Abstract
Several vertebrates display the ability to regenerate parts of their body after amputation. During this process, differentiated cells reenter the cell cycle and proliferate to generate a mass of undifferentiated cells. Repatterning mechanisms act on these cells to eventually shape a regenerated tissue or organ that replaces the amputated one. Experiments with regenerating limbs/fins in newts and zebrafish have shown that members of the Msx family of homeodomain-containing transcription factors play key roles during blastema formation and patterning. Here we show that adult zebrafish have a remarkable capacity to regenerate the heart in a process that involves up-regulation of msxB and msxC genes. We present evidence indicating that heart regeneration involves the execution of a specific genetic program, rather than redeployment of a cardiac development program. Preceding Msx activation, there is a marked increase in the expression of notch1b and deltaC, which we show are also up-regulated during fin regeneration. These data suggest a role for the Notch pathway in the activation of the regenerative response. Taken together, our results underscore the use of zebrafish as a model for investigating the process of regeneration in particular and the biology of stem cells in general. Advances in these fields will undoubtedly aid in the implementation of strategies for regenerative medicine.
Regeneration is a complex biological process by which parts of the body plan are restored after injury or amputation. This process requires the concerted action of mechanisms inducing and regulating dedifferentiation, pattern generation, and, in certain instances, transdifferentiation events. Although embryos from most vertebrates show a remarkable capacity to regenerate damaged structures, this ability plummets as development proceeds, such that adults generally display very limited regenerative capacity. Notable exceptions to this rule are urodele amphibians such as the newt and axolotl, which can regenerate tissues and even entire organs during their lifetime (reviewed in refs. 1 and 2). The regeneration of limbs in these organisms has been particularly well studied (3).
The process of limb regeneration in newts, as well as that of fin regeneration in zebrafish, can be divided into three main phases: (i) wound healing, in which epithelial cells migrate and cover the amputation site; (ii) dedifferentiation of cells in the surrounding tissue to give rise to a proliferating mass of undifferentiated cells known as a blastema; and (iii) redevelopment, in which patterning of the blastema results in the generation of a new limb/fin (4).
The molecular bases that underlie blastema formation and regeneration are not known. In the well studied model of limb regeneration in newts and, more recently, fin regeneration in zebrafish, special attention has been paid to homeodomain-containing transcription factors of the Msx family. Members of this family have distinct roles in patterning limb and fin structures during embryonic development, as well as during regeneration in the adult tissues. Precisely because of these similarities, the phase of blastemal growth and pattern formation during regeneration has been proposed to recapitulate embryonic development to some extent (5, 6), although specific differences have also been reported (7, 8). Several studies have linked the expression of Msx genes to the repression of differentiation during limb or fin development and regeneration (7, 9-11). Furthermore, expression of Msx-1 is sufficient to induce dedifferentiation of mouse myotubes in culture (12).
In addition to limbs, newts also regenerate large portions of the heart after amputation (13, 14). However, the molecular mechanisms regulating this process have not been addressed. Here we provide evidence demonstrating the capacity of zebrafish heart to regenerate. Heart regeneration in zebrafish is accompanied by up-regulation of components of the Notch pathway, followed by members of the Msx family. These genes are not expressed during zebrafish heart development, indicating that regeneration involves the execution of a specific genetic program, rather than redeployment of a developmental program. Finally, we show that components of the Notch pathway are also up-regulated during zebrafish fin regeneration, suggesting that this pathway may play a general role in the activation of regenerative processes.
Materials and Methods
Zebrafish. Wild-type zebrafish (AB line) were maintained at 28.5°C by standard methods (15), unless otherwise indicated. The generation of myosin light-chain 2a (mlc2a)-enhanced GFP (EGFP) transgenic zebrafish will be reported elsewhere. For the CARP-EGFP transgenic line, a DNA construct was prepared by fusing 10 kb of mouse CARP genomic DNA (a generous gift from P. Ruiz-Lozano, University of California at San Diego, La Jolla) to EGFP. The linearized construct was injected into one-cell-stage zebrafish embryos by using a Femtojet microinjector and Micromanipulator 5171 (Eppendorf). F0 founder fish were identified by EGFP expression analysis of the F1 embryos. We identified eight independent transgenic lines of the CARP-EGFP zebrafish. Only adult fish from the F1 generation of line no. 4 were used for heart regeneration experiments.
Cardiac Amputation. Adult fish (>1 yr old) were anesthetized in 0.65 mM Tricaine and secured ventral side up in a slotted sponge. Watchmaker forceps were used to remove the surface scales and penetrate the skin, muscle, and pericardial sac. The ventricle, which is easily visible once the skin has been penetrated, was gently pulled at the apex and cut with iridectomy scissors. Intense bleeding occurred immediately after cutting but quickly stopped due to the rapid formation of a blood clot. It was not necessary to suture the treated fish. After surgery, fish were immediately returned to system water. Typically 20-30% of the ventricle was removed at the apex, and >80% of the fish survived the procedure. For heart amputation during embryonic development, 24-h postfertilization (hpf) mlc2a-EGFP embryos were anesthetized in 0.65 mM Tricaine, secured in agarose wells, and manually dechorionated. A fine tungsten needle was used to tear open the pericardial sac and amputate the posterior half of the atrium. Manipulations were performed under a dissecting microscope equipped with a mercury lamp and EGFP filters. The embryos were allowed to develop at 28.5°C and were processed for in situ hybridization analysis at various time points.
