30-min centrifugation at 12,000 xg. The RNA pellets were resuspended in RNAsecure (Ambion, Foster City, CA). Up to 10 µg of RNA for each sample was treated with Turbo DNase (Ambion, Foster City, CA) for 1 h at 37 °C to remove any residual genomic DNA. All samples were evaluated using UV spectrophotometry and gel electrophoresis. Total RNA concentration was calculated by determining absorbance at 260 nm (Spectramax Plus 384 spectrophotometer, Molecular Devices, Sunnyvale, CA) in 10 mM Tris-HCl. All samples had A260/A280 ratio greater than 2.0. Low molecular weight (LMW) RNA (~0–200 nucleotides) was purified from total RNA by size fractionation on YM-100 ultrafilteration columns (Millipore) and further purified on RNeasy MinElute columns (Qiagen, Valencia, CA) using a small RNA isolation Protocol. The LMW RNA samples were 3’-end labeled with Alexa-647 fluorescent dye using the Rapid Labeling Kit (Invitrogen, Carlsbad, CA). All RNA samples underwent a quality control check, and were labeled using the same standardized protocols.
Global miRNA expression profiling
Global miRNA profiling was conducted using custom microarrays at Ocean Ridge Biosciences (ORB, Palm Beach Gardens, FL). The microarray, manufactured by Microarrays Inc. (Huntsville, Alabama), consisted of epoxide glass substrates spotted in triplicate with each probe. They contain a total of 1295 probes including 854 probes against human mature microRNA sequences from Sanger 12.0 mirBASE, 379 probes against novel small RNAs from the Invitrogen Ncode Version 3.0 probe set (Invitrogen, Carlsbad, CA) and 59 control probes. Labeled LMW RNA samples were hybridized to the miRNA microarrays according to the Rapid Labeling Kit manual (Invitrogen, Carlsbad, CA). Microarrays were scanned using an Axon Genepix 4000B scanner (Molecular Devices Corp., Sunnyvale, CA), and data was extracted using GenePix V4.1 software (Molecular Devices Corp., Sunnyvale, CA).
Data preprocessing
Spot intensities were obtained for the 3906 features on each microarray by subtracting the median local background from the median local foreground for each spot. Detection thresholds for each array were determined by calculating the mean intensity of the negative control spots and adding 5X the standard deviation of the background (non-spot area). The spot intensities and the threshold (T) were transformed by taking the log (base 2) of each value. The normalization factor (N) for each microarray was determined by obtaining the average spot intensity for novel small RNAs probes from Invitrogen (IVGN-Novel).19 The log2-transformed spot intensities for all features were normalized by subtracting N from each spot intensity, and scaled by adding the grand mean of N across all microarrays. The mean probe intensities for each probe on each of the 40 arrays were then determined by averaging the triplicate spot intensities. Spots flagged as poor quality during data extraction were omitted prior to averaging.
Confirmatory qRT-PCR experiment
A confirmatory qRT-PCR experiment to validate microarray-based measurements was conducted for selected miRNAs. The selection, limited by availability of primers, was based on microarray study findings and previous reports of potential significance in preeclampsia or related pathophysiologic processes. Low molecular weight RNA was reverse-transcribed using miRNA-specific primers (Applied Biosystems, Foster City, CA). The cDNA was amplified by qRT-PCR using universal Taqman mix and miRNA-specific primers according to the manufacturer’s protocol. Reactions were run on an ABI Step1 Plus Real Time PCR machine (Applied Biosystems, Foster City, CA) using the default cycling conditions. All reactions were analyzed by using the 2−ΔΔCT calculation procedure.20 For normalization, we used ΔCT values calculated by subtraction of CT values for expression of the hsa-miRNA-525-5p, a housekeeping miRNA.
Statistical analysis
Analysis was conducted on normalized and log2-transformed data for the set of probes showing signal above T in at least 10% of the samples. Differences between cases and controls were evaluated by 1-way ANOVA using National Institute of Ageing (NIA) Array Analysis software.21 An exploratory Principal Component Analysis (PCA) was performed using the subroutine built in to NIA Array Analysis Software.21 The purpose of the PCA was to reduce multidimensionality of the expression data and identify potential outliers. Absolute fold change greater than 1.5 and p-value less than 0.05 were used to identify differentially expressed miRNA(s). In addition, we recalculated p-values of differentially expressed miRNAs using the ranking based Benjamin and Hochberg false discovery rate correction method.22 Data for the human non-control probes were clustered using Gene Cluster 3.0 software. In the hierarchical clustering, Centered Correlation was used as the similarity metric and Average Linkage as the clustering method.23 TargetScan (www.targetscan.org), a database that employs both conserved and non-conserved seed pairing algorithms, was used to identify putative miRNA targets of differentially expressed miRNA. Functions and functional relationships of genes that are targeted by 2 or more miRNAs were evaluated using KEGG and Ingenuity Pathway Analysis (IPA, Ingenuity, Redwood, CA). In order to identify KEGG pathways enriched with gene targets, we performed the Gene Set Analysis using WEB-based GEne SeT AnaLysis Toolkit (WebGestalt). Statistical significance of pathways were determined on the basis of a p value <0.05 and the presence of at least 2 target genes in the pathway. In IPA, each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes were overlaid onto a global molecular network developed from information contained in the IPKB. Network enrichment was then assessed using a network score (negative log of p-values of Fisher tests).
