Abstract
The existence of a postnatal prostate stem cell is supported by several types of evidence. Withdrawal of androgen leads to involution of the gland, but readdition can rapidly stimulate regeneration. Tissue fragments derived from mouse or rat prostatic epithelia from midgestation embryos or adult mice, when combined with tissue fragments from urogenital sinus mesenchyme and grafted under the kidney capsule, can regenerate prostatic structures. Indirect evidence supports that the stem cell population is contained within the basal layer. Purified prostatic stem cell preparations would be useful to define the physical and functional properties required for regeneration and to compare with cells that accumulate during abnormal growth states, like prostate cancer. We have developed a regeneration system using dissociated cell populations of postnatal prostate epithelia and embryonic urogenital sinus mesenchyme. Efficient in vivo regeneration of prostatic structures in the subcapsular space of the kidney was observed within 4-8 wk with as few as 103 epithelial cells from prostates derived from donors 10 d to 6 wk of age. The regenerated structures show a branching tubular epithelial morphology, with expression of a panel of markers consistent with prostate development. Donor epithelial populations can be readily infected with GFP expressing lentiviral vectors to provide integration markers and easy visualization. The cell preparations of urogenital sinus mesenchyme can be expanded in short-term in vitro culture while their inductive capabilities are retained. Further definition of the subpopulation of prostate epithelial cells containing the regeneration activity should be possible with such technologies.
A variety of postnatal stem cells have been defined by cell isolation procedures and reconstitution strategies, including those for hematopoietic, neural, skeletal, and epidermal tissues (1-3). All must share the property of balanced self renewal and differentiation that depends on the particular physiological state of the tissue or organism (4). Some recent studies suggest that stem cells from very different types of tissues may contain a common set of gene transcripts and help define the “stemness” property (5-7).
The nonsenescent phenotype of cancer resembles that of stem cells in many ways. An intriguing hypothesis is that the genetic damage and epigenetic changes involved in producing the cancer phenotype accumulate within a stem cell, and that this cell is essential for the establishment and maintenance of the cancer, even if the majority of cells acquire a more differentiated phenotype as they proliferate (4, 8, 9). Additionally, effective cancer therapies would have to eliminate or regulate this cancer stem cell pool, or growth of the cancer after the end of a therapeutic regime will occur (4). Although prostate cancers in humans generally express markers of mature epithelial cells, like prostate-specific antigen and initial sensitivity to androgen deprivation, the precise cell type in which the cancer originates has been not directly determined but largely inferred from marker protein studies (10, 11). Our long-term goal is to define the subpopulation of cells in the prostate that can function as stem cells in reconstitution experiments and as potential target cells for oncogenic insults.
The prostate is formed through epithelial budding from the urogenital sinus (UGS) around days 17-18 of gestation in the mouse (12, 13). The gland undergoes extensive ductal outgrowth and branching, which continue for several weeks after birth (12). In humans, budding of the prostatic epithelium is seen at ≈10 wk of gestation (14). The structures of the normal human and rodent prostate are distinct. In humans, the mature gland is an acorn-shaped structure lacking distinct lobular organization, which is generally described as having three morphological regions: the peripheral zone, transitional zone, and central area. Benign prostatic hypertrophy occurs mainly within the transitional zone, whereas prostatic cancer is most frequent in the peripheral zone. In mice and rats, the normal prostate consists of four distinct lobular areas named anterior (or coagulating gland), ventral, dorsal, and lateral (10). The dorsal and lateral regions are considered counterparts to the human peripheral zone (15).
Histologically, the tubules of the prostate are comprised of three major cell types with distinct morphologies and functions (10). The predominant cell type is the secretory or luminal cell, which depends on androgen for its differentiated status and viability and is the major producer of prostatic secretory proteins. The basal cell type is located between the secretory cells and the basement membrane. These cells are thought to be the precursor population for the luminal cells and to contain the stem or regenerative cell populations within the prostate (10, 11, 16). In humans, the basal cells form a nearly continuous layer beneath the luminal cells, whereas in rodents, the basal cell layer is usually discontinuous (10). A final cell type is the neuroendocrine component, which is thought to produce paracrine factors that regulate the secretory epithelium. A variety of evidence from immunohistochemical analyses monitoring differential expression of cytokeratins and other markers suggests that transitional cell types between the basal and luminal types can be defined as important intermediates in this developmental sequence (17-19).
Prostatic stem cells in the adult mouse or rat can be defined by their ability to survive acute androgen withdrawal and to differentiate and reform a prostate on androgen add back (18). This cycle can be repeated a remarkable number of times. It is generally assumed that the stem cell population responsible for the regeneration is contained within the basal cells, and they would be regulated by controls similar to those operating during prostate development during gestation (10). Two interesting regulators important in prostate development are NKX 3.1, a homeobox gene, and P63, a transcriptional regulator. Loss-of-function mutants of NKX 3.1 show abnormal prostatic development histologically and enhanced susceptibility to prostatic cancer (20-22), whereas knockouts of P63 cannot develop a prostate (23, 24). P63 is expressed strongly in the basal cell component of human and mouse prostatic tubules (25).
A number of studies have used in vitro culture methods to define subpopulations of prostatic cells with characteristics expected of stem or progenitor cell types, including enhanced clonal expansion, higher replating efficiency, and ability to create 3D structures reminiscent of tubular architecture in matrigel-type cultures (26-28). Selective expression of cytokeratin 5, the antiapoptotic molecule Bcl-2, and α2-integrin was observed in the progenitor-like populations in selected studies (26, 28). Analysis of mouse and human prostate populations for such stem-like cells showed similar frequencies in the 1-2% range in studies from different groups (27, 28).
Work from other systems, including the hair follicle and gastrointestinal tract, strongly supports a model in which stem cells must reside in a specialized niche that provides an essential balance of regulatory cell types and factors to enable the stem cell to match its division and differentiation to the needs of the system (29-31). Coordinated cell division and movement are required to create the population of transient amplifying cells needed to produce mature epithelia (29). Some recent evidence suggests that cells with stem-like characteristics, like long-term retention of BrdUrd label, and higher levels of telomerase components are concentrated at the proximal (closest to the urethra) region of the prostatic tubules, and that after castration and testosterone add back, a fraction of the stem cells will divide and migrate to the more distal ends of the ducts where cell division is most pronounced (32).