Fin Amputation. Zebrafish 6-9 mo of age were used for caudal fin amputations. Fish were anesthetized in 0.65 mM Tricaine, and amputations were performed by using a razor blade. The distal region, from two to three segments above the first fin ray bifurcation points, was removed. Immediately after amputation, fish were allowed to regenerate for 24, 48, and 72 h in system water at 32°C. The temperature of 32°C facilitates more rapid regeneration than that of 28.5°C (16), commonly used for maintaining fish. At the appropriate time points, fish were anesthetized, and the caudal fin regenerate was removed and processed for in situ hybridization analysis.
In Situ Hybridization. Hearts were fixed in 4% paraformaldehyde overnight at 4°C, washed several times over 5 h in PBS, equilibrated in 30% sucrose in PBS, and frozen for cryosectioning. Fourteen-micrometer sections were prepared through the entire ventricle, and slides were dried at room temperature overnight. In situ hybridizations on the cryosections were performed essentially as described (17). Probes were obtained by RT-PCR and/or screening of zebrafish cDNA libraries. Whole-mount in situ hybridizations on zebrafish embryos and fins were performed essentially as described (18), except that a 30-min proteinase K digestion was used for the fins.
BrdUrd Incorporation. Fish were anesthetized in 0.65 mM Tricaine, and 20 μlofa50 μg/ml solution of BrdUrd (in PBS) was injected into the abdominal cavity once every 24 h for 7 d. In addition, immediately after each injection, fish were incubated in a 50 μg/ml solution of BrdUrd (in system water) for 4 h. After 7 d, hearts were removed and fixed in 4% paraformaldehyde overnight at 4°C, washed several times over 4 h in PBS, equilibrated in 30% sucrose in PBS, and frozen for cryosectioning. Fourteen-micrometer sections were prepared through the entire ventricle. Slides were dried at room temperature overnight. All subsequent procedures were performed at room temperature, unless otherwise indicated. Slides were submerged in -20°C acetone for 10 s, followed by three rinses in PBS plus 0.1% Tween 20 (PBT). Slides were then treated with 4% paraformaldehyde in PBS for 15 min and immersed in PBS for 15 min. Then slides were treated with 10 μg/ml proteinase K (Roche Applied Science) in 10 mM Tris, pH 8.0, for 10 min. After four washes in PBT, slides were treated with 3% H2O2 in PBS for 5 min, followed by three rinses in PBS. Slides were then incubated in 2 M HCl for 30 min, followed by four rinses in PBT. After blocking in PBT-FCS (PBT plus 2% FCS) for 20 min, slides were treated with 10 μg/ml DNase in PBT-FCS for 40 min and rinsed five times in PBT-FCS. Incubation with primary antibody to BrdUrd (mouse; Sigma) at a dilution of 1:50 was performed for 3 h at 37°C in PBT-FCS. After five washes in PBT, slides were incubated for 30 min at 37°C in a 1:200 dilution of anti-mouse secondary antibody conjugated to horseradish peroxidase (goat; Pierce). Slides were washed five times in PBT, stained with diaminobenzidine (Sigma), and counterstained with eosin.
Results and Discussion
Heart Regeneration in Zebrafish. To evaluate the regenerative potential of zebrafish heart, we amputated the ventricular apex of adult fish and analyzed the hearts immediately after manipulation and at different time points, from 1 d to 4 mo. In an initial series of experiments, we used a transgenic zebrafish line expressing the enhanced version of GFP (EGFP) under the control of the mlc2a promoter, to facilitate visualization of cells of the cardiomyocyte lineage. Amputation of 20-30% of the ventricle results in intense but very transient bleeding that stops within 1 min, mainly due to the extremely efficient formation of a blood clot surrounding the open wound (Fig. 1 A and B). In contrast to newts (13), extensive myocardium contraction is not evident at the site of resection (Fig. 1B) and therefore is not likely to play an important role in the initial control of the hemorrhage. Also, in contrast to heart amputation in newts, no circulatory stasis is observed in zebrafish, and rhythmic cardiac beating is maintained after the surgical resection. The initial blood clot organizes into a dense fibrin clot that fills the resected area and seals the cardiac wall as early as 1 d postamputation (dpa; Fig. 1 C and D). This dense fibrin clot further compacts during the first week of recovery (Fig. 1 E and F), after which it is progressively invaded by mlc2a-positive cells (Fig. 1 E, G, I, and K). One month after amputation, the resected myocardium is completely replaced by de novo-regenerated tissue that exhibits beating activity comparable to the surrounding tissue and from that of control hearts (data not shown). The regenerated myocardium also displays histological characteristics indistinguishable from the surrounding tissue and comprises cells that express mlc2a (Fig. 1K) and other late markers of myocardial differentiation (see below). It is interesting to note that ventricular myocardium displays histological characteristics of hypertrophy at 21 and 31 dpa, most likely reflecting compensatory reaction to the hemodynamic overload subsequent to myocardial loss. The regenerative process proceeds, however, achieving a complete repatterning of the myocardium, evident 2 (Fig. 1 M and N) and 4 (Fig. 1 O and P) mo postamputation.
