Abstract
Freshwater planarian flatworms are capable of regenerating complete organisms from tiny fragments of their bodies; the basis for this regenerative prowess is an experimentally accessible stem cell population that is present in the adult planarian. The study of these organisms, classic experimental models for investigating metazoan regeneration, has been revitalized by the application of modern molecular biological approaches. The identification of thousands of unique planarian ESTs, coupled with large-scale whole-mount in situ hybridization screens, and the ability to inhibit planarian gene expression through double-stranded RNA-mediated genetic interference, provide a wealth of tools for studying the molecular mechanisms that regulate tissue regeneration and stem cell biology in these organisms. Here we show that, as in Caenorhabditis elegans, ingestion of bacterially expressed double-stranded RNA can inhibit gene expression in planarians. This inhibition persists throughout the process of regeneration, allowing phenotypes with disrupted regenerative patterning to be identified. These results pave the way for large-scale screens for genes involved in regenerative processes.
As young students, most of us learn about the amazing abilities of certain animals (e.g., urodele amphibians) to regenerate missing body parts. Despite the obvious biomedical ramifications of understanding how such organisms are capable of replacing lost parts, we remain largely in the dark about the mechanisms involved (1). One of the classic models for studying animal regeneration, the freshwater planarian (2), has become the focus of recent efforts to investigate the mechanisms underlying regenerative processes. The introduction of the powerful techniques of molecular biology has reinvigorated interest in these fascinating organisms and provided new tools for studying this fundamental problem of developmental biology (3, 4).
X-irradiation and elegant transplantation experiments showed that stem cells, referred to as neoblasts, serve as the source of new tissue during planarian regeneration (2, 5). These stem cells can be specifically labeled and followed through the process of regeneration and differentiation (6). To initiate the molecular analysis of regeneration and stem cell regulation in planarians, a collection of ≈3,000 unique ESTs has been generated from a clonal line of the planarian Schmidtea mediterranea (7). Whole-mount in situ hybridization techniques in planarians (8) have been adapted to allow large-scale screens for gene expression patterns in the whole organism, providing markers for nearly all of the differentiated cell types (7). Most importantly, double-stranded RNA (dsRNA)-mediated genetic interference (RNAi) (9) can be used to inhibit specifically gene expression in planarians (10), allowing gene function to be dissected in these animals (11-13). For example, RNAi knockdowns of the gene noudarake, which encodes a fibroblast growth factor receptor-related protein, result in ectopic brain formation throughout the planarian (13), providing an initial glimpse into the mechanisms underlying inhibitory signals first suggested by transplantation experiments (2).
With the availability of thousands of planarian ESTs, and the potential for using RNAi to identify genes involved in regenerative processes, we sought to develop a method for delivering dsRNA that would facilitate large-scale screening efforts. Large-scale RNAi-based screens in Caenorhabditis elegans have been expedited by a method devised by Timmons and Fire (14) for using Escherichia coli to express and deliver dsRNA to the nematode via feeding (15). This approach has been used to screen for phenotypes resulting from the inhibition of all ORFs encoded on C. elegans chromosome I (16) and for more recent genomewide screens (17, 18). Here we show that ingestion of bacteria that express dsRNA triggers the planarian RNAi response, leading to specific gene inhibition. This inhibition is long lasting, enabling gene expression to be inhibited throughout the process of regeneration. After amputation of animals fed appropriate targeting constructs, defects in the regenerative process can be observed with specific molecular markers. These results indicate that large-scale RNAi screens are possible in planarians; such screens will facilitate a systematic dissection of the mechanisms that regulate regeneration in these organisms.
Materials and Methods
Planarian Culture. A clonal, asexual strain (7) of S. mediterranea (19) was used for the experiments and maintained as described (6).