A very useful system for the analysis of prostate development is the tissue recombination procedure developed by Cunha and Lung (33). Tissue fragments dissected from midgestation rat or mouse UGS mesenchyme (UGSM) provide a strong inductive influence when combined with tissue fragments from prostatic epithelium of embryonic or postnatal origin and grafted under the kidney capsule of immune-defective mice. This system is capable of extensive proliferation and regeneration of prostatic branching tubular architecture. Appropriate markers, like species-specific antibody, for defining the origin of the epithelium enable analysis of the resulting tissue grafts for the contribution of different tissue components. This system has been particularly useful in defining the necessity for specific gene expression, like the androgen receptor, within the mesenchyme or epithelial compartment, for effective reconstitution (33, 34). It has also been useful in analyzing the effects of specific genes, like loss-of-function mutations of NKX3.1 or the retinoblastoma protein, in altering normal tissue architecture and progression to a cancerous phenotype during regeneration (22, 35).
Our long-term goal is to define in a physical and functional manner the subpopulation of cells in the rodent prostate with the essential characteristics of stem cells. We needed a system in which dispersed cell populations rather than tissue fragments could be analyzed to eventually define surface antigens and other markers to fractionate the population, enrich for cells capable of tissue structure reconstitution, develop appropriate methods for clonally marking individual cells, and genetically manipulate the epithelial and mesenchymal populations.
Materials and Methods
Preparation of Dissociated Prostate Epithelial Cells. Ten-day-old or 6- to 8-wk-old β-actin GFP transgenic mice [C57BL/6-TgN(ACTbEGFP)1Osb, The Jackson Laboratory] were killed by carbon dioxide inhalation. Prostates were dissected, minced into small pieces with a steel blade, and digested with 0.8 mg/ml collagenase (GIBCO, 226 units/mg) in 10 ml of DMEM 10% FBS (GIBCO) at 37°C for 90 min. Cells were passed through 100-μm nylon mesh (Becton Dickinson), washed twice with 20 ml of DMEM 10% FBS, resuspended in 1 ml of DMEM 10% FBS, and counted.
Preparation of Lentivirus and Infection of Prostate Cells. The lentivirus transfer vector FUGW expressing enhanced GFP and packaging vectors VSVg and Δ8.9 were kindly provided by David Baltimore (California Institute of Technology, Pasadena) and Inder Verma (Salk Institute for Biological Sciences, La Jolla, CA). The lentivirus was prepared as described (36, 37). Prostate cells were infected by using the centrifugation method. Basically, 1-2 × 105 prostate cells made as described were suspended in 0.5 ml of DMEM 10% FBS in an Eppendorf tube. A tube of 0.5 ml of frozen lentivirus stock (titer = 3 × 107, titer determined by infecting NIH 3T3 cells with serially diluted virus followed by FACS analysis for GFP expression) was quickly thawed in a 37°C water bath and mixed with the prostate cells; 0.8 mg/ml polybrene (Sigma) was added to the mixture to a final concentration of 8 μg/ml. Cells were infected by centrifuging at 1,500 rpm with a Beckman GS-6R centrifuge (Beckman Coulter) for 3 h at room temperature and washed twice with 1 ml of DMEM 10% FBS. All procedures were performed under University of California, Los Angeles, safety regulations for lentivirus usage.
Prostate Regeneration. Mouse prostate regeneration was adapted from previous reports (38). Seven to eight 16-d-pregnant C57BL/6 mice were killed by carbon dioxide inhalation and embryos collected and put in a 100-mm cell culture dish with 10-15 ml of DMEM 10% FBS. The UGS is located near the caudal part of the embryo and is composed of the rostral, pelvic, and caudal parts, which will develop into the urethra, prostate, and bladder, respectively. Fine forceps were used to blunt dissect away the embryonic UGS. The rostral and caudal parts of UGS tissues and the Wolffian duct were cut off with fine scissors (Fisher) to retrieve only the pelvic part of the UGS, as shown in Fig. 1. Pelvic UGS was placed on a concave plate (Fisher) and digested with ≈250 μl 1% trypsin (Invitrogen) at 4°C for 90 min. Trypsin was removed carefully, and digestion was stopped with 250 μl of DMEM 20% FBS followed by two washes with the same amount of medium. The digested UGS was sticky because of the release of the genomic DNA. It was treated with about 1 mg of DNAseI powder (Roche Diagnostics) briefly, washed twice with 250 μl of DMEM 20% FBS, and resuspended in 250 μl of DMEM 20% FBS. The UGS epithelia (UGSE) were located centrally, whereas the mesenchyme surrounds the epithelia outside. Under the microscope, the UGSE appears more translucent, whereas the UGS mesenchyme (UGSM) is more opaque. The UGSM was dissected by fine needles and then digested with 0.8 mg/ml collagenase (GIBCO, 226 units/mg) in 10 ml of DMEM 10% FBS at 37°C for 90 min. Single cells were passed through 100 μm of nylon mesh (Becton Dickinson), washed twice with 20 ml of DMEM 10% FBS, resuspended in 1 ml of DMEM 10% FBS, and counted. We recovered 1 × 105 UGSM cells from 10 embryos. UGSM was cultured in Bfs media (DMEM/5% FBS/5% Nu-serum/5 μg/ml insulin/10-8 M 5α dihydrotestosterone) (39). UGSM cells were viable by Trypan blue staining and were cultured for up to 2 wk. UGSM (1 × 105) cells and variable numbers of prostate epithelial cells prepared as described above were resuspended in 40-50 μl of 1% type I rat-tail collagen either separately or together and incubated overnight at 37°C/8% CO2 in a six-well plate in Bfs medium. Rat-tail collagen was prepared as described (40) and kept ice cold during all procedures. Collagen was neutralized before use with setting solution (0.34 M NaOH: any 10× medium = 1:1.9, volume ratio). The color of the collagen solution should change from yellow (acidic) to medium red (neutral). The next day, cells were grafted under the kidney capsule of CB.17SCID/SCID mice simultaneously with a s.c. testosterone pellet (12.5 mg of androgen per pellet, 90-d release; Innovative Research of America). All surgical procedures were performed under Division of Laboratory Animal Medicine regulations of the University of California, Los Angeles.
Fig. 1.