Fig. 1.
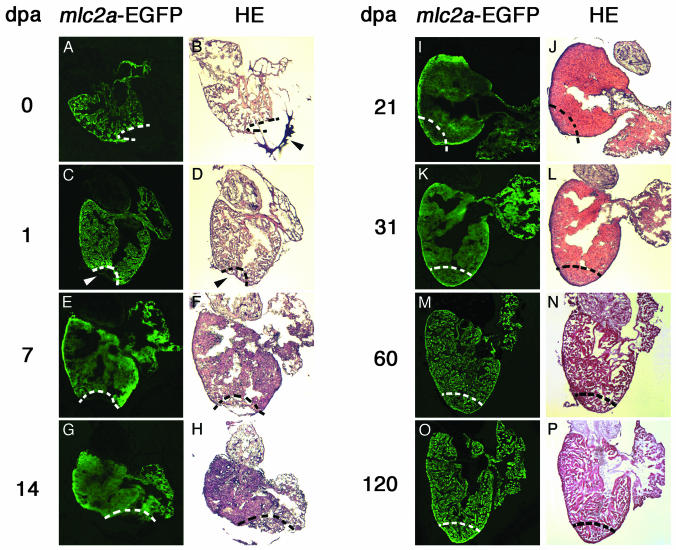
Visualization of heart regeneration in mlc2a-EGFP transgenic zebrafish. Zebrafish heart regeneration was monitored by examining mlc2a-EGFP expression (A, C, E, G, I, K, M, and O) and histological characteristics of hematoxylin/eosin (HE)-stained sections (B, D, F, H, J, L, N, and P) for 0 (A and B),1(C and D),7(E and F), 14 (G and H), 21 (I and J), 31 (K and L), 60 (M and N), and 120 (O and P) dpa. Immediately after amputation, a blood clot forms near the open wound (arrowhead in B). By 1 dpa, this region has organized into a dense fibrin clot (arrowhead in D) devoid of EGFP expression (arrowhead in C). Progressive invasion of the fibrin clot by mlc2a-EGFP-positive cells occurs (E, G, I, and K), such that by 31 dpa, the amputated myocardium is completely replaced by regenerated tissue (K). Shown are sagittal sections through the midventricle. Dotted lines mark the amputation plane in A-H and the estimated amputation plane in I-P, for comparison.
Heart regeneration in newts involves the reentry of differentiated cardiomyocytes into the cell cycle and their subsequent proliferation to repattern the amputated tissue (19). Our finding that heart regeneration in zebrafish is carried out from early time points by mlc2a-positive cells suggests this may also be the case for zebrafish. To address this point, we used BrdUrd to label cells undergoing mitosis during heart regeneration. Although control hearts show a small number of cells that have incorporated BrdUrd, we observed a significant accumulation of BrdUrd-positive cells in the area surrounding the lesion at 7 dpa (Fig. 2 A and B). These cells display morphological characteristics of cardiomyocytes when analyzed at higher magnification (Fig. 2 C and D). These results indicate that adult zebrafish display an extraordinary capacity to regenerate extensive portions of the heart after surgical amputation. During the preparation of this paper, evidence demonstrating the regenerative capacity of zebrafish heart was also published (20).
Fig. 2.
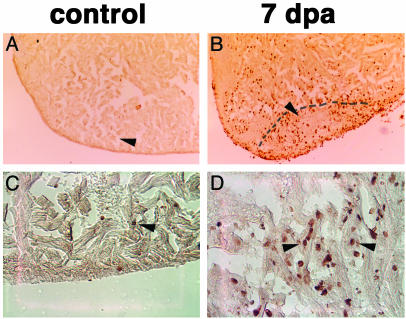
Proliferation of cardiomyocytes is associated with zebrafish heart regeneration. Control (A and C) and amputated (B and D) zebrafish were labeled with BrdUrd for 7 d to detect cells undergoing mitosis during heart regeneration. Although an increase in BrdUrd incorporation occurs throughout the amputated hearts, a significant accumulation is observed near the amputation plane and in the tissue immediately beneath the regenerated epicardium (B). Arrowheads in A and B point to the area examined at higher magnification (×400) in C and D. We could identify BrdUrd-positive cells displaying morphological characteristics of cardiomyocytes (arrowhead in C; arrowheads and neighboring cells in D). Shown are sagittal sections through the midventricle stained for BrdUrd and counterstained with eosin. Dotted line marks the amputation plane.