Finally, we investigated the correlation between microarray based expression measurement fold change differences between cases and controls and similar differences based on the qRT-PCR expression measurements for selected miRNA. The correlation coefficient (R2) of fold changes values was used as an indicator of overall consistency between the two measurements.
Results
Study population characteristics are described in Table 1. Mean maternal age of preeclampsia cases and controls were 32.8 and 30.4 years, respectively. Preeclampsia cases delivered early and were more likely to deliver by cesarean section than controls (both statistically significant, p<0.05). Preeclampsia cases and controls were similar with regards to maternal race/ethnicity, nulliparity and history of labor.
Table 1.
Study population characteristics
| Characteristics | Preeclampsia Cases (N=20) |
Normotensive Controls (N=20) |
|---|---|---|
| Age, yearsa | 32.8 (7.4) | 30.4 (5.6) |
| ≥35 years, % | 50.0 | 40.0 |
| Gestational age, weeksa | 36.0 (0.9) | 38.8 (0.3)b |
| Pre-pregnancy body mass indexa | 26.7 (1.4) | 26.3 (2.0) |
| ≥25 kg/m2, % | 50.0 | 25.0 |
| ≥30 kg/m2, % | 30.0 | 20.0 |
| Race, % | ||
| White | 60.0 | 70.0 |
| Black | 5.0 | 10.0 |
| Hispanic | 15.0 | 15.0 |
| Asian | 5.0 | 5.0 |
| Systolic blood pressure, mmHg | 119.5 (8.1) | 114.8 (7.8) |
| Diastolic blood pressure, mmHg | 76.0 (6.5) | 68.0 (6.5)b |
| Severe preeclampsia, %c | 35 | 0 |
| Mode of delivery, % | ||
| Cesarean section | 55.0 | 35.0b |
| Vaginal | 45.0 | 65.0 |
| Infant birth weight, kgsa | 2.7 (1.0) | 3.2 (0.4)b |
| <2.5 kgs, % | 45.0 | 5.0b |
| Labor, % | 65 | 70 |
| Nulliparous, % | 65 | 60 |
mean (standard deviation)
p-value < 0.05 (Student’s T-test and Chi-square comparisons for continuous and discrete variables, respectively, of cases and controls)
Severe preeclampsia diagnosed by presence of preeclampsia and additional complications (e.g HEELP syndrome, severe fetal growth restriction)
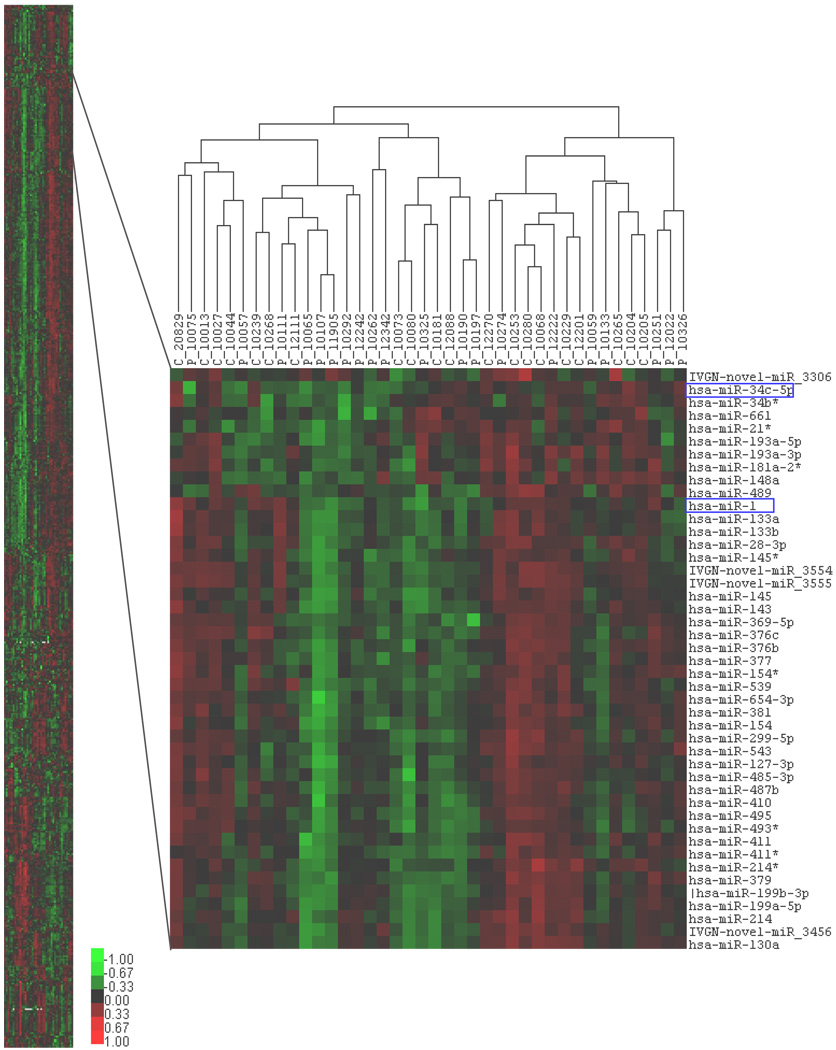
A total of 611 human non-control probes (representing 361 Sanger 12.0 miRNAs and 250 Invitrogen Novel probes) showed hybridization signal above detectable threshold in at least 10% of samples. In PCA, no significant outliers were detected among the samples (not shown). The hierarchical clustering (heat map) of participants and all miRNA expression measurements indicated that preeclampsia cases and controls could not be separated in to distinct groups based solely on miRNA expression patterns (Figure 1).