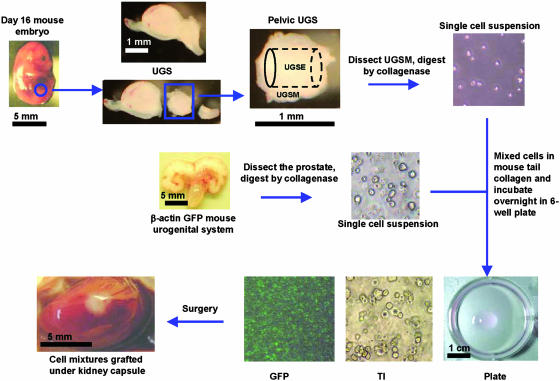
Outline of the prostate regeneration system. Mid-UGS was dissected from day-16 C57BL/6 mouse embryos. Pelvic UGS was collected as described in Materials and Methods and trypsinized for 90 min in 4°C with 1% trypsin (Invitrogen), and UGSM was peeled off from the UGSE under a dissecting microscope. UGSM was digested with 0.8 mg/ml collagenase (GIBCO, 226 units/mg) in 10 ml of DMEM 10% FBS for 90 min at 37°C to produce single-cell populations. Prostate tissues were dissected from β-actin GFP mice, blade minced, and collagenase digested as for UGSM to produce single-cell suspensions. Varying numbers of UGSM cells and prostate cells were mixed in 40-50 μl of 1% rat-tail collagen (40) and incubated in a six-well plate in Bfs media (see Materials and Methods) at 37°C overnight (Plate). Under the transilluminating microscope (TI), the mixed cells were confirmed to be single-cell populations, whereas under the fluorescent microscope, a subportion was green. The next day, the collagen gel was implanted under the kidney capsule of an adult male CB.17SCID/SCID mouse. A testosterone pellet (12.5 mg per pellet, 90-d release, Innovative Research of America) was s.c. implanted at the same time. Refer to Materials and Methods for more details of reagents and procedures. Tissue photos were taken with a Leica MZFLIII dissecting microscope (×10). Bar sizes are indicated in each picture. Cell photos were taken with a Nikon Diaphot phase contrast microscope (×100).
RNA Expression Analysis. Total RNAs from the adult mouse prostate, seminal vesicle, urethra, bladder, liver, and regenerated prostate were extracted with an RNeasy Mini kit (Qiagen, Valencia, CA). RNAs were reverse-transcribed by oligo(dT) primer and SuperScript reverse transcriptase (Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions. The resulting cDNA was subjected to PCR. The length of the expected product and sequences of the primer set used were as follows: actin, 310 bp, 5′-GACTACCTCATGAAGATCCT-3′ and 5′-GCGGATGTCCACGTCACACT-3′; pro-basin, 480 bp, 5′-ATCATCCTTCTGCTCACACTGCATG-3′ and 5′-ACAGTTGTCCGTGTCCATGATACGC-3′; prostate stem cell antigen (PSCA), 400 bp, 5′-TTCTCCTGCTGGCCACCTAC-3′ and 5′-CCAGGTCATCCCTTGAGAAT-3′; NKX3.1, 480 bp, 5′-CCACCA AGTATCCGGCATAG-3′ and 5′-CTACCAGAAAGATGGATGCC-3′; Cdh-1, 520 bp, 5′-ATCCTCGGAATCCTTGGAGG-3′ and 5′-CTAGTCGTCCTCACCACCGC-3′; Sca-1, 390 bp, 5′-ATGGACACTTCTCACACTACAAAG-3′ and 5′-TCAGAGCAAGGTCTGCAGGAGGACTG-3′; P63, 240 bp, 5′-TTGTACCTGGAAAACAATG-3′ and 5′-TCGAAGCTGTGTGGGCCCGGG-3′; androgen receptor, 600 bp, 5′-AGACCTATCGAGGAGCGTTC-3′ and 5′-CTGCTGCCTTCGGAGATTAC-3′; estrogen receptor, α 600 bp, 5′-TCTACGGCCAGTCGGGCATC-3′ and 5′-CTGACGCTTGTGCTTCAACAT-3′; estrogen receptor β, 500 bp, 5′-CCAGACCTCGT TCTGGACAG-3′ and 5′-GCGTGTGAGCATTCAGCATCT-3′; CD44, 400 bp, 5′-GATCGAGACTCATCCAAGGA-3′ and 5′-TTCATGTCCACACTCTGCAG-3′; albumin, 600 bp, 5′-GATGAGCATGCCAAATTAGTGC-3′ and 5′-CCTTGTTGACTTTGGTCAGGTC-3′; and cytokeratin5, 300 bp, 5′-ACAGGAAGCTGCTGGAGGGC-3′ and 5′-GGTGGAGACAAATTTGACACTGG-3′. PCR reactions were performed under the following conditions: 94°C for 5 min; 94°C for 30 sec, 50°C for 30 sec, and 72°C for 40 sec for 40 cycles; and 72°C for 5 min. PCR products were analyzed by DNA electrophoresis in 1% agarose gel in 0.5× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3).
Southern Blot. The methods used for DNA isolation, Southern blotting, and hybridization were as described (41). Genomic DNAs were extracted from NIH/3T3 cells infected with GFP lentivirus and regenerated prostate tissues from GFP lentivirus infected and uninfected prostate epithelial cells. Ten to fifteen micrograms of NotI-digested genomic DNA were separated by 0.8% agarose gel electrophoresis, blotted onto nylon membranes, and hybridized to the [α-32P]dATP-labeled DNA probe. The probe was made by the Prime-It II random labeling kit (Stratagene) by using 750 bp of the GFP gene from the FUGWGFP plasmid as the template.
Immunohistochemistry. CB.17SCID/SCID mice bearing prostate grafts under the kidney capsule were killed 1-3 mo after surgery. Transilluminating or fluorescent pictures were taken with a dissecting microscope. Grafts were fixed in 10% buffered formalin overnight and embedded with paraffin. Sections were stained with hematoxylin/eosin (42). For immunohistochemistry, serial 4-μm-thick sections were deparaffinized, and endogenous peroxidase activity was blocked with 3% H2O2/methanol. Antigen was retrieved by steam heating in 0.01 M citrate buffer (pH 6.0) for 40 min in a commercial vegetable steamer. Non-specific binding was blocked with 5% goat serum in low-salt TBST medium [0.02 M Tris/0.13 M NaCl/0.02% Tween 20 (pH 7.8)], and whole-tissue sections were incubated overnight at 4°C with polyclonal rabbit anti-GFP (Abcam, Cambridge, U.K., 1:300 dilution), polyclonal rabbit antiandrogen receptor (Santa Cruz Biotechnology, 1:200 dilution), polyclonal antiserum anti-secretions of mouse dorsolateral prostate (1:5,000 dilution) (43), or monoclonal hamster anti-PSCA developed in our laboratory, 2 μg/ml (44). Subsequently, sections were processed for immunohistochemistry by using the EnVision+ system (DAKO) or ABC system (DAKO) according to the manufacturer's instructions. For P63 staining, monoclonal mouse antimouse p63 (4A4, Santa Cruz Biotechnology, 1:200 dilution) and an ARK peroxidase kit (DAKO) were used to stain the slides. Specificity of staining was confirmed by replacing the primary antibody with corresponding IgG control. Sections were counterstained with hematoxylin, dehydrated, and mounted.