Delivery of dsRNA Via Bacterial Feeding. Restriction fragments (≈400-700 bp) from the targeted cDNAs (central marker B2, GenBank accession no. AY068367; central marker B10, GenBank accession no. AY068369; gut marker D14, GenBank accession no. AY066311; photoreceptor marker E30, GenBank accession no. AY066982) were ligated into the double T7 vector pL4440 double T7 script II (15) and transfected into competent DH10B cells. A Gateway (Invitrogen)-compatible L4440 vector was constructed by ligating Gateway Cassette C into SmaI-digested L4440. Clone H.108.3a (GenBank accession no. AY067513) was cloned into the resulting L4440 Gateway vector by PCR amplification and cloning into pDONR221 (Invitrogen), and subsequent transfer by using an LR cloning reaction into L4440 Gateway. The L4440 unc-22 plasmid, pLT61, was provided by Andy Fire (Carnegie Institution of Washington, Baltimore). The resulting recombinant plasmids were introduced into competent (20) HT115(DE3) cells that lack RNase III and can be induced to express T7 polymerase in the presence of isopropyl β-d-thiogalactoside (IPTG) (14, 15). For the experiments shown in Fig. 2, 2-ml cultures in LB/carbenicillin were started with a 1:10 dilution of an overnight culture (grown in LB/carbenicillin plus tetracycline) and incubated for 90 min at 37°C with shaking. The cultures were induced with 0.4 mM IPTG and incubated for an additional 4 h at 37°C. For the experiments using the H.108.3a construct, 4-ml cultures were induced with 1 mM IPTG for 2 h at 37°C. The cultures were then centrifuged and resuspended in 50-100 μl of an artificial food mix consisting of liver homogenate, ultra-low gelling agarose, and food coloring (6). Planarians commenced eating almost immediately on exposure to the food and finished eating within 15-30 min. Preliminary experiments suggested that the most consistent inhibition was observed after three feedings of the L4440 constructs. After the initial feeding, the vast majority of planarians would not eat the artificial food mixture on a daily feeding regimen. Thus, feedings were repeated every other day for a total of three times; in the case of H.108.3a, feedings were repeated every third day. In situ hybridization was used as described below to monitor reduction in transcript levels after ingestion of bacterially expressed dsRNA, in both intact and regenerating animals.
Fig. 2.
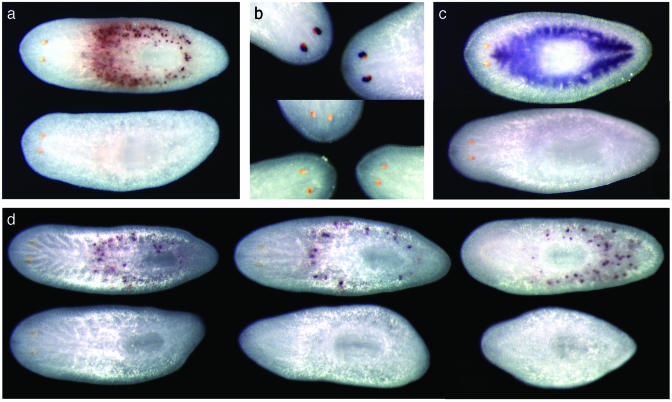
Inhibition of gene expression in planarians by ingestion of bacterially expressed dsRNA. (a) Specific inhibition of a centrally expressed MP. Both animals were fed bacteria expressing a construct targeting an astacin-like MP (B10). One animal (Upper) was hybridized to detect the transcript encoded by another centrally expressed MP (collagenase-like B2; positive signal in 10/10), whereas another animal (Lower) was hybridized to detect B10 (no detectable expression in 32/36; significantly reduced expression in 3/36). When MP B2 was targeted, and animals were fixed and hybridized to detect B2 transcripts, no signal was detected in 39/47 samples; 8/47 showed a strong reduction of transcript (data not shown). (b) Inhibition of photoreceptor expression of arrestin E30. (Upper) The animals were fed bacteria targeting a gut-specific gene; photoreceptor expression of E30 is normal (7/7). (Lower) The animals were fed a construct targeting photoreceptor-specific arrestin (reduction observed in 17/17). (c) Inhibition of expression in the gastrovascular system. Hybridization with gastrovascular-specific probe D14.38. (Upper) The animal was fed a construct targeting a photo-receptor-specific gene (4/4 positive). (Lower) The animal was fed a construct targeting D14.38 (no signal in 44/44 worms). (d) Inhibition persists throughout the regenerative process. (Upper) The animals were fed a control bacterial strain, containing the vector alone. (Lower) The animals were fed bacteria targeting astacin-like MP B10. Three days after feeding, planarians were cut into three parts and allowed to regenerate for 9 days [12 days after the last feeding; animals are regenerated from head fragments (Left), trunk fragments (Center), and tail fragments (Right)]. The control worms reestablished expression of B10 (positive signal in 6/6 treated worms), whereas in the experimental worms B10 expression was inhibited below detectable levels (no detectable expression in 14/14 worms).