Figure 1. Hierarchical clustering of participants and microRNA expression.
Clustering (Gene Cluster 3.0) of expression measures using Centered Correlation as the similarity metric and Average Linkage as the clustering method. Left panel: clustering using all 611 human miRNA probes. Right panel: an enlarged view of a representative section of the clustered image. Columns: samples (P=preeclampsia case and C=control). Rows: miRNA. Differentially expressed miRNA are highlighted. Scales: normalized miRNA expression values for each sample with the sum of squares of all expression values for each miRNA equal to 1.
Based on fold change (absolute fold change > 1.5) and ANOVA p-value (< 0.05) evaluations, 8 miRNAs were differentially expressed among preeclampsia cases compared with controls (Table 2 and Figure 2). These included the up regulated miR-210 as well as seven down regulated miRNAs (miR-328, miR-584, miR-139-5p, miR-500, miR-1247, miR-34c-5p and miR-1).
Table 2.
List of differentially expressed microRNAs (miRNAs)
| MicroRNA | Location | Sanger 12.0 mirBASE sequence | P-valuea | Corrected P-valueb |
|---|---|---|---|---|
| hsa-miR-328 | 16q22.1 | CUGGCCCUCUCUGCCCUUCCGU | 0.00033 | 0.00033 |
| hsa-miR-584 | 5q33.1 | UUAUGGUUUGCCUGGGACUGAG | 0.00188 | 0.00189 |
| hsa-miR-210 | 11p15.5 | CUGUGCGUGUGACAGCGGCUGA | 0.00276 | 0.00279 |
| hsa-miR-139-5p | 11q13.4 | UCUACAGUGCACGUGUCUCCAG | 0.00354 | 0.00365 |
| hsa-miR-500 | Xp11.23 | UAAUCCUUGCUACCUGGGUGAGA | 0.00539 | 0.00563 |
| hsa-miR-1247 | 14q32.31 | ACCCGUCCCGUUCGUCCCCGGA | 0.00581 | 0.00609 |
| hsa-miR-34C-5p | 11q23.1 | AGGCAGUGUAGUUAGCUGAUUGC | 0.01059 | 0.01166 |
| hsa-miR-1 | 20q13.33 | UGGAAUGUAAAGAAGUAUGUAU | 0.01284 | 0.01431 |
ANOVA 1-Way p-values comparing expression among preeclampsia cases and controls.
Benjamini and Hochberg false discovery rate corrected p-value.
Figure 2. Fold change comparison* of differentially expressed microRNAs.
Fold change comparison of placental miRNA expression between preeclampsia cases and controls. Positive values (green) represent up regulation in preeclampsia while negative values (red) represent down regulation.
In Targetscan based evaluation of putative targets of these miRNAs, 76 genes were targeted by two or more differentially expressed miRNAs from our list. Of these, most genes (70) were targeted by 2 miRNAs, while 6 genes (AFF2, AZIN1, HNRNPU, NUFIP2, RSBN1L and SFRS1) were targeted by 3 miRNAs. Results of target gene enrichment analysis based on functions and functional relationships (using KEGG and IPA) are shown in Table 3. Target genes of the 8 significant miRNAs were over represented in the colorectal cancer, focal adhesion, adherens junction, and cell adhesion molecules KEGG pathways. In IPA analysis (Table 3 and Figure 3), networks identified in organ system development (cardiovascular system and reproductive system), immune dysfunction, cell cycle and cell signaling were over represented.
Table 3.