Results
Single-Cell Populations of UGSM and Postnatal Prostate Epithelial Cells Can Combine to Regenerate Prostatic-Like Structures in Vivo. Self-renewal, clonogenicity, and tissue regeneration are essential characteristics of stem cells. To eventually isolate, quantify, and define the physical nature and biological activities of prostatic stem cells, we sought to develop a reconstitution system with dissociated cell preparations. Using collagenase digestion, we prepared single-cell suspensions from midgestation UGSM to provide the critical paracrine factors needed for induction of prostatic epithelial stem cell growth and development (Fig. 1). To differentially mark the origin of the epithelial stem cells, we used postnatal day-10 prostate tissue harvested from mice transgenic for the β-actin promotor-driven expression of GFP (β-actin GFP) to prepare dissociated cell preparations. Postnatal day 10 is a time of active branching morphogenesis and expansion of the prostate gland in a relatively low androgen environment. The mesenchymal and epithelial cell preparations are combined in a collagen gel overnight before transfer under the kidney capsule of host mice. Grafts are allowed to develop for periods between 1 and 2 mo before harvest and analysis (Fig. 1).
Grafts composed solely of UGSM are small and fibrous in appearance. Microscopically, they show a mixture of stromal cell types without clear epithelial structures (Fig. 2A). If UGSM is combined with cell suspensions prepared from UGSE, they form large translucent grafts, which on microscopic examination show tubular epithelial structures with fluid-filled lumens. Because the UGSE was derived from a nontransgenic mouse, there is no green fluorescence of the graft or detection of GFP by immunohistochemistry (Fig. 2B). If the grafts were prepared with UGSM cell preparations and postnatal day-10 β-actin GFP mouse prostate cell suspensions as a source of epithelial cells, they are translucent in appearance, show bright green fluorescence, and have an epithelial morphology with GFP protein detectable in the luminal epithelial cells of lining the tubules (Fig. 2C).
Fig. 2.
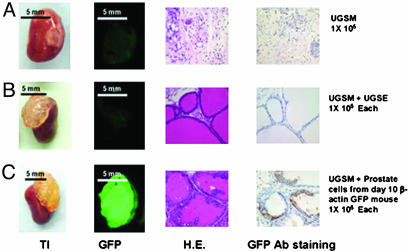
Dissociated prostate epithelial cells combined with UGSM can regenerate the prostatic-like tissue. UGSE (1 × 105) or 1 × 105 10-d-old β-actin GFP mouse prostate epithelial cells were mixed with 1 × 105 UGSM, and the regeneration protocol was carried out as described in Materials and Methods and Fig. 1. Mice were killed 3 mo after the surgery, and regenerated tissues from the kidney capsules were collected. (A) UGSM (1 × 105) alone was used as a negative control. (B) Graft initiated with 1 × 105 UGSM and 1 × 105 UGSE. (C) Graft composed of 1 × 105 UGSM and 1 × 105 dissociated 10-d-old β-actin GFP prostate epithelial cells. (A and B) From left to right: transilluminated image (TI) and green fluorescent image (GFP) of regenerated grafts; hematoxylin/eosin staining (H.E.), and the GFP antibody staining (GFP Ab staining) of regenerated tissue sections. GFP protein was detected by immunohistochemistry with rabbit polyclonal antibody against GFP (Abcam, 1:300 dilution) with the Envision+ system (DAKO). Photos were taken with a Leica MZFLIII dissecting microscope (×10). (Bars = 5 mm.)
Regenerated Tissue Grafts Express a Panel of Prostatic Markers. The branching tubular morphology of the regenerated tissues resembles that expected for prostate but could be representative of other tissues from the urogenital region. We compared a series of markers at the mRNA level by RT-PCR (Fig. 3A) or in some cases by immunohistochemistry (Fig. 3B) to define whether the pattern of expression most closely resembled prostate tissue. The overall pattern was similar for the regenerated prostate compared with seminal vesicle, urethra, and bladder, but in each pairwise comparison at least one or two differences could be found. For example, regenerated prostate expresses androgen receptor, as does normal prostate, urethra, and bladder, but this is not detected in seminal vesicle. The pattern of expressed mRNAs was indistinguishable between normal prostate and regenerated prostate. The most diagnostic marker is NKX3.1, which is seen only in normal prostate and regenerated prostate in this series (Fig. 3A).
Fig. 3.
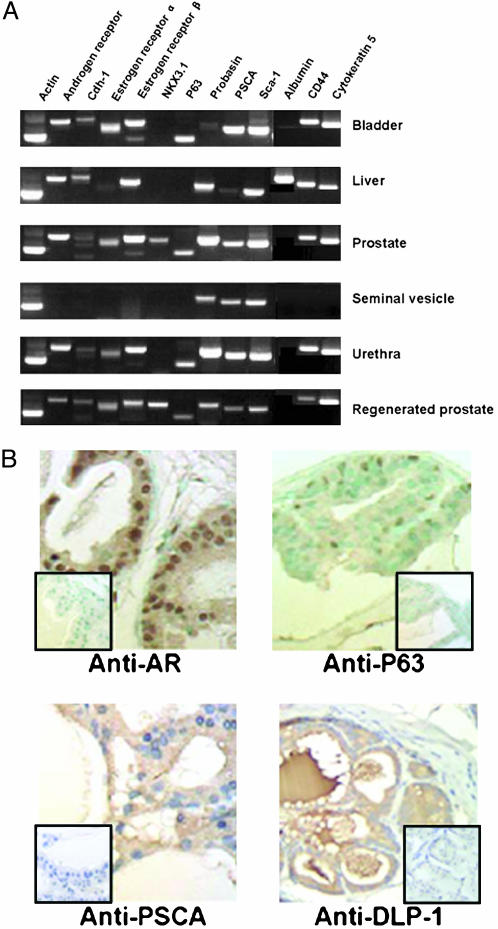
Regenerated prostate has the same gene expression profile as normal prostate. (A) RT-PCR analysis of RNAs from regenerated prostate, 4-wk-old C57/BL6 mouse prostate, bladder, seminal vesicle, urethra, and liver. Gene expression analysis of different tissues for the indicated genes was performed by RT-PCR. RNA preparation, RT-PCR conditions, product length, and primer sequences are described in Materials and Methods. Actin was amplified to test the quality of the RNAs; albumin is a hepatocyte marker protein, and other proteins were previously reported to be expressed in prostate tissue (10, 42, 44). Note that NKX3.1 is detected only in the prostate as well as regenerated prostate. Sca-1, stem cell antigen-1. (B) Immunohistochemistry analysis of the expression of androgen receptor, P63, PSCA, and DLP-1 in regenerated prostate tissue. Tissue sections from regenerated prostate were stained with the polyclonal rabbit anti-androgen receptor (Santa Cruz Biotechnology, 1:200 dilution), polyclonal antiserum anti-secretions of mouse dorsolateral prostate (1:5,000 dilution) (43), or monoclonal hamster anti-PSCA, 2 μg/ml (44). Subsequently, sections were processed for immunohistochemistry by using the EnVision+ system (DAKO) or the ABC system (DAKO). P63 was detected with polyclonal mouse anti-mouse p63 (4A4, Santa Cruz Biotechnology, 1:200 dilution) and an ARK peroxidase kit (DAKO). (Inset) Lack of staining when using a corresponding control isotype IgG. (×400).
