Abstract
The Alu element has been a major source of new exons during primate evolution. Thousands of human genes contain spliced exons derived from Alu elements. However, identifying Alu exons that have acquired genuine biological functions remains a major challenge. We investigated the creation and establishment of Alu exons in human genes, using transcriptome profiles of human tissues generated by high-throughput RNA sequencing (RNA-Seq) combined with extensive RT-PCR analysis. More than 25% of Alu exons analyzed by RNA-Seq have estimated transcript inclusion levels of at least 50% in the human cerebellum, indicating widespread establishment of Alu exons in human genes. Genes encoding zinc finger transcription factors have significantly higher levels of Alu exonization. Importantly, Alu exons with high splicing activities are strongly enriched in the 5′-UTR, and two-thirds (10/15) of 5′-UTR Alu exons tested by luciferase reporter assays significantly alter mRNA translational efficiency. Mutational analysis reveals the specific molecular mechanisms by which newly created 5′-UTR Alu exons modulate translational efficiency, such as the creation or elongation of upstream ORFs that repress the translation of the primary ORFs. This study presents genomic evidence that a major functional consequence of Alu exonization is the lineage-specific evolution of translational regulation. Moreover, the preferential creation and establishment of Alu exons in zinc finger genes suggest that Alu exonization may have globally affected the evolution of primate and human transcriptomes by regulating the protein production of master transcriptional regulators in specific lineages.
Keywords: transcriptome evolution, transposable element, alternative splicing, deep sequencing, uORF
Alu elements have emerged as a major contributor to gene regulation and genome evolution in primates (1–3). Created ∼60 million years ago during primate evolution, Alu is the most abundant type of mobile elements in the human genome, with more than 1 million copies occupying ∼10% of the human genomic DNA (3). Historically, Alu elements were regarded as “junk DNA” with no apparent function. However, studies in the past decade have revealed diverse roles for Alu elements in gene regulation and genome evolution (1–3).
The exonization of Alu elements is a major mechanism for de novo exon creation in primate and human genomes (4). The consensus sequence of Alu harbors sites that resemble the 5′ and 3′ splice site signals (5, 6). After the insertion of an Alu element into the intronic region of an existing gene, subsequent mutations could activate these splice sites or introduce additional splicing regulatory signals, leading to the creation of a new exon (6). Although the vast majority of Alu exons are rarely incorporated into transcripts (7), the analyses of expressed sequence tags (ESTs) and exon array data recently have revealed a small number of Alu exons with high splicing activities or tissue-specific splicing profiles (8, 9). In a few genes, the functional impact and evolutionary history of Alu exons have been characterized experimentally (9–12). These data suggest that Alu exonization contributed to the adaptive evolution of primates and humans.
In this work, we carried out a genome-wide analysis of Alu exon splicing using transcriptome profiles generated by deep RNA sequencing (RNA-Seq) (13). Previous studies of Alu exons using ESTs and exon arrays were limited by the incomplete exon coverage and low resolution of these technologies. By contrast, deep RNA-Seq enabled an unbiased analysis of Alu exonization events and allowed us to estimate quantitatively the transcript inclusion levels of Alu exons. Importantly, we found that Alu exons with high splicing activities were strongly enriched in the 5′-UTR, and a large fraction of 5′-UTR Alu exons significantly altered mRNA translational efficiency. These results suggest an important role for Alu exonization in the evolution of translational regulation in primates and humans.
Results
RNA-Seq Analysis of Alu Exons in the Human Brain Transcriptome.
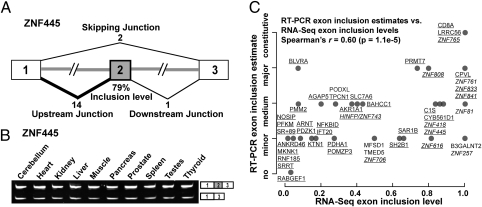
To investigate the splicing activities of Alu exons, we analyzed a deep RNA-Seq data set of the human cerebellum totaling 123 million single-end reads (13). Using the University of California, Santa Cruz (UCSC) Known Genes annotation (14), we extracted 627 Alu-derived internal exons whose flanking exons were constitutively spliced. For each Alu exon, we collected a set of three exon–exon junction sequences corresponding to its upstream, downstream, and skipping junctions (Fig. 1A). We mapped the RNA-Seq reads to these exon–exon junctions (Materials and Methods). Because the flanking exons of most Alu exons were non–repeat-derived, we were able to map exon–exon junction reads unambiguously. In total, 287 Alu-derived exons had at least one read mapped to one of the three junctions, including 127 exons with at least five reads and 82 exons with at least 10 reads mapped to one of the three junctions. The majority of the 287 exons (197, 69%) had at least one read mapped to the upstream or downstream junction, indicating exon inclusion in the transcripts. For each of the 287 exons, we calculated its transcript inclusion level in the cerebellum (i.e., the percentage of transcripts including the exon among all transcripts including or excluding the exon) using its junction read counts (see the formula in Materials and Methods). For example, the Alu exon in zinc finger protein 445 (ZNF445) had an estimated inclusion level of 79% in the human cerebellum (Fig. 1A).
Fig. 1.
RNA-Seq analysis and RT-PCR validation of Alu exons in the cerebellum. (A) Cerebellum RNA-Seq data of an Alu exon (exon 2) in ZNF445. The numbers of reads that map uniquely to the upstream, downstream, and skipping exon–exon junctions and the estimated Alu exon inclusion level are indicated. (B) RT-PCR analysis of an Alu exon in ZNF445. (C) Correlation between the cerebellum Alu exon inclusion levels estimated by RNA-Seq (x axis) and by RT-PCR (y axis). The corresponding gene symbols of Alu exons with at least five reads mapped to one of the three exon–exon junctions are underlined. ZNF genes are shown in italics.