Target gene enrichment network analysis results
| Network/pathway | Gene number | Target genes | Enrichment P-value |
|---|---|---|---|
| KEGG a | |||
| Colorectal cancer | 3 | IGF1R, MET, FZD7 | 0.00056 |
| Focal adhesion | 3 | IGF1R, ITGB8, MET | 0.00653 |
| Adherens junction | 2 | IGF1R, MET | 0.00942 |
| Cell adhesion molecules (CAMs) | 2 | NEGR1, ITGB8 | 0.0256 |
| Ingenuity pathway analysis b | |||
| Genetic Disorder, Immunological Disease, Cardiovascular System Development and Function |
16 |
ASH1L, BAZ2A, DHX15, DMD, ELL2, ETS1, GABBR2, HIPK2, IGF1R, IGFBP5, MARCKS (includes EG:4082), MECP2, MET, PDGFA, SMARCA4, THRB |
30 |
| Organ Morphology, Reproductive System Development and Function, Cellular Development |
13 |
C7ORF42, CAPRIN1, CNOT1, DENR (includes EG:8562), GREM2, ITGB8, LARP4, NAB1, PHF15, SPOPL, TCF12, ZIC1, ZRANB2 |
25 |
| Cell Signaling, Cancer, Cell Cycle | 12 |
AFF2, AFF4, AKIRIN2, DDX3X, FOXN3, PTPRD, PTPRS (includes EG:5802), SFRS1, SGIP1, TNPO1, YTHDC1, ZNF148 |
23 |
| Organismal Development, Gene Expression, Cancer |
12 |
ARHGEF3, CUL3, FZD7, GATAD2B, GPR85,MNT, PDIK1L, PRDM16, SOX4, SOX11, ZBTB34, ZFHX4 |
22 |
| Cell Morphology, Cellular Assembly and Organization, Cell Cycle |
9 |
AAK1, AZIN1, GPR158, HNRNPU,NEGR1, NUFIP2, PXMP3, RNF165, WDR40A |
16 |
Putative gene targets from Targetscan (http://www.targetscan.org) that are related to 2 or more differential expressed miRNAs were analyzed using web-based GEne SeT AnaLysis Toolkit (WebGestalt). Significance of the pathways was determined by a p value <0.05 and presence of at least 2 target genes in the pathway.
The networks were generated through the use of Ingenuity Pathways Analysis ngenuity® Systems, www.ingenuity.com). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB) and overlaid onto a global molecular network developed from information contained in the IPKB. Enrichment p-values correspond to network score (negative log of p-values of Fisher test). Genes listed are genes that are targeted by two or more microRNAs that are differentially expressed in preeclamptic placenta in our study.
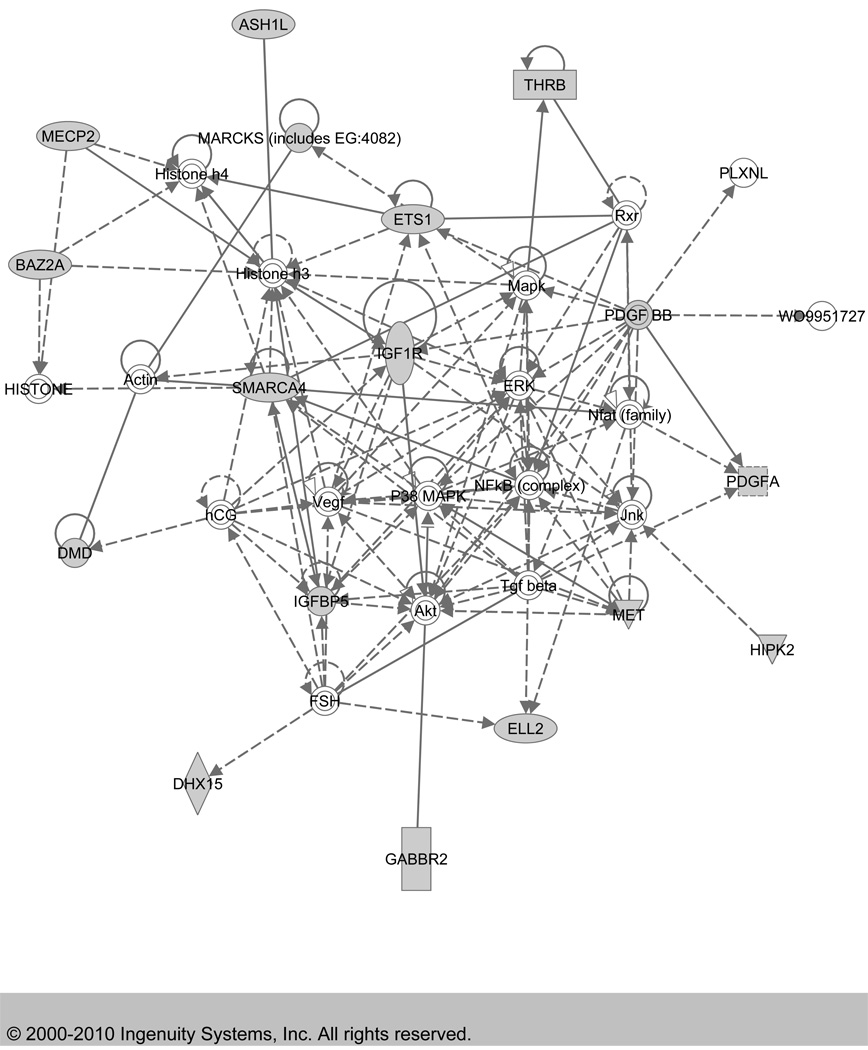
Figure 3. Top Network (Score=30) identified by Ingenuity Pathway Analysis.