Some of these markers were evaluated by immunohistochemistry to analyze tissue and subcellular distribution. The regenerated prostate grafts show strong nuclear staining of the androgen receptor in luminal cells throughout the structure (Fig. 4B). P63-reactive antibodies stain a subpopulation of basal-appearing cells, similar to patterns reported in normal prostates, whereas anti-murine PSCA stains more generally throughout the structure (Fig. 4B). A complex antiserum produced against the secretions of dorsolateral prostate (45) strongly stains the apical cells and the fluid-filled lumen of the reconstituted structures.
Fig. 4.
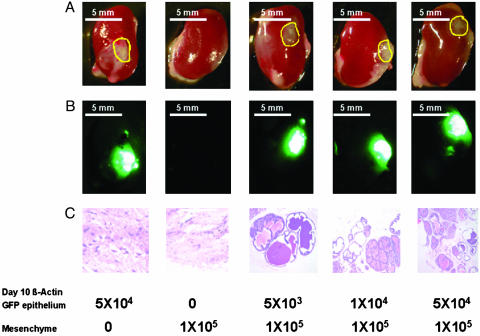
Number of postnatal day 10 epithelial cells required for effective regeneration. UGSM cells (1 × 105) were mixed with 10-d-old β-actin GFP mouse prostate epithelial cells ranging from 5 × 103 to 5 × 104, and the regeneration protocol was carried out as described in Materials and Methods and Fig. 1. Mice were killed 1 mo after surgery, and regenerated tissues from the kidney capsules were collected. UGSM alone and β-actin GFP mouse prostate epithelial cells alone were used as controls. (A) Transillumination. (B) Green fluorescence image of the regenerated tissues under kidney capsules. (C) Hematoxylin/eosin staining of the regenerated tissue sections. Numbers below the panels indicate the cell number of the UGSM and the prostate epithelial cells. Yellow circles in A show the positions of the regenerated tissues. A and B photos were taken with a Leica MZFLIII dissecting microscope (×10). (Bars = 5 mm.) (C) Photo was taken with a Nikon Diaphot phase contrast microscope (×100).
Limiting Dilution Analysis of Prostatic Regeneration Activity in Epithelial Preparations. The eventual goal of defining stem-like cells in the postnatal prostate depends on our ability to subdivide epithelial populations and reconstitute from small numbers of cells. To begin such an analysis, we evaluated dose ranges between 5,000 and 50,000 postnatal day 10 cells harvested from actin-GFP mice and combined with a constant number of UGSM cells (Fig. 4). Even at the lowest dose tested in this experiment, a clear GFP signal and branching epithelial morphology were seen on histological sections.
Because adult rodent prostates can undergo multiple cycles of involution after androgen withdrawal and regeneration on androgen addition, we wanted to evaluate prostate tissue harvested from older animals for this regenerative activity. The larger size of such glands would simplify some technical aspects of the process. We used 6-wk-old animals as donors for the prostate epithelium and UGSM, as in previous experiments. In this case, cell numbers ranging from 1 × 105 to as low as 3 × 103 were evaluated. Even with the lowest number of total prostate cells, there was clear reconstitution of the GFP signal and the histological appearance of branching tubular epithelial structures (Fig. 5 and additional data not shown). At the highest dose of prostate cells used (105) in the absence of UGSM cells, there were small GFP-positive grafts with foci of epithelial structures (Fig. 5).
Fig. 5.
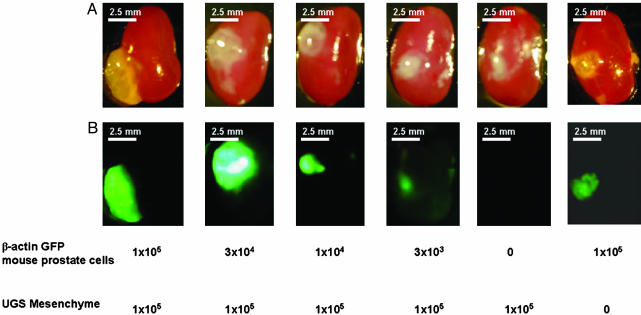
Regeneration by using adult mice prostate epithelial cells. UGSM cells (1 × 105) were mixed with 6-wk-old β-actin GFP mouse prostate epithelial cells ranging from 3 × 103 to 1 × 105, and the regeneration experiment was done as described in Materials and Methods. Grafts composed solely of UGSM or UGSE cells were used as controls. Mice were killed 2 mo after surgery, and regenerated tissues under the kidney capsules were collected. Transillumination (A) and green fluorescence (B) images of the regenerated tissues under the kidney capsule. Numbers below the panels indicate the cell number of UGSM and prostate epithelial cells. Photos were taken with a Leica MZFLIII dissecting microscope (×10). (Bars = 2.5 mm.)
Efficient Marking of Donor Epithelial Populations by Lentiviral Vector Infection. We envision the need to mark the population of donor cells in such a manner that individual clones and their progeny can be identified by visual and molecular techniques. Previous studies of murine hematopoietic stem cells using marking with retroviral or lentiviral vectors with ecotrophic or amphotrophic envelopes have been extremely useful in defining clonal or pauciclonal reconstitution when small numbers of putative stem cells are used (46). We have prepared and tested a lentiviral vector expressing GFP in the prostate reconstitution system. The dissociated epithelial cell preparations are highly susceptible to lentiviral infection and clearly withstand the in vitro culture period and washing required for these procedures, as evidenced by the intense GFP signal and detection of integrated genome (Fig. 6C, lane 3).
Fig. 6.