Microinjection of dsRNA. dsRNA was prepared from H.108.3a by using the MegaScript in vitro transcription system (Ambion, Austin, TX). Intact planarians were injected three times with 32 nl dsRNA (or H2O for negative controls) by using a Drummond Nanoject II microinjector (Broomall, PA) (10). Injections were repeated for 3 consecutive days, and the worms were amputated, allowed to regenerate, and then fixed and processed for immunofluorescence as described below.
Whole-Mount in Situ Hybridization and Immunofluorescence. To detect differences in transcript levels after dsRNA feeding, intact and regenerating planarians were fixed and processed for whole-mount in situ hybridization as described (7). To monitor photoreceptor axonal projections, planarians were fixed and processed for whole-mount immunostaining as described (10). mAb VC-1 (21), a generous gift of Kiyokazu Agata (RIKEN, Kobe, Japan), was used (1:5,000-1:10,000 dilution) to label the photoreceptor axons; goat anti-mouse Alexa 488 (Molecular Probes) was used at 1:400.
Results
Bacterially Expressed dsRNA Inhibits Gene Expression in Planarians.
Genomewide RNAi-based screens have been conducted in C. elegans by feeding nematodes E. coli that express dsRNA (17, 18). To test the feasibility of using this methodology to inhibit gene expression in planarians, we designed constructs to target several abundantly expressed, cell type-specific genes: two centrally expressed metalloproteinases (MPs), B2 (a collagenase III-like MP) and B10 (an astacin-like MP); a digestive system marker (D14.38); and a photoreceptor-specific arrestin (E30) (7). Unlike C. elegans, planarians do not normally ingest bacteria. Therefore, we resuspended dsRNA-expressing E. coli cells in a planarian artificial food mixture containing homogenized liver, ultra-low gelling temperature agarose, and food coloring (Fig. 1). This artificial food was then fed to the planarians three times over the course of 5 days (see Materials and Methods). Planarians that ingested the bacteria/food mixture were viable, and we observed no perturbation of the regenerative process after feeding of a control bacterial strain containing the vector alone. However, when the bacteria expressing specific dsRNAs were fed to the flatworms, we observed inhibition of the targeted genes (Fig. 2). The inhibition was quite specific: targeting of one centrally expressed MP did not affect transcript levels of the other centrally expressed MP (Fig. 2a), nor were effects observed on gene expression in other cell types, like the gut or photoreceptors. Similar specificity was observed when gut-specific or photoreceptor-specific genes were targeted (Fig. 2 b and c). Thus, the observed inhibition is not caused by a general reduction in transcript levels, but is specific to the targeted RNA.
Fig. 1.
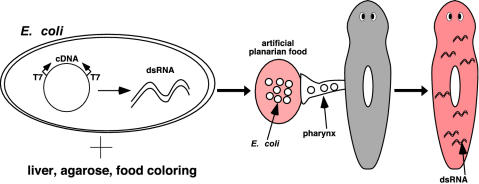
Method for feeding bacteria expressing dsRNA to planarians. dsRNA synthesis is induced in E. coli by using a vector developed for use in C. elegans (15) that contains two T7 RNA polymerase promoters flanking the cDNA sequence of interest. After induction of dsRNA synthesis, bacteria are mixed with homogenized liver (normal planarian food), ultra-low gelling temperature agarose, and red food coloring (for visualization of eating). This mixture solidifies, and animals that ingest this artificial food become red and are exposed to the dsRNA.
Inhibition of gene expression could be observed as early as 1-2 days after the third feeding, and the effects were relatively long lasting, with reduced levels still observed in intact animals 24 days postfeeding (data not shown). Complete regeneration takes between 1 and 2 weeks in this species, depending on the site of amputation. To test whether the inhibitory effect of the ingested bacterially expressed dsRNA would perdure throughout the regenerative process, we amputated planarians after their third feeding, allowed them to regenerate, and then processed them for whole-mount in situ hybridization. As illustrated in Fig. 2d, inhibition of expression of the astacin-like MP B10 persists after amputation and regeneration of new tissue. Similar results were obtained after targeting the other centrally expressed MP (B2) or genes expressed in other cell types (sensory neurons, gastrodermal cells), demonstrating the feasibility of using this method to inhibit genes expressed in many cell types throughout the process of regeneration.