Heart Regeneration vs. Heart Development. Probably the most complex form of regeneration is that occurring after limb or lens amputation in newts. The formation of the blastema that regenerates these tissues requires successive processes of dedifferentiation, transdifferentiation, and pattern formation (reviewed in refs. 1 and 2). For instance, during limb regeneration, differentiated muscle cells give rise to both muscle and cartilage structures (21). To do so, they dedifferentiate to a point from where they can deploy either a muscle or a cartilage development program. We reasoned that if this were the case for zebrafish heart regeneration, we would be able to detect molecular markers of cardiac differentiation during this process. To address this point, we analyzed the expression pattern of a group of genes known to be activated during heart development.
Nkx2.5 is the earliest known marker of cardiac lineage (22). Expression of this gene, although at low levels, persists in myocardium during adult life (23). In zebrafish, low levels of nkx2.5 expression can be detected in the myocardium of control hearts (Fig. 3A). However, no significant alterations in the levels of nkx2.5 transcripts could be observed at any time point during heart regeneration in zebrafish (Fig. 3 D, G, and J). To confirm these results, we analyzed the levels of nkx2.5 transcripts by RT-PCR. As was the case with the in situ hybridization experiments, no changes were observed in the amount of transcripts amplified from control hearts vs. those allowed to regenerate for 7 days (data not shown). Tbx5 is also expressed very early during heart development (24) and continues to be expressed at low levels in the adult myocardium (25). We detected tbx5 expression in the adult zebrafish myocardium, but no changes were apparent during heart regeneration (data not shown).
Fig. 3.

Markers of early cardiac development are not up-regulated during zebrafish heart regeneration. Low levels of nkx2.5 expression are present throughout the myocardium of adult fish (A). However, no significant alterations in the expression of nkx2.5 are observed in regenerating hearts 7 (D), 14 (G), or 21 (J) dpa. Areas shown in D, G, and J illustrate the boundary between nonamputated myocardium and de novo formed (i.e., regenerated) tissue. Expression of the CARP-EGFP transgene, which is observed in the hearts of 24- (M), 36- (N), and 72- (O) hpf developing embryos but not adult control hearts (B) is also not induced during heart regeneration 7 (E), 14 (H), or 21 (K) dpa. Heart regeneration was confirmed in sections consecutive to those shown in B, E, H, and K by analyzing the expression of ventricular myosin heavy chain (vmhc) in control (C) and at 7 (F), 14 (I), or 21 (L) dpa. Shown are sagittal sections through the midventricle of adult hearts. Dotted lines mark the amputation plane. Sections in B, E, H, and K were overexposed (five times longer than required for the EGFP images in Fig. 1) so the background tissue autofluorescence could be seen.
It is possible that a very transient expression of these genes may not have been uncovered by our in situ hybridization approach. To overcome this caveat, we made use of a transgenic zebrafish line expressing EGFP under the control of the CARP promoter. CARP is a direct target of Nkx2.5 whose expression is limited to heart structures during cardiac development (26). Embryos from this line display intense fluorescence, specifically in the heart tube, readily visible from 24 hpf (Fig. 3 M-O). Importantly, this fluorescence is not present in the adult heart (Fig. 3B). We reasoned that if the cardiac development program were redeployed during heart regeneration, the transgene would be expressed. Even though this expression was very transient, the stability of EGFP would allow us to detect that such an event had occurred. Adult CARP-EGFP transgenic fish were subjected to apical ventricular amputation and allowed to regenerate for 7, 14, or 21 d, after which their hearts were harvested and examined for EGFP expression. We were unable to detect fluorescence by visual inspection of the hearts under a dissecting scope at any time point analyzed (data not shown). Sections of the hearts comprising the regenerating area were thoroughly examined, yielding similar negative results (Fig. 3 E, H, and K). As a control, we analyzed consecutive sections for the presence of ventricular myosin heavy chain (vmhc), which confirmed the extent of ventricular regeneration (Fig. 3 C, F, I, L).
Our inability to detect clear up-regulation of early markers of cardiac development during zebrafish heart regeneration indicates that de novo produced cardiomyocytes are unlikely to be derived from undifferentiated stem cells. Together with the results from mlc2a expression and BrdUrd incorporation experiments, our findings suggest a scenario in which differentiated cardiomyocytes reenter the cell cycle and proliferate in response to heart injury, thus providing the cellular basis for epimorphic regeneration in the zebrafish heart.