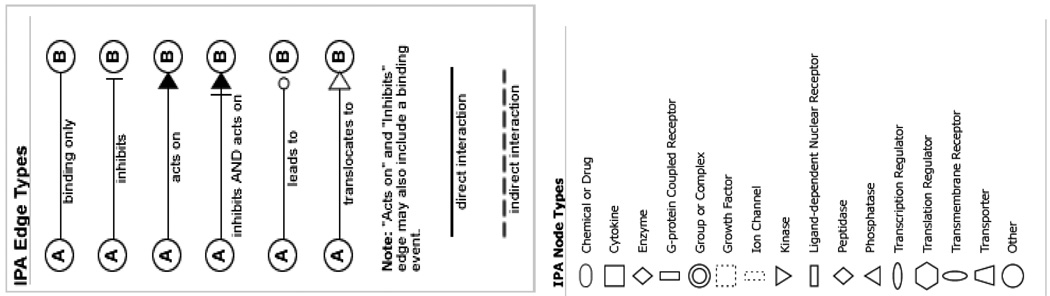
The network was generated through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Identifier of each gene was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB) and overlaid onto a global molecular network developed from information contained in the IPKB. Shaded genes are genes that are targeted by two or more microRNAs that are differentially expressed in preeclamptic placenta in our study. The IPA edge type and node type descriptions illustrate the type of relationship between the genes and their functions.
Results from confirmatory qRT-PCR experiments are shown in Table 4. Expression fold change differences between preeclampsia cases and controls for selected miRNA in qRT-PCR experiments were similar to fold change differences from our microarray experiment (overall correlation coefficient=0.94).
Table 4.
Comparison of microarray and qRT-PCR expression measurements
| MicroRNA | Fold change differences | Overall validation | |
|---|---|---|---|
| Microarray | QRT-PCR | 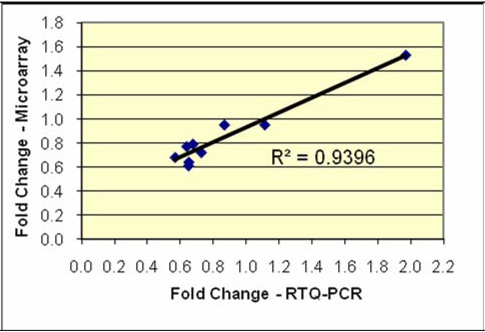 |
|
| hsa-miR-518c* | 0.950 | 0.866 | |
| hsa-miR-1 | 0.640 | 0.649 | |
| hsa-miR-103 | 0.790 | 0.674 | |
| hsa-miR-15a | 0.950 | 1.112 | |
| hsa-miR-584 | 0.610 | 0.647 | |
| hsa-miR-324-5p | 0.720 | 0.725 | |
| hsa-miR-200b | 0.680 | 0.566 | |
| hsa-miR-210 | 1.530 | 1.970 | |
| hsa-miR-154 | 0.770 | 0.636 | |
Comments
In this microarray study, we evaluated differential placental miRNA expression in preeclampsia using 611 probes that represented both characterized and novel miRNAs. We found 8 miRNAs differentially expressed (1 up regulated and 7 down regulated) among preeclampsia cases compared with controls. Differentially expressed miRNAs included those that were previously associated with preeclampsia (miR-210, miR-1 and miRNA clusters in the 14q32.31 chromosomal region) and others that are novel (miR-584 and miR-34c-5p). Target genes of these differentially expressed miRNAs participated in diverse pathophysiological processes including organ/system development (cardiovascular and reproductive system), immunologic dysfunction, cell adhesion, cell cycle and signaling.