RNAi by Feeding or Injection Can Produce Similar Phenotypes. Because RNAi has been applied only recently to studies of planarian biology, only five RNAi-induced phenotypes have been reported to date, and all of these have used microinjection to deliver the dsRNA (10-13). To address the question of whether ingestion of bacterially expressed dsRNA could also produce phenotypes comparable to those observed after microinjection of dsRNAs, we tested the efficacy of RNAi for inhibiting a planarian gene (H.108.3a) (7) that is predicted to play a role in axon guidance based on its similarity to the netrin receptor encoded by frazzled (22). Whole-mount in situ hybridization analysis of numerous CNS-specific genes in the planarian Dugesia japonica showed that a planarian netrin gene is expressed in the brain in the presumptive visual center to which the photoreceptor axons project (23). The planarian photoreceptors send axonal projections across the midline to the visual center of the brain; these cells can be visualized with mAb VC-1 (Fig. 3 a and e) (21). Given the role of netrin in axonal guidance in other organisms (24-29), it is reasonable to predict that it plays a similar role in guiding the photoreceptor axons to their proper targets in the planarian brain.
Fig. 3.
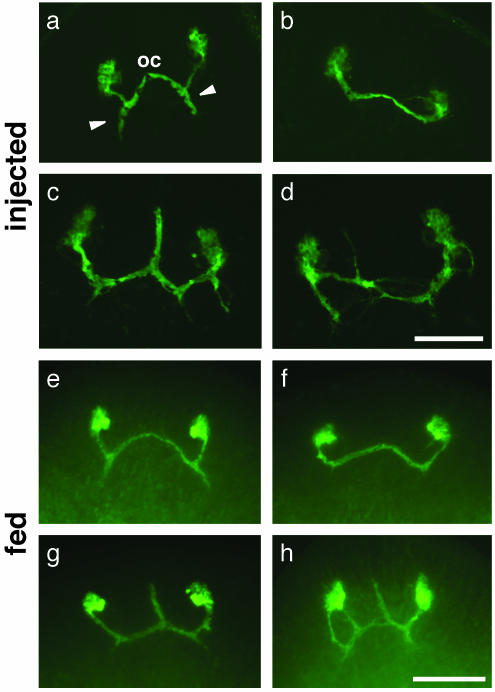
Introduction of H.108.3a dsRNA by injection (a-d) or feeding (e-h) results in similar defects in axonal guidance after regeneration. (a) Control injections; 16/18 regenerated animals display normal axonal trajectories; 2/18 show minor defects distinct from those shown below. (b-d) Phenotypes observed after injection of H.108.3a dsRNA. (b) Regenerated photoreceptor neurons failed to extend axons posteriorly from the optic chiasmata toward the visual center of the brain (8/28 animals). (c) Regenerated photoreceptor neurons had abnormal anterior projections from the optic chiasmata (11/28 animals). (d) Regenerated photoreceptors had ectopic projections or patterning defects (9/28 animals). (e) Control RNAi by feeding; animals were fed bacteria expressing dsRNA from the C. elegans unc-22 gene. After amputation and regeneration, 0/40 animals displayed any abnormalities in axon path-finding. (f-h) Phenotypes observed after feeding of bacterially expressed H.108.3a dsRNA. (f) Regenerated photoreceptor neurons failed to extend axons posteriorly from the optic chiasmata (18/48 animals). (g) Regenerated photoreceptors had abnormal anterior projections from the optic chiasmata (10/48 animals). (h) Regenerated photoreceptors had ectopic projections or patterning defects (4/48). The remainder (16/48) had no observable defects in the pattern of axonal projections (data not shown). oc, optic chiasmata. Arrowheads, posterior axon projections from optic chiasmata to brain. The view is anterior to the top in all frames. (Scale bars: a-d, 100 μm; e-h, 125 μm.)
Planarians injected with H.108.3a dsRNA were amputated and allowed to regenerate; in all cases, these animals regenerated photoreceptor axons that did not target the brain appropriately (Fig. 3 b-d). A range of phenotypes was observed: in approximately one-third of the fragments, the photoreceptor axons appeared to approach or cross the midline but did not extend posteriorly toward the visual center (Fig. 3b); in another one-third, ectopic anterior projections were observed, usually running along the midline (Fig. 3c); in the remaining samples, the axonal projections were highly disorganized, with numerous ectopic projections (Fig. 3d). These results clearly implicate H.108.3a in photoreceptor axonal targeting in planarians.