Up-Regulation of Msx Transcription Factors During Heart Regeneration. Because several lines of evidence have linked the expression of members of the Msx family of homeodomain-containing transcription factors with repression of differentiation during development and regeneration (7, 9-11, 27), we next asked whether members of this family have a role in zebrafish heart regeneration. At least five Msx genes have been identified in zebrafish (msx A-E), of which only msxB and msxC are expressed in the regenerating fin blastema (7). We used antisense riboprobes representing these two genes to hybridize sections of adult zebrafish hearts and could not detect expression of either transcript (Fig. 4 A and F). However, strong expression is detected in regenerating hearts, starting as early as 3 dpa, in the myocardial areas surrounding the lesion area (Fig. 4 B and G). Both transcripts can be detected in the regenerating myocardium reaching a peak of expression by week two after amputation (Fig. 4 C and H). After this, the expression diminishes (Fig. 4 D and I) such that almost no msxB nor msxC transcripts are detected once heart regeneration is complete, 1 mo after amputation (Fig. 4 E and J).
Fig. 4.
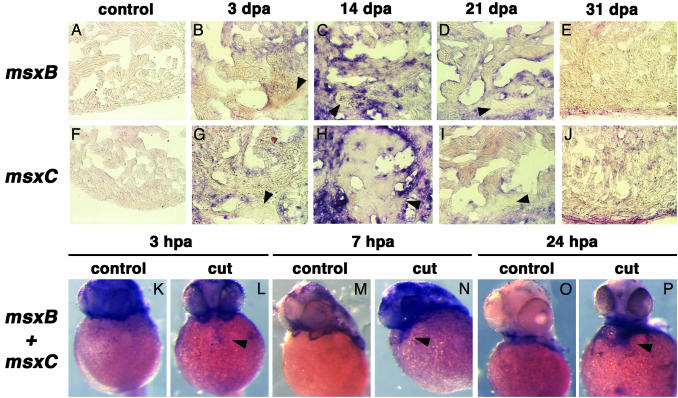
msxC and msxB are expressed in the regenerating zebrafish heart. (A-J) The expression of msxB (A-E) and msxC (F-J) was analyzed in control adult hearts (A and F)and3(B and G), 14 (C and H), 21 (D and I), and 31 (E and J) dpa. At 3 dpa, both genes are up-regulated in myocardial tissue surrounding the lesion area (B and G) and reach a peak of expression by 14 dpa (C and H). Transcripts are no longer detected when heart regeneration is complete at 31 dpa (E and J). Shown are sagittal sections through the midventricle of adult hearts. Areas shown in B-E and G-J illustrate the boundary between nonamputated myocardium and de novo formed (i.e., regenerated) tissue. Arrowheads point to fibrin clot remnants in the lesion area. (K-P) Neither msxB nor msxC is expressed in the hearts of 24- to 48-hpf embryos (K, M, and O). However, both genes are dramatically up-regulated 3 (L), 7 (N), and 24 (P) h after removal of ≈50% of the developing atrium. Embryo views are frontal, anterior to the top. Arrowheads mark gene expression in the damaged embryonic hearts.
For several reasons, that Msx genes are up-regulated during zebrafish heart development is particularly interesting. First, to our knowledge, this is the first time that myocardial regeneration is linked to Msx up-regulation. Second, in contrast to what occurs during limb/fin development, neither msxB nor msxC is expressed during development of the zebrafish heart (Fig. 4 K, M, and O). Therefore, our findings provide additional evidence for a clear distinction between the molecular mechanisms at work during regeneration and development. Finally, these results underscore the notion that Msx transcription factors are general tissue-independent markers of the regenerative response.
Our findings indicate that genes expressed during cardiac development are not up-regulated in regenerating hearts, whereas genes expressed during cardiac regeneration are not expressed in the developing heart. This fact prompted us to study whether regeneration and embryonic development rely on mutually exclusive genetic programs. To address this issue, we attempted to induce regeneration in hearts of developing zebrafish embryos. For this purpose, ≈50% of the prospective atrium was removed from 24-hpf embryos, a stage in which both prospective ventricle and atrium are readily distinguishable within the looping heart tube. Embryos were allowed to develop for an additional 3, 7, or 24 h, and msxB-msxC expression was analyzed by in situ hybridization. A strong up-regulation of Msx transcripts was evident in the remnants of the cardiac tube as early as 3 h postamputation and persisted through the time points analyzed (Fig. 4 L, N, and P). These results clearly indicate that the expression of molecular markers of regeneration is not an exclusive property of adult cells, but that embryonic cardiac cells are also competent to activate the genetic program of regeneration.
Up-Regulation of Notch Pathway Components. As part of our ongoing effort to elucidate the molecular mechanisms of regeneration in zebrafish, we are performing a large-scale in situ hybridization screening of factors specifically showing altered expression during early heart regeneration and have identified a homologue of Drosophila Notch. Although this signaling pathway has not been previously implicated in epimorphic regeneration, up-regulation of Notch pathway components has been reported to occur after injury of teeth (28) and arteries (29). Therefore, we decided to investigate in more detail the possible implication of this pathway in heart regeneration of zebrafish. The sequence of the clone was confirmed to be identical to notch1b, which most likely represents one of the two duplicated zebrafish orthologues of Notch1 (30). We first analyzed the pattern of expression of this gene in adult zebrafish. notch1b is weakly expressed in the ventricle of control hearts, lining the myocardial fibers, in a pattern compatible with endocardial expression (Fig. 5A). This is noteworthy in light of the fact that a role for Notch signaling has been reported in arterial-venous differentiation during zebrafish development (31, 32). We next investigated the expression of notch1b during heart regeneration in zebrafish. notch1b expression is dramatically up-regulated in regenerating hearts very early after amputation. The increased expression of notch1b is evident throughout the endocardium of the affected ventricle as soon as 1 dpa, although up-regulation appears stronger in the area surrounding the lesion (Fig. 5B). notch1b expression continues to be up-regulated during the first week postamputation, after which it declines until reaching control levels around 2 wk postamputation (Fig. 5 C-F).