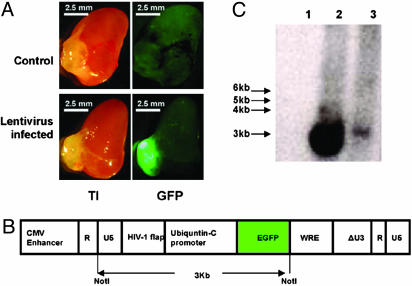
Prostate regeneration with GFP-lentivirus-infected prostate epithelial cells. Six-week-old C57/BL6 mouse prostate epithelial cells were infected by GFP-expressing lentivirus stock as described in Materials and Methods. Six-week-old prostate epithelial cells (1 × 105) infected by GFP-lentivirus or control cells were mixed with 1 × 105 UGSM cells and processed for regeneration as described in Materials and Methods and Fig. 1. Mice were killed 2 mo after surgery, and the regenerated tissues from the kidney capsules were collected. (A) Transillumination (TI, Left) and green fluorescence (GFP, Right) images of regenerated prostate using uninfected prostate epithelial cells (Control, Upper) and regenerated prostate using GFP-lentivirus infected prostate epithelial cells (Lentivirus infected, Lower). Photos were taken with a Leica MZFLIII dissecting microscope (×10). (Bars = 2.5 mm.) (B) Schematic diagram of GFP-lentivirus genomic backbone. The expression of GFP is driven by the ubiquitin-C promoter (36, 37). On NotI digestion, a 3-kb band is released from the lentivirus backbone (diagram adapted from ref. 36). (C) Southern blot analysis of the integration of lentivirus in the regenerated prostate. A 3-kb band can be released on NotI digestion of the genomic DNA. Southern blot was performed as described in Materials and Methods. NIH/3T3 cells infected with GFP-lentivirus were used as positive control. A band of ≈3 kb was detected in the positive control (lane 2, 15 μg of genomic DNA) and the regenerated prostate by using GFP-lentivirus infected prostate epithelial cells (lane 3, 10 μg of genomic DNA) but not in the regenerated prostate by using uninfected epithelial cells (lane 1, 10 μg of genomic DNA).
UGSM Cell Populations Expanded by Short-Term Culture Retain Their Inductive Properties for Prostate Epithelial Cells. The technical demands to harvest and process UGSM are substantial. It requires timed pregnancies, laborious dissections, and processing time for collagenase digestion and cell recovery. Coordination with other tissue-sample processing can also present a significant technical impediment. To improve our ability to coordinate tissue harvests from different animals, we have evaluated the ability of dissociated cell populations of UGSM to survive in culture before their use in reconstitution experiments. We were pleased to see that over a period of 1 wk, such UGSM preparations not only remained viable in culture but also expanded up to 8- to 10-fold in total cell number and could support vigorous engraftment of prostate epithelial cells from 4-wk-old donors (Fig. 7). UGSM cell populations expanded in culture for 2 wk could also function similarly (data not shown). This simple technical step enables a greater number of graft experiments to be accomplished, while somewhat easing the technical demands of coordinating all of the tissue components on a single day.
Fig. 7.
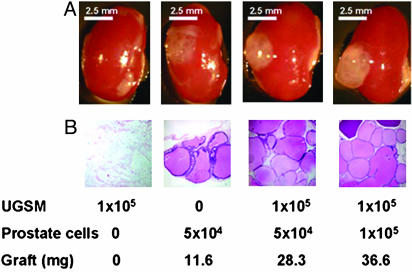
In vitro cultured UGSM cells are able to regenerate prostate. UGSM cells were cultured in Bfs media as described in Materials and Methods for 1 wk. Four-week-old prostate epithelial cells (1 × 105 or 5 × 104) were mixed with 1 × 105 UGSM, and the regeneration protocol was done as described in Materials and Methods. Grafts composed of UGSM only or epithelial cells were used as controls. Mice were killed 2 mo after surgery, and regenerated tissues under the kidney capsules were collected. (A) Transillumination image of regenerated prostate under kidney capsule. (B) Hematoxylin/eosin staining of regenerated tissue sections. Numbers below the panels indicate the number of the UGSM cells and the prostate epithelial cells used and the weight of the grafts. Note that the graft composed of only epithelial cells grew to a small extent compared with the prostate regenerated from UGS and epithelial cells; its weight and size are much smaller, and the number of branching tubular epithelial structures is lower. (A) Photos were taken with a Leica MZFLIII dissecting microscope (×10). (Bars = 2.5 mm.) (B) Photos were taken with a Nikon Diaphot phase contrast microscope (×100).
Discussion
In this work, we have defined the experimental conditions to regenerate prostate tissue from dissociated cell populations of UGS-derived mesenchyme and prostate epithelial populations taken from postnatal animals from 10 d to 6 wk of age. These single-cell suspensions allow for accurate quantitation, uniform introduction of visual markers like GFP by viral vector technologies, and eventually selectable markers or genes, which may influence the growth and developmental phenotype of the regenerated tissue. The regenerated tissues have the branching epithelial morphology and marker gene expression pattern expected for murine prostate tissue.
Further work must be done to define the regenerative activity in this system as a true stem cell. Our current efforts are centered on using the lentiviral system to mark individual cells and assessing the ability of cells to sequentially reconstitute prostate tissue from one graft to another in a clonally marked fashion. Such strategies have been very useful in defining the properties of the hematopoietic stem cell (46-48). One major technical limitation of the system is the labor-intensive harvesting and preparation of UGSM. Our short-term (1- to 2-wk) culture step does help expand the number of cells and hence number of experiments able to be carried out, but currently we are evaluating a variety of continuous cell lines for their ability to support prostate regeneration in this system (49). Certain growth factors, like FGF7 and FGF10, have been shown to be important for the action of the UGSM to induce prostate epithelial development (50, 51). Augmented levels of selected factors may prove useful in improving the efficiency of the regeneration system.
Our long-term goal is to physically isolate and functionally define the stem cell fraction of postnatal prostate. Such cell preparations will be useful for a variety of functional studies with regard to control of differentiation and abnormal patterns of growth in response to defined oncogenic stimuli. Physical parameters of cell shape, metabolic activity, and patterns of surface antigen expression have all been used to fractionate complex tissue mixtures for the isolation of stem cells (2, 52). Several groups have defined cell-surface antigens displayed on prostatic epithelium, and many gene expression studies using microarrays provide additional candidates to use in such separations (5, 28). Liu et al. (53-55) have provided a detailed list of CD antigens present on human prostate epithelial cells. Other groups have shown that specific isoforms of integrins are enhanced in expression on prostate cells with adhesive and growth properties expected of stem-like cells (28). Although the information from studies on normal human prostate epithelium and human cancer is not always transferable to the study of mouse tissues because of differences in expression or reagent availability, there are many candidate surface antigens and reagents to use. The challenge is deciding on the sequence and combination of reagents, because each experimental condition must be tested in a relatively lengthy and cumbersome assay in vivo. One possible strategy is to define a surrogate in vitro colony assay that quantitatively measures the same cell population or its descendants that functions in vivo for tissue regeneration. Limiting dilution colony assays on specific feeder cell lines for hematopoietic stem cells to produce cells of myeloid and lymphoid lineages has been particularly useful in this setting (56). Several groups have reported such short-term colony-like assays for prostate epithelial cells, and these should be evaluated for their ability to enumerate or even expand cells with regenerative activity in vivo (26-28).