The dramatic phenotypes observed after injection of H.108.3a dsRNA provide a useful measure by which to compare the effects of dsRNA injections and feeding. Planarians were fed bacterially expressed H.108.3a dsRNA; they were then amputated and allowed to regenerate. In two-thirds of the samples, the regenerated photoreceptor axons were misrouted: the phenotypic classes produced after RNAi by feeding were strikingly similar to those produced after RNAi by injection. Regenerated photoreceptor neurons failed to send projections posteriorly from the optic chiasmata (Fig. 3f), had abnormal optic chiasmata (Fig. 3g), or had additional projections and patterning defects (Fig. 3h). None of these phenotypes was observed in animals fed bacteria expressing control dsRNA from the C. elegans unc-22 gene, a negative control for bacteria expressing dsRNA not expected to affect any S. mediterranea gene (Fig. 3e). Thus, delivery of dsRNA by feeding is capable of generating phenotypes comparable to those observed after microinjection of dsRNA, albeit with lower penetrance.
Discussion
The above results demonstrate that feeding bacterially expressed dsRNA is an effective method for reducing gene expression in planarians. This method is specific, affecting only the gene targeted for disruption, and can be used to target genes expressed in a variety of cell types, including secretory cells, cells of the gastrovascular system, and neurons. The sensitivity of planarian neurons to microinjected dsRNA has already been demonstrated for sensory neurons (10, 11) and cells of the CNS (13). This sensitivity to dsRNA is in marked contrast to C. elegans, in which mature neurons are largely refractory to RNAi, independent of whether the dsRNA is delivered via injection, feeding, or soaking (15).
The inhibitory effect of ingested dsRNA in planarians is relatively long lasting, with inhibition observed over a period of several weeks (up to at least 24 days; we have not yet tested later time points) after feeding. This result is consistent with the 3-week inhibition reported after dsRNA injection in the planarian Girardia tigrina (11). Critically, when planarians that have ingested bacterially expressed dsRNA are amputated and allowed to regenerate, target gene expression in numerous different cell types remains reduced throughout the process of regeneration. As we show by targeting a putative netrin receptor homologue, this inhibition can yield specific regeneration-defective phenotypes similar to those observed after injection of dsRNA. The lower penetrance with which these phenotypes are observed likely reflects the need for further optimization of the feeding protocol.
The results reported here establish RNAi by bacterial feeding as a method for inhibiting gene function in planarians. This feeding methodology will allow much larger RNAi screens to be conducted in planarians than would have been possible by using the more technically demanding and time-consuming microinjection. When combined with molecular markers to visualize specific cell types (7), such large-scale RNAi-based screens will expedite discovery of genes required for the formation and patterning of planarian cells, tissues, and organs and will facilitate dissection of the molecular basis of regeneration in these fascinating organisms.
Acknowledgments
We thank Lisa Timmons and Andy Fire for sharing the bacterial strains and plasmids used in this work, many helpful discussions, and once again making it respectable to feed worms with RNA; Kiyokazu Agata for providing the VC-1 mAb; Sumita Saha for help with the initial feeding experiments; and our colleagues at the Carnegie Institution of Washington's Department of Embryology for their support. This work has been supported by National Institutes of Health Grants RO-1 GM57260 (to A.S.A) and RO-1 HD043403 (to P.A.N.) and National Research Service Award F32-GM19539 (to P.A.N.). P.W.R. is a fellow of the Helen Hay Whitney Foundation; F.C. is a Visiting Fulbright Scholar of the Generalitat of Catalunya; and P.A.N. is a Damon Runyon Scholar supported by the Damon Runyon Cancer Research Foundation (Grant DRS 33-03).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Regenerative Medicine,” held October 18-22, 2002, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: dsRNA, double-stranded RNA; RNAi, RNA-mediated genetic interference; MP, metalloproteinase.
References
- 1.Sánchez Alvarado, A. (2000) BioEssays 22, 578-590. [DOI] [PubMed] [Google Scholar]
- 2.Brøndsted, H. V. (1969) Planarian Regeneration (Pergamon, London).