Fig. 5.
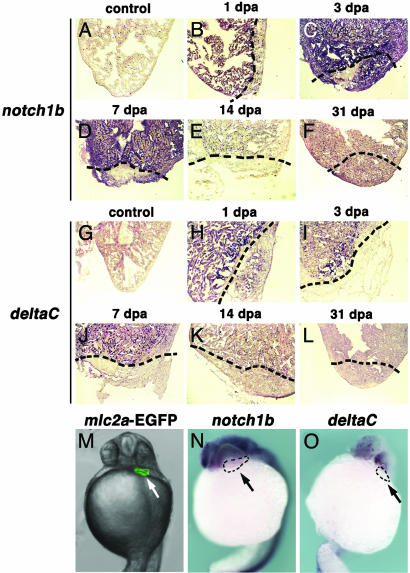
Up-regulation of notch1b and deltaC during zebrafish heart regeneration. (A-L) Expression of notch1b (A-F) and deltaC (G-L) was monitored in control adult hearts (A and G)and1(B and H),3(C and I),7(D and J), 14 (E and K) and 31 (F and L) dpa. Only weak expression of notch1b is detected in the ventricle of control adult hearts (A) but is dramatically up-regulated by 1 dpa (B) and persists until 3 dpa (C). By 14 dpa (C), expression of notch1b returns to control levels (E and F). Similarly, weak expression of deltaC is observed throughout the ventricle of control adult hearts (G), in a pattern similar to notch1b, but is dramatically up-regulated within 1 dpa (H) and returns to control levels after 7 dpa (J-L). Shown are sagittal sections through the midventricle. Dotted lines mark the amputation plane. (M-O) In contrast to regenerating adult hearts, neither notch1b (N) nor deltac (O) is expressed in the developing hearts of 24-hpf embryos. For comparison, M shows the location of the 24-hpf embryonic heart, as monitored by expression of an mlc2a-EGFP transgene. Arrows indicate the location of the embryonic heart (outlined in N and O). Embryo views are frontal, anterior to the top.
We next asked whether any Notch ligand is similarly expressed during heart regeneration. Because deltaC colocalizes with notch1b in the endothelium of arterial vessels in zebrafish (33), we analyzed its expression pattern in normal and regenerating hearts. A weak and widespread expression of deltaC is observed in zebrafish endocardium, in a pattern very similar to the expression of notch1b (Fig. 5G). Also, analogous to the case of notch1b, expression of deltaC is up-regulated very early after heart amputation (Fig. 5H) and starts to decrease after 1 wk postamputation (Fig. 5 I-L). It is interesting to note that neither notch1b (Fig. 5N and data not shown) nor deltaC (Fig. 5O and data not shown) appear to be expressed in the developing heart. This is consistent with our previous findings (Figs. 3 and 4), indicating that heart regeneration and development are carried out by means of different genetic programs.
Our finding that components of the Notch pathway are up-regulated in response to heart amputation may be interpreted as because of an endothelial response to injury (29) or as truly related to the regenerative process. To gain further insight into this mechanism, we analyzed the expression pattern of notch1b and deltaC during zebrafish caudal fin regeneration, a well established model of epimorphic regeneration. Neither notch1b nor deltaC is expressed in nonregenerating fins (data not shown). However, 24 h after fin amputation, obvious expression of both transcripts can be visualized in the early blastema (Fig. 6 A and D), in an area that also expresses msxB and msxC (Fig. 6 G and J). As regeneration proceeds, msxB and msxC expression is restricted to the distal part of the blastema (refs. 7 and 11 and Fig. 6 H, I, K, and L). Similarly, expression of notch1b and deltaC is confined to the distal-most part of the blastema at 48 and 72 h postamputation (Fig. 6 B, C, E, and F). These results indicate that components of the Notch pathway are up-regulated very early during regeneration and further suggest that this signaling pathway may play a role during both heart and fin regeneration.
Fig. 6.