One surprising aspect of the present work is the efficiency with which a tissue structure can be regenerated after dissociation of the mesenchyme and epithelium into single-cell preparations. The concept of a specialized cell environment or niche for stem cell residence and function is well established for a variety of tissues. If the number of distinct cell types required to recreate the niche is high, it would be unlikely that the relatively dilute cell suspensions recombined in a collagen gel as used here could provide the cell-cell contact imagined to be essential for this inductive influence. If, however, stem or progenitor elements and the needed UGSM cell types are more limited in number or can function at a distance with soluble factors, the regeneration process may be initiated at lower cell densities with less demand for multiple cell-cell interactions. The process of regeneration observed in these studies may use a distinct or overlapping set of molecular cues compared with those used in the natural process of prostate development or regeneration from castration. However, the method should prove useful to define functional subpopulations of rodent prostate epithelial cells and for their utilization in studies of the progression of normal to neoplastic epithelium, as occurs in the development of prostate cancer.
Acknowledgments
O.N.W. is an investigator and L.X. is an associate of the Howard Hughes Medical Institute. We thank Simon Hayward, Annemarie Donjacour, and Gerry Cunha and members of the Cunha laboratory for introducing us to the surgical techniques essential for this study, the area of prostate regeneration by tissue transplantation, and the polyclonal antiserum anti-secretions of mouse dorsolateral prostate. We thank Matthew Au for help in lentiviral preparations. Portions of this work were supported by funds from the Association for the Cure of Cancer of the Prostate (CaP CURE).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Regenerative Medicine,” held October 18-22, 2002, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: UGS, urogenital sinus; UGSE, UGS epithelia; UGSM, UGS mesenchyme; PSCA, prostate stem cell antigen.
References
- 1.Weissman, I. L., Anderson, D. J. & Gage, F. (2001) Annu. Rev. Cell Dev. Biol. 17, 387-403. [DOI] [PubMed] [Google Scholar]
- 2.Morrison, S. J., Hemmati, H. D., Wandycz, A. M. & Weissman, I. L. (1995) Proc. Natl. Acad. Sci. USA 92, 10302-10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson, K. A., Majka, S. M., Wulf, G. G. & Goodell, M. A. (2002) J. Cell Biochem. 38, Suppl., 1-6. [DOI] [PubMed] [Google Scholar]
- 4.Reya, T., Morrison, S. J., Clarke, M. F. & Weissman, I. L. (2001) Nature 414, 105-111. [DOI] [PubMed] [Google Scholar]
- 5.Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. (2002) Science 298, 597-600. [DOI] [PubMed] [Google Scholar]
- 6.Burns, C. E. & Zon, L. I. (2002) Dev. Cell 3, 612-613. [DOI] [PubMed] [Google Scholar]
- 7.Stappenbeck, T. S., Mills, J. C. & Gordon, J. I. (2003) Proc. Natl. Acad. Sci. USA 100, 1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu, S. M., Lin, S. H. & Logothetis, C. J. (2002) Lancet Oncol. 3, 508-513. [DOI] [PubMed] [Google Scholar]
- 9.Taipale, J. & Beachy, P. A. (2001) Nature 411, 349-354. [DOI] [PubMed] [Google Scholar]
- 10.Abate-Shen, C. & Shen, M. M. (2000) Genes Dev. 14, 2410-2434. [DOI] [PubMed] [Google Scholar]
- 11.Foster, C. S., Dodson, A., Karavana, V., Smith, P. H. & Ke, Y. (2002) J. Pathol. 197, 551-565. [DOI] [PubMed] [Google Scholar]
- 12.Sugimura, Y., Cunha, G. R. & Donjacour, A. A. (1986) Biol. Reprod. 34, 961-971. [DOI] [PubMed] [Google Scholar]
- 13.Sciavolino, P. J., Abrams, E. W., Yang, L., Austenberg, L. P., Shen, M. M. & Abate-Shen, C. (1997) Dev. Dyn. 209, 127-138. [DOI] [PubMed] [Google Scholar]
- 14.Kellokumpu-Lehtinen, P., Santti, R. & Pelliniemi, L. J. (1980) Anat. Rec. 196, 263-273. [DOI] [PubMed] [Google Scholar]
- 15.Abate-Shen, C. & Shen, M. M. (2002) Trends Genet. 18, S1-S5. [DOI] [PubMed] [Google Scholar]
- 16.Robinson, E. J., Neal, D. E. & Collins, A. T. (1998) Prostate 37, 149-160. [DOI] [PubMed] [Google Scholar]
- 17.Wang, Y., Hayward, S., Cao, M., Thayer, K. & Cunha, G. (2001) Differentiation (Berlin) 68, 270-279. [DOI] [PubMed] [Google Scholar]
- 18.Isaacs, J. T. (1985) Prog. Clin. Biol. Res. 185A, 383-405. [PubMed] [Google Scholar]
- 19.Tran, C. P., Lin, C., Yamashiro, J. & Reiter, R. E. (2002) Mol. Cancer Res. 1, 113-121. [PubMed] [Google Scholar]
- 20.Bhatia-Gaur, R., Donjacour, A. A., Sciavolino, P. J., Kim, M., Desai, N., Young, P., Norton, C. R., Gridley, T., Cardiff, R. D., Cunha, G. R., et al. (1999) Genes Dev. 13, 966-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowen, C., Bubendorf, L., Voeller, H. J., Slack, R., Willi, N., Sauter, G., Gasser, T. C., Koivisto, P., Lack, E. E., Kononen, J., et al. (2000) Cancer Res. 60, 6111-6115. [PubMed] [Google Scholar]