- 3.Agata, K. & Watanabe, K. (1999) Semin. Cell Dev. Biol. 10, 377-383. [DOI] [PubMed] [Google Scholar]
- 4.Newmark, P. A. & Sánchez Alvarado, A. (2002) Nat. Rev. Genet. 3, 210-219. [DOI] [PubMed] [Google Scholar]
- 5.Baguñà, J., Saló, E. & Auladell, C. (1989) Development (Cambridge, U.K.) 107, 77-86. [Google Scholar]
- 6.Newmark, P. & Sánchez Alvarado, A. (2000) Dev. Biol. 220, 142-153. [DOI] [PubMed] [Google Scholar]
- 7.Sánchez Alvarado, A., Newmark, P. A., Robb, S. & Juste, R. (2002) Development (Cambridge, U.K.) 129, 5659-5665. [DOI] [PubMed] [Google Scholar]
- 8.Umesono, Y., Watanabe, K. & Agata, K. (1997) Dev. Growth Differ. 39, 723-727. [DOI] [PubMed] [Google Scholar]
- 9.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- 10.Sánchez Alvarado, A. & Newmark, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 5049-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pineda, D., Gonzalez, J., Callaerts, P., Ikeo, K., Gehring, W. J. & Salo, E. (2000) Proc. Natl. Acad. Sci. USA 97, 4525-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bueno, D., Fernandez-Rodriguez, J., Cardona, A., Hernandez-Hernandez, V. & Romero, R. (2002) Dev. Biol. 252, 188-201. [DOI] [PubMed] [Google Scholar]
- 13.Cebria, F., Kobayashi, C., Umesono, Y., Nakazawa, M., Mineta, K., Ikeo, K., Gojobori, T., Itoh, M., Taira, M., Sánchez Alvarado, A. & Agata, K. (2002) Nature 419, 620-624. [DOI] [PubMed] [Google Scholar]
- 14.Timmons, L. & Fire, A. (1998) Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- 15.Timmons, L., Court, D. L. & Fire, A. (2001) Gene 263, 103-112. [DOI] [PubMed] [Google Scholar]
- 16.Fraser, A. G., Kamath, R. S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M. & Ahringer, J. (2000) Nature 408, 325-330. [DOI] [PubMed] [Google Scholar]
- 17.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231-237. [DOI] [PubMed] [Google Scholar]
- 18.Ashrafi, K., Chang, F. Y., Watts, J. L., Fraser, A. G., Kamath, R. S., Ahringer, J. & Ruvkun, G. (2003) Nature 421, 268-272. [DOI] [PubMed] [Google Scholar]
- 19.Benazzi, M., Ballester, R., Baguña, J. & Puccinelli, I. (1972) Caryologia 25, 59-68. [Google Scholar]
- 20.Chung, C. T., Niemela, S. L. & Miller, R. H. (1989) Proc. Natl. Acad. Sci. USA 86, 2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agata, K., Soejima, Y., Kato, K., Kobayashi, C., Umesono, Y. & Watanabe, K. (1998) Zool. Sci. 15, 433-440. [DOI] [PubMed] [Google Scholar]
- 22.Kolodziej, P. A., Timpe, L. C., Mitchell, K. J., Fried, S. R., Goodman, C. S., Jan, L. Y. & Jan, Y. N. (1996) Cell 87, 197-204. [DOI] [PubMed] [Google Scholar]
- 23.Cebria, F., Nakazawa, M., Mineta, K., Ikeo, K., Gojobori, T. & Agata, K. (2002) Dev. Growth Differ. 44, 135-146. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, T. E., Serafini, T., de la Torre, J. R. & Tessier-Lavigne, M. (1994) Cell 78, 425-435. [DOI] [PubMed] [Google Scholar]
- 25.Serafini, T., Kennedy, T. E., Galko, M. J., Mirzayan, C., Jessell, T. M. & Tessier-Lavigne, M. (1994) Cell 78, 409-424. [DOI] [PubMed] [Google Scholar]
- 26.Harris, R., Sabatelli, L. M. & Seeger, M. A. (1996) Neuron 17, 217-228. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell, K. J., Doyle, J. L., Serafini, T., Kennedy, T. E., Tessier-Lavigne, M., Goodman, C. S. & Dickson, B. J. (1996) Neuron 17, 203-215. [DOI] [PubMed] [Google Scholar]
- 28.de la Torre, J. R., Hopker, V. H., Ming, G. L., Poo, M. M., Tessier-Lavigne, M., Hemmati-Brivanlou, A. & Holt, C. E. (1997) Neuron 19, 1211-1224. [DOI] [PubMed] [Google Scholar]
- 29.Wadsworth, W. G. (2002) Trends Neurosci. 25, 423-429. [DOI] [PubMed] [Google Scholar]