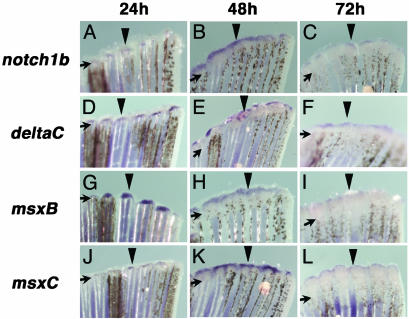
notch1b, deltaC, msxB, and msxC are expressed during caudal fin regeneration. Expression of notch1b (A-C), deltaC (D-F), msxB (G-I), and msxC (J-L) were analyzed after 24 (A, D, G, and J), 48 (B, E, H, and K), or 72 (C, F, I, and L) h postamputation of caudal fins. Both msxB and msxC have been reported to be expressed during fin regeneration (7) and are shown for comparison. notch1b is expressed in the blastema formation stage (24 h postamputation) (A), and the signal becomes prominent in the distal blastema 48 h postamputation (B) and is down-regulated at 72 h postamputation (C). deltaC is expressed in the blastema at 24 h postamputation (D) and is detected in the distal blastema at 48 (E) and 72 (F) h postamputation. Both msxB (G) and msxC (J) are expressed in the blastema at 24 h postamputation. The signals are detected in the distal blastema 48 h postamputation (H and K) and are down-regulated and more distally restricted at 72 h postamputation (I and L). Arrows indicate the level of amputation, and arrowheads point to representative gene expression.
The data reported here provide evidence that adult zebrafish display an extraordinary capacity to regenerate extensive portions of the heart after surgical amputation. Thus, the zebrafish joins urodele amphibians as the only vertebrates described so far that can regenerate their hearts. That zebrafish can also regenerate fins and other structures and their amenability to genetic manipulations make this organism a valuable model to investigate the molecular mechanisms underlying epimorphic regeneration (34). We provide evidence that specific Msx transcription factors are up-regulated during heart regeneration in zebrafish but not during cardiac development. We also demonstrate that, before Msx activation, there is marked up-regulation of notch1b and deltaC expression, suggesting a previously unreported role for the Notch signaling pathway during heart regeneration. Furthermore, the same components of the Notch pathway are also up-regulated during fin regeneration, suggesting that this pathway plays a role in the process of regeneration itself rather than being a heart-specific response to injury. In this respect, Notch activation is at the base of the decision-making event for proliferation/differentiation in a number of resident stem cells, including those of hematopoietic (35), neural (36), gastrointestinal (37), and skeletal muscle (38) lineages. Whether such cells exist in the zebrafish heart and whether they play a role in the regenerative response remain to be elucidated. The involvement of the Notch pathway during regeneration is of biological and biomedical importance and warrants further investigation.
Acknowledgments
We are indebted to May-Fun Schwarz, Harley Pineda, and Reiko Aoki for excellent technical assistance and to Ilir Dubova for expertise with zebrafish. We thank Lorraine Hooks for assistance in preparation of this manuscript. Á.R. is partially supported by a postdoctoral fellowship from the Ministerio de Educación, Cultura y Deporte, Spain. C.M.K. is partially supported by a postdoctoral fellowship from the Canadian Institutes of Health Research. T.I. is supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad. This work was supported by grants from BioCell, the National Institutes of Health, the March of Dimes, and the G. Harold and Leila Y. Mathers Charitable Foundation.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Regenerative Medicine,” held October 18-22, 2002, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: dpa, days postamputation; EGFP, enhanced GFP; hpf, hours postfertilization; mlc2a, myosin light-chain 2a; PBT, PBS plus 0.1% Tween 20.
References
- 1.Tsonis, P. A. (2000) Dev. Biol. 221, 273-284. [DOI] [PubMed] [Google Scholar]
- 2.Brockes, J. P. & Kumar, A. (2002) Nat. Rev. Mol. Cell Biol. 3, 566-574. [DOI] [PubMed] [Google Scholar]
- 3.Brockes, J. P. (1997) Science 276, 81-87. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, S. V., Endo, T. & Gardiner, D. M. (2002) Int. J. Dev. Biol. 46, 887-896. [PubMed] [Google Scholar]
- 5.Gardiner, D. M. & Bryant, S. V. (1996) Int. J. Dev. Biol. 40, 797-805. [PubMed] [Google Scholar]
- 6.Gardiner, D. M., Endo, T. & Bryant, S. V. (2002) Semin. Cell Dev. Biol. 13, 345-352. [DOI] [PubMed] [Google Scholar]
- 7.Akimenko, M. A., Johnson, S. L., Westerfield, M. & Ekker, M. (1995) Development (Cambridge, U.K.) 121, 347-357. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, M. R., Bryant, S. V. & Gardiner, D. M. (1998) J. Exp. Zool. 282, 715-723. [DOI] [PubMed] [Google Scholar]
- 9.Song, K., Wang, Y. & Sassoon, D. (1992) Nature 360, 477-481. [DOI] [PubMed] [Google Scholar]
- 10.Koshiba, K., Kuroiwa, A., Yamamoto, H., Tamura, K. & Ide, H. (1998) J. Exp. Zool. 282, 703-714. [DOI] [PubMed] [Google Scholar]
- 11.Nechiporuk, A. & Keating, M. T. (2002) Development (Cambridge, U.K.) 129, 2607-2617. [DOI] [PubMed] [Google Scholar]
- 12.Odelberg, S. J., Kollhoff, A. & Keating, M. T. (2000) Cell 103, 1099-1109. [DOI] [PubMed] [Google Scholar]
- 13.Becker, R. O., Chapin, S. & Sherry, R. (1974) Nature 248, 145-147. [DOI] [PubMed] [Google Scholar]
- 14.Oberpriller, J. O. & Oberpriller, J. C. (1974) J. Exp. Zool. 187, 249-253. [DOI] [PubMed] [Google Scholar]
- 15.Westerfield, M. (2000) The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) (Univ. of Oregon Press, Eugene).