- 22.Kim, M. J., Bhatia-Gaur, R., Banach-Petrosky, W. A., Desai, N., Wang, Y., Hayward, S. W., Cunha, G. R., Cardiff, R. D., Shen, M. M. & Abate-Shen, C. (2002) Cancer Res. 62, 2999-3004. [PubMed] [Google Scholar]
- 23.Mills, A. A., Zheng, B., Wang, X. J., Vogel, H., Roop, D. R. & Bradley, A. (1999) Nature 398, 708-713. [DOI] [PubMed] [Google Scholar]
- 24.Yang, A., Schweitzer, R., Sun, D., Kaghad, M., Walker, N., Bronson, R. T., Tabin, C., Sharpe, A., Caput, D., Crum, C., et al. (1999) Nature 398, 714-718. [DOI] [PubMed] [Google Scholar]
- 25.Signoretti, S., Waltregny, D., Dilks, J., Isaac, B., Lin, D., Garraway, L., Yang, A., Montironi, R., McKeon, F. & Loda, M. (2000) Am. J. Pathol. 157, 1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawicki, J. A. & Rothman, C. J. (2002) Prostate 50, 46-53. [DOI] [PubMed] [Google Scholar]
- 27.Hudson, D. L., O'Hare, M., Watt, F. M. & Masters, J. R. (2000) Lab. Invest. 80, 1243-1250. [DOI] [PubMed] [Google Scholar]
- 28.Collins, A. T., Habib, F. K., Maitland, N. J. & Neal, D. E. (2001) J. Cell Sci. 114, 3865-3872. [DOI] [PubMed] [Google Scholar]
- 29.Spradling, A., Drummond-Barbosa, D. & Kai, T. (2001) Nature 414, 98-104. [DOI] [PubMed] [Google Scholar]
- 30.Oshima, H., Rochat, A., Kedzia, C., Kobayashi, K. & Barrandon, Y. (2001) Cell 104, 233-245. [DOI] [PubMed] [Google Scholar]
- 31.Bjerknes, M. & Cheng, H. (1999) Gastroenterology 116, 7-14. [DOI] [PubMed] [Google Scholar]
- 32.Tsujimura, A., Koikawa, Y., Salm, S., Takao, T., Coetzee, S., Moscatelli, D., Shapiro, E., Lepor, H., Sun, T. T. & Wilson, E. L. (2002) J. Cell Biol. 157, 1257-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunha, G. R. & Lung, B. (1978) J. Exp. Zool. 205, 181-193. [DOI] [PubMed] [Google Scholar]
- 34.Chung, L. W., Anderson, N. G., Neubauer, B. L., Cunha, G. R., Thompson, T. C. & Rocco, A. K. (1981) Prog. Clin. Biol. Res. 75A, 177-203. [PubMed] [Google Scholar]
- 35.Wang, Y., Hayward, S. W., Donjacour, A. A., Young, P., Jacks, T., Sage, J., Dahiya, R., Cardiff, R. D., Day, M. L. & Cunha, G. R. (2000) Cancer Res. 60, 6008-6017. [PubMed] [Google Scholar]
- 36.Lois, C., Hong, E. J., Pease, S., Brown, E. J. & Baltimore, D. (2002) Science 295, 868-872. [DOI] [PubMed] [Google Scholar]
- 37.Pfeifer, A., Ikawa, M., Dayn, Y. & Verma, I. M. (2002) Proc. Natl. Acad. Sci. USA 99, 2140-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, T. C., Southgate, J., Kitchener, G. & Land, H. (1989) Cell 56, 917-930. [DOI] [PubMed] [Google Scholar]
- 39.Rowley, D. R. (1992) In Vitro Cell Dev. Biol. 28A, 29-38. [DOI] [PubMed] [Google Scholar]
- 40.Hallowes, R. C., Bone, E. J. & Jones, W. A. (1980) A New Dimension in the Culture of Human Breast (Pergamon, Oxford).
- 41.Le, L. Q., Kabarowski, J. H., Weng, Z., Satterthwaite, A. B., Harvill, E. T., Jensen, E. R., Miller, J. F. & Witte, O. N. (2001) Immunity 14, 561-571. [DOI] [PubMed] [Google Scholar]
- 42.Ide, H., Seligson, D. B., Memarzadeh, S., Xin, L., Horvath, S., Dubey, P., Flick, M. B., Kacinski, B. M., Palotie, A. & Witte, O. N. (2002) Proc. Natl. Acad. Sci. USA 99, 14404-14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donjacour, A. A., Rosales, A., Higgins, S. J. & Cunha, G. R. (1990) Endocrinology 126, 1343-1354. [DOI] [PubMed] [Google Scholar]
- 44.Dubey, P., Wu, H., Reiter, R. E. & Witte, O. N. (2001) Cancer Res. 61, 3256-3261. [PubMed] [Google Scholar]
- 45.Donjacour, A. A. & Cunha, G. R. (1995) Development (Cambridge, U.K.) 121, 2199-2207. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, J. M., Danos, O., Grossman, M., Raulet, D. H. & Mulligan, R. C. (1990) Proc. Natl. Acad. Sci. USA 87, 439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capel, B., Hawley, R., Covarrubias, L., Hawley, T. & Mintz, B. (1989) Proc. Natl. Acad. Sci. USA 86, 4564-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nolta, J. A., Dao, M. A., Wells, S., Smogorzewska, E. M. & Kohn, D. B. (1996) Proc. Natl. Acad. Sci. USA 93, 2414-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salm, S. N., Takao, T., Tsujimura, A., Coetzee, S., Moscatelli, D. & Wilson, E. L. (2002) Prostate 51, 175-188. [DOI] [PubMed] [Google Scholar]
- 50.Sugimura, Y., Foster, B. A., Hom, Y. K., Lipschutz, J. H., Rubin, J. S., Finch, P. W., Aaronson, S. A., Hayashi, N., Kawamura, J. & Cunha, G. R. (1996) Int. J. Dev. Biol. 40, 941-951. [PubMed] [Google Scholar]
- 51.Thomson, A. A. & Cunha, G. R. (1999) Development (Cambridge, U.K.) 126, 3693-3701. [DOI] [PubMed] [Google Scholar]
- 52.Goodell, M. A., Brose, K., Paradis, G., Conner, A. S. & Mulligan, R. C. (1996) J. Exp. Med. 183, 1797-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, A. Y., LaTray, L. & van Den Engh, G. (2000) Prostate 44, 303-312. [DOI] [PubMed] [Google Scholar]
- 54.Liu, A. Y. & True, L. D. (2002) Am. J. Pathol. 160, 37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu, A. Y., Nelson, P. S., van den Engh, G. & Hood, L. (2002) Prostate 50, 92-103. [DOI] [PubMed] [Google Scholar]
- 56.Weilbaecher, K., Weissman, I., Blume, K. & Heimfeld, S. (1991) Blood 78, 945-952. [PubMed] [Google Scholar]