Only a few published studies reported investigations of placental miRNA expression in relation to preeclampsia. Pineles et al investigated placental expression of 157 miRNA among women with complicated pregnancies (preeclampsia and small for gestational age) using qRT-PCR.13 Among 2 miRNAs that were reported to be up regulated among preeclampsia cases compared with controls was miR-210, similar to our findings. The other miRNA that was upregulated, miR-182, was not differentially expressed in our study. Zhu et al, in a microarray based study of placental miRNA expression, investigated preeclampsia cases (mild and severe) and controls who had elective cesarean section.14 They reported that 34 miRNAs were differentially expressed (11 over expressed and 23 down regulated) in preeclamptic placenta, notably in several miRNA clusters that include a region around 14q32.31 (a human imprinted region) (miR-411, -377, and -154*). In our study, we found supportive evidence for preeclampsia related down regulation of a miRNA in the 14q32.31 chromosomal region (miR-1247). Zhu et al also reported preeclampsia related up regulation of miR-210 and down regulation of miR-1 similar to our findings. There was no other overlap between their findings and ours. In addition, miR-584 was up regulated in their study while it was down regulated in our study. In a recent report, Hu et al conducted staged (screening microarray and validating qRT-PCR) investigations of placental miRNA expression and risk of severe preeclampsia.15 In their study, 27 miRNAs were differentially expressed (20 up regulated and 7 down regulated) among preeclamptic placenta. There was no overlap of identified miRNAs between their study and ours. There was also no overlap between their list of differentially expressed miRNAs with those of Pineles et al, while two miRNAs (miR-181a and miR-195) from their list were also differentially expressed in the study by Zhu et al.
In our study, we provided evidence supporting previously reported differential expression of miR-210, miR-1 and a miRNA in the 14q32.31chromosomal cluster region. Hypoxia related up regulation of miR-210 plays roles in endothelial cell response to hypoxia, formation of capillary-like structures, vascular endothelial growth factor-driven cell migration, cell differentiation and survival, events that are integral to preeclampsia pathogenesis.24–25 MiR-1 has been shown to influence calcium signaling through negative regulations of the calmodulin coding mRNAs, Mef2a and Gata4 mainly in smooth muscle cells.26 Preeclampsia has been associated with abnormal calcium metabolism and related consequences and miR-1 may influence risk of preeclampsia through its effect on calcium signaling.27 Previous reports have suggested associations of chromosomal regions (including chromosomal rearrangements) with preeclampsia.28 The conserved chromosomal region in 14q32.31 imprinted domain has more than 40 miRNAs that are mostly organized as large clusters.29 These clusters potentially help facilitate the coordinated regulation of functions of individual miRNAs in embryonic and/or placental growth.29 Interestingly, expression (in placenta and other tissues) of these miRNAs are regulated by an intergenic germline-derived differentially methylated region located ~200 kb upstream from the miRNA cluster.29 Further studies aimed at elucidating the role and risk factors of DNA methylation in this region of the genome are warranted.
In addition to previously described candidate miRNAs, we identified a number of novel miRNAs of potential importance in the pathogenesis of preeclampsia including miR-584 and miR-34c-5p. A conserved region complementary to the seed region of miR-584 has been identified within the lactoferrin receptor mRNA-3’-untranslated region.30 Lactoferrin receptors play critical roles in mediating multiple functions of lactoferrin that include immune activation and platelet aggregation, events closely associated with preeclampsia.31–33 Functional studies of miRNAs that belong to the miR-34b and miR-34c family, including miR-34c-5p, have shown that these miRNAs are mediators of p53 dependent suppression of endometrial proliferation.34 This regulation of the cell cycle has been demonstrated in the pathogenesis of endometriosis and can potentially be important in preeclampsia.34–35
Differentially expressed miRNAs have been related to target genes encode proteins that have been implicated in the pathogenesis of preeclampsia such as the insulin-like growth factor protein family.36 Functional relationship analyses of target genes of differentially expressed miRNAs indicate the role of miRNAs in pathways that are known to be affected in preeclampsia including cell adhesion, immune system, organ/system development (cardiovascular and reproductive system), signaling and cell cycle.8, 37–38 Several genes that have been previously demonstrated to play major roles in preeclampsia pathogenesis (such as VEGF and NFKB) are central in these pathways.8
Our study was conducted post diagnosis and study findings may have been influenced by expression changes that are consequences of disease and/or treatment. Future studies that examine early pregnancy miRNA expression profiles in placental samples (obtained during chorionic villi sampling procedures) or peripheral tissue (e.g. whole blood) with risk of preeclampsia could address this limitation. Differences in mode of delivery and presence/absence of labor may result in miRNA expression differences unrelated to preeclampsia. We preferred not to match cases and controls (one-to-one) on mode of delivery to avoid over-representation of non-preeclamptic complicated pregnancies that lead to cesarean deliveries among our controls. In post-hoc sensitivity analyses (Table 5), we confirmed that demonstrated miRNA expression differences between our preeclampsia cases and controls were similar within strata defined by either mode of delivery (C-section/vaginal), presence/absence of labor or parity (nulliparous/multiparous). We did not match on gestational age, a potential confounder, to avoid over representation of complicated pregnancies that result in preterm deliveries among controls. These preterm deliveries may result from pathophysiologic changes (e.g. infection and inflammation) that manifest in expression profiles that are similar to what is expected in preeclampsia.39 Heterogeneity of cases and related differences in pathophysiological processes may have limited the power of our study to identify significant miRNA expression differences between cases and controls. However, in post-hoc analysis, using additional information on standard deviation of expression differences between the two groups, we determined that our study had a >80% power to detect an absolute fold change difference >1.5. Finally, discordance among previous reports of list of differentially expressed miRNAs13–15 may be attributed to false positives/negatives, differences in study populations, variations in the distribution of severity of preeclampsia cases, and experimental methods. In addition, the rapidly evolving identification, characterization and description of miRNAs, in this relatively young area of research, may have contributed to differences in study findings.