- 16.Johnson, S. L. & Weston, J. A. (1995) Genetics 141, 1583-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jowett, T. (1997) Tissue in Situ Hybridization: Methods in Animal Development (Wiley, New York).
- 18.Hammerschmidt, M., Pelegri, F., Mullins, M. C., Kane, D. A., van Eeden, F. J., Granato, M., Brand, M., Furutani-Seiki, M., Haffter, P., Heisenberg, C. P., et al. (1996) Development (Cambridge, U.K.) 123, 95-102. [DOI] [PubMed] [Google Scholar]
- 19.Oberpriller, J. O., Oberpriller, J. C., Matz, D. G. & Soonpaa, M. H. (1995) Ann. N.Y. Acad. Sci. 752, 30-46. [DOI] [PubMed] [Google Scholar]
- 20.Poss, K. D., Wilson, L. G. & Keating, M. T. (2002) Science 298, 2188-2190. [DOI] [PubMed] [Google Scholar]
- 21.Lo, D. C., Allen, F. & Brockes, J. P. (1993) Proc. Natl. Acad. Sci. USA 90, 7230-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz, R. J. & Olson, E. N. (1999) Development (Cambridge, U.K.) 126, 4187-4192. [DOI] [PubMed] [Google Scholar]
- 23.Kasahara, H., Wakimoto, H., Liu, M., Maguire, C. T., Converso, K. L., Shioi, T., Huang, W. Y., Manning, W. J., Paul, D., Lawitts, J., et al.(2001) J. Clin. Invest. 108, 189-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begemann, G. & Ingham, P. W. (2000) Mech. Dev. 90, 299-304. [DOI] [PubMed] [Google Scholar]
- 25.Hatcher, C. J., Goldstein, M. M., Mah, C. S., Delia, C. S. & Basson, C. T. (2000) Dev. Dyn. 219, 90-95. [DOI] [PubMed] [Google Scholar]
- 26.Zou, Y., Evans, S., Chen, J., Kuo, H. C., Harvey, R. P. & Chien, K. R. (1997) Development (Cambridge, U.K.) 124, 793-804. [DOI] [PubMed] [Google Scholar]
- 27.Odelberg, S. J. (2002) Semin. Cell Dev. Biol. 13, 335-343. [DOI] [PubMed] [Google Scholar]
- 28.Mitsiadis, T. A., Fried, K. & Goridis, C. (1999) Exp. Cell Res. 246, 312-318. [DOI] [PubMed] [Google Scholar]
- 29.Lindner, V., Booth, C., Prudovsky, I., Small, D., Maciag, T. & Liaw, L. (2001) Am. J. Pathol. 159, 875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kortschak, R. D., Tamme, R. & Lardelli, M. (2001) Dev. Genes Evol. 211, 350-354. [DOI] [PubMed] [Google Scholar]
- 31.Lawson, N. D., Scheer, N., Pham, V. N., Kim, C. H., Chitnis, A. B., Campos-Ortega, J. A., Weinstein, B. M., Villa, N., Walker, L., Lindsell, C. E., et al. (2001) Development (Cambridge, U.K.) 128, 3675-3683. [DOI] [PubMed] [Google Scholar]
- 32.Zhong, T. P., Childs, S., Leu, J. P. & Fishman, M. C. (2001) Nature 414, 216-220. [DOI] [PubMed] [Google Scholar]
- 33.Smithers, L., Haddon, C., Jiang, Y. & Lewis, J. (2000) Mech. Dev. 90, 119-123. [DOI] [PubMed] [Google Scholar]
- 34.Poss, K. D., Keating, M. T. & Nechiporuk, A. (2003) Dev. Dyn. 226, 202-210. [DOI] [PubMed] [Google Scholar]
- 35.Radtke, F., Wilson, A., Ernst, B. & MacDonald, H. R. (2002) Immunol. Rev. 187, 65-74. [DOI] [PubMed] [Google Scholar]
- 36.Gaiano, N. & Fishell, G. (2002) Annu. Rev. Neurosci. 25, 471-490. [DOI] [PubMed] [Google Scholar]
- 37.Brittan, M. & Wright, N. A. (2002) J. Pathol. 197, 492-509. [DOI] [PubMed] [Google Scholar]
- 38.Conboy, I. M. & Rando, T. A. (2002) Dev. Cell 3, 397-409. [DOI] [PubMed] [Google Scholar]