Table 5.
Differential expression of microRNAs stratified by type of delivery, history of labor or parity
| MicroRNA | Whole cohort (20/20)a |
Cesarean section (11/7)a |
Vaginal delivery (9/13)a |
No labor (5/7)a |
Labor (15/13)a |
Nulliparous (13/12)a |
Multiparous (7/8)a |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FCb | P-valuec | FCb | P-valuec | FCb | P-valuec | FCb | P-valuec | FCb | P-valuec | FCb | P-valuec | FCb | P-valuec | |
| hsa-miR-328 | 0.66 | <0.000 | 0.64 | 0.007 | 0.69 | 0.022 | 0.61 | 0.032 | 0.70 | 0.004 | 0.73 | 0.019 | 0.54 | 0.002 |
| hsa-miR-584 | 0.61 | 0.003 | 0.73 | 0.082 | 0.56 | 0.048 | 0.82 | 0.248 | 0.56 | 0.011 | 0.67 | 0.040 | 0.51 | 0.048 |
| hsa-miR-210 | 1.53 | 0.004 | 1.65 | 0.020 | 1.31 | 0.198 | 1.73 | 0.014 | 1.42 | 0.081 | 1.30 | 0.137 | 2.09 | 0.008 |
| hsa-miR-139-5p | 0.44 | 0.009 | 0.37 | 0.079 | 0.53 | 0.102 | 0.29 | 0.136 | 0.55 | 0.042 | 0.56 | 0.129 | 0.28 | 0.037 |
| hsa-miR-500 | 0.66 | 0.008 | 0.68 | 0.080 | 0.65 | 0.085 | 0.64 | 0.097 | 0.69 | 0.052 | 0.74 | 0.115 | 0.52 | 0.037 |
| hsa-miR-1247 | 0.66 | 0.008 | 0.63 | 0.054 | 0.63 | 0.041 | 0.53 | 0.075 | 0.71 | 0.056 | 0.75 | 0.159 | 0.50 | 0.127 |
| hsa-miR-34C-5p | 0.65 | 0.016 | 0.63 | 0.115 | 0.65 | 0.117 | 0.62 | 0.193 | 0.68 | 0.086 | 0.66 | 0.046 | 0.64 | 0.184 |
| hsa-miR-1 | 0.64 | 0.020 | 0.44 | 0.006 | 0.94 | 0.801 | 0.36 | 0.005 | 0.83 | 0.406 | 0.78 | 0.241 | 0.45 | 0.041 |
Cases/controls
FC: fold change
Students T-Test p-value comparing expression among preeclampsia cases and controls.
In summary, we have shown that differential placental miRNA expression is associated with preeclampsia. We also identified novel candidate miRNAs (and pathways they regulate) that may be of etiologic relevance in the pathogenesis of preeclampsia. Further investigations on post-transcriptional regulation in preeclampsia as well as experimental studies to evaluate biologic effects of identified miRNAs (including confirmations of miRNA and target gene interactions) are warranted.
Acknowledgments
The authors are indebted to the participants of the Omega and Placenta MicroArray studies for their cooperation. They are also grateful for the technical expertise of staffs of the Center for Perinatal Studies, Swedish Medical Center and Oceanridge Biosciences.
This work was supported by grants from the National Institute of Child Health and Human Development, National Institutes of Health (HD/HL R01-32562); the March of Dimes (#1 FY08-425); and a gift from the Sarah and Chuck Genuardi through the Mourning Dove Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sibai B, Dekker G, Kupferminc M. Pre-eclampsia. Lancet. 2005;365:785–799. doi: 10.1016/S0140-6736(05)17987-2. [DOI] [PubMed] [Google Scholar]
- 2.Redman CW, Sargent IL. Latest advances in understanding preeclampsia. Science. 2005;308:1592–1594. doi: 10.1126/science.1111726. [DOI] [PubMed] [Google Scholar]
- 3.Grill S, Rusterholz C, Zanetti-Dällenbach R, et al. Potential markers of preeclampsia--a review. Reprod Biol Endocrinol. 2009;14:70. doi: 10.1186/1477-7827-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppertz B. Placental origins of preeclampsia: challenging the current hypothesis. Hypertension. 2008;51:970–975. doi: 10.1161/HYPERTENSIONAHA.107.107607. [DOI] [PubMed] [Google Scholar]
- 5.Oudejans CB, van Dijk M. Placental gene expression and pre-eclampsia. Placenta. 2008;29:S78–S82. doi: 10.1016/j.placenta.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Sitras V, Paulssen RH, Grønaas H, et al. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30:424–433. doi: 10.1016/j.placenta.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Founds SA, Conley YP, Lyons-Weiler JF, Jeyabalan A, Hogge WA, Conrad KP. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009;30:15–24. doi: 10.1016/j.placenta.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. Am J Obstet Gynecol. 2008;199:e1–e11. doi: 10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol Genomics. 2008;33:139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
- 11.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 12.Bartell DP. MicroRNAs: genomics biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Pineles BL, Romero R, Montenegro D, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196:e1–e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200:e1–e7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y, Li P, Hao S, Liu L, Zhao J, Hou Y. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin Chem Lab Med. 2009;47:923–929. doi: 10.1515/CCLM.2009.228. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Diao Z, Su L, et al. MicroRNA-155 contributes to preeclampsia by down-regulating CYR61. Am J Obstet Gynecol. 2010;202:e1–e7. doi: 10.1016/j.ajog.2010.01.057. [DOI] [PubMed] [Google Scholar]
- 17.Report of the National High Blood Pressure Education Program Working Group report on high blood pressure in pregnancy. Am J Obstet Gynecol. 2000;183:S1–S22. [PubMed] [Google Scholar]
- 18.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 19.Eickhoff B, Korn B, Schick M, Poustka A, van der Bosch J. Normalization of array hybridization experiments in differential gene expression analysis. Nucleic Acids Res. 1999;27:e33. doi: 10.1093/nar/27.22.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statistic Soci B. 1995;57:289–300. [Google Scholar]
- 23.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soleymanlou N, Jurisica I, Nevo O, et al. Molecular evidence of placental hypoxia in preeclampsia. J Clin Endocrinol Metab. 2005;90:4299–4308. doi: 10.1210/jc.2005-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasanaro P, D'Alessandra Y, Di Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeda S, He A, Kong SW, et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193–2204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thway TM, Shlykov SG, Day MC, et al. Antibodies from preeclamptic patients stimulate increased intracellular Ca2+ mobilization through angiotensin receptor activation. Circulation. 2004;110:1612–1619. doi: 10.1161/01.CIR.0000142855.68398.3A. [DOI] [PubMed] [Google Scholar]
- 28.Vaiman D, Mondon F, Garcès-Duran A, et al. Hypoxia-activated genes from early placenta are elevated in preeclampsia, but not in Intra-Uterine Growth Retardation. BMC Genomics. 2005;6:111. doi: 10.1186/1471-2164-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaillé J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao Y, Lönnerdal B. miR-584 mediates post-transcriptional expression of lactoferrin receptor in Caco-2 cells and in mouse small intestine during the perinatal period. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.07.019. in press. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki YA, Lopez V, Lönnerdal B. Mammalian lactoferrin receptors: structure and function. Cell Mol Life Sci. 2005;62:2560–2575. doi: 10.1007/s00018-005-5371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito S, Shiozaki A, Nakashima A, Sakai M, Sasaki Y. The role of the immune system in preeclampsia. Mol Aspects Med. 2007;28:192–209. doi: 10.1016/j.mam.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Askie LM, Duley L, Henderson-Smart DJ, Stewart LA PARIS Collaborative Group. Antiplatelet agents for prevention of pre-eclampsia: a meta-analysis of individual patient data. Lancet. 2007;369:1791–1798. doi: 10.1016/S0140-6736(07)60712-0. [DOI] [PubMed] [Google Scholar]
- 34.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 35.Burney RO, Hamilton AE, Aghajanova L, et al. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol Hum Reprod. 2009;15:625–631. doi: 10.1093/molehr/gap068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ning Y, Williams MA, Vadachkoria S, Muy-Rivera M, Frederick IO, Luthy DA. Maternal plasma concentrations of insulinlike growth factor-1 and insulinlike growth factor-binding protein-1 in early pregnancy and subsequent risk of preeclampsia. Clin Biochem. 2004;37:968–973. doi: 10.1016/j.clinbiochem.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Ozkan S, Vural B, Filiz S, Coştur P, Dalçik H. Placental expression of insulin-like growth factor-I, fibroblast growth factor-basic, and neural cell adhesion molecule in preeclampsia. J Matern Fetal Neonatal Med. 2008;21:831–838. doi: 10.1080/14767050802251024. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Lu S, Liu C, et al. Expressional and epigenetic alterations of placental matrix metalloproteinase 9 in preeclampsia. Gynecol Endocrinol. 2009 doi: 10.3109/09513590903184100. in press. [DOI] [PubMed] [Google Scholar]
- 39.Urakubo A, Jarskog LF, Lieberman JA, Gilmore JH. Prenatal exposure to maternal infection alters cytokine expression in the placenta, amniotic fluid, and fetal brain. Schizophr Res. 2001;47:27–36. doi: 10.1016/s0920-9964(00)00032-3. [DOI] [PubMed] [Google Scholar]