To confirm the RNA-Seq estimates of Alu exon inclusion levels, we randomly selected 46 exons with at least one junction read indicating exon inclusion for RT-PCR analysis. Of these 46 exons, 34 exons (74%) originated completely from Alu elements; the remaining 12 exons (26%) were derived from mergers of Alu and non-Alu sequences. We examined their splicing patterns in the human cerebellum using semiquantitative RT-PCR (see the list of 46 exons in Table S1 and their gel pictures in Fig. S1). For example, the Alu exon in ZNF445 was confirmed to be included in the majority of its transcript products (Fig. 1B). Of all 46 exons tested, 45 (98%) showed exon inclusion in the cerebellum (Fig. S1A). From these 45 exons, we further selected 17 exons with weak exon inclusion PCR bands and confirmed their splicing into transcripts by additional RT-PCR experiments with one primer within the Alu exon and the other primer in a flanking constitutive exon (Fig. S1B). We classified the 46 exons into five categories ranging from no exon inclusion to constitutive splicing (i.e., 100% exon inclusion), based on the RT-PCR results. We observed a significant positive correlation between the exon inclusion levels estimated by RNA-Seq and by RT-PCR (Spearman's r = 0.60; P = 1.1e-5; Fig. 1C). This correlation was stronger among Alu exons with at least five reads mapped to one of the three exon–exon junctions (r = 0.64) (underlined gene symbols in Fig. 1C). This result was expected, because the increased RNA-Seq read coverage allows more reliable estimates of exon inclusion levels (15). For the remainder of this paper, all analyses involving exon inclusion levels are restricted to Alu exons with at least five reads mapped to one of the three junctions. Of the 127 Alu exons meeting this criterion, 36 (28%) had an estimated exon inclusion level of at least 50% (referred to as “highly included” Alu exons). These results indicate that a significant portion of Alu exons have acquired strong splicing signals to be spliced into the majority of their genes’ transcripts in the cerebellum.
Ubiquitous or Tissue-Specific Splicing Patterns of Alu Exons.
To assess whether the cerebellum-spliced Alu exons are ubiquitously spliced in a broad range of tissues or specifically spliced in the cerebellum, we analyzed a second RNA-Seq data set of the human liver with a total of 90 million reads (13, 16). We found 85 Alu exons with at least five reads mapped to one of the three exon–exon junctions in both data sets. For these 85 exons, the estimated transcript inclusion levels in the cerebellum and liver were strongly correlated (Pearson's r = 0.61; P = 4.8e-10), indicating that most cerebellum-spliced Alu exons also were spliced in the liver. To examine directly the variations of Alu exon splicing among human tissues, for the 46 exons tested by RT-PCR in the cerebellum, we expanded our RT-PCR analysis to nine additional tissues (Materials and Methods). Of the 45 Alu exons spliced in the cerebellum, five displayed notable changes in splicing patterns in various human tissues; the rest had similar splicing patterns in all tissues analyzed (Fig. S1 and Table S1). For example, the Alu exon in ZNF445 had high inclusion levels in all 10 tissues (Fig. 1B).
Alu Exonization Events Are Strongly Enriched in Zinc Finger Transcription Factors.
The expanded list of Alu exons and the ability to estimate transcript inclusion levels by RNA-Seq allowed us to identify gene families and functional categories enriched for the creation and establishment of Alu exons. We compared 35 genes with highly included Alu exons with a background list of 16,530 cerebellum-expressed genes (Materials and Methods). Using the functional annotation tool Database for Annotation, Visualization and Integrated Discovery (DAVID) (17), we identified two significantly enriched Gene Ontology (GO) terms with a Benjamini-corrected false discovery rate (FDR) of <0.05: “KRAB box transcription factor” (P = 1.8e-4; FDR = 3.6e-3) and “zinc finger transcription factor” (P = 1.4e-3; FDR = 1.4e-2). Both GO terms refer to zinc finger (ZNF) transcription factors, a large family of transcription factors in the human genome. These transcription factors typically are characterized by an N-terminal protein interaction domain, most commonly the Kruppel-associated box (KRAB) domain and the C2H2 ZNF DNA-binding domain in the C-terminal region (18). Interestingly, ZNF genes underwent rapid expansion and adaptive evolution during primate and human evolution (18, 19). Therefore, they have been considered key contributors to lineage-specific transcriptome regulation in primates and humans (18, 20).
To confirm the splicing of Alu exons in ZNF genes, we conducted RT-PCR analysis of 12 exons that had an estimated inclusion level of ∼50% or higher (Table S2). All 12 exons were validated as being spliced into the transcripts, including nine exons with at least medium inclusion level in the cerebellum, according to RT-PCR. To avoid misinterpretation of RT-PCR results because of nonspecific amplification of paralogous ZNF genes, the identities of all PCR products were confirmed by sequencing. Although the specific biological functions of most ZNF genes remain obscure (18), some of the Alu-exon–containing ZNF genes have been implicated in disease or gene regulation. For example, ZNF445 has been shown to activate the transcriptional activity of activator protein 1 (AP1) (21). Zinc finger protein 706 (ZNF706) has a sex-specific gene expression pattern (22) and is up-regulated in larynx tumors (23). Zinc finger protein 81 (ZNF81) is one of the three X-chromosome ZNF genes associated with nonsyndromic X-linked mental retardation (24). Histone H4 transcription factor (HINFP, also known as zinc finger protein 743, ZNF743) encodes a ZNF protein that interacts with methyl-CpG-binding protein 2 (MBD2) and is involved in transcriptional and epigenetic regulation (25).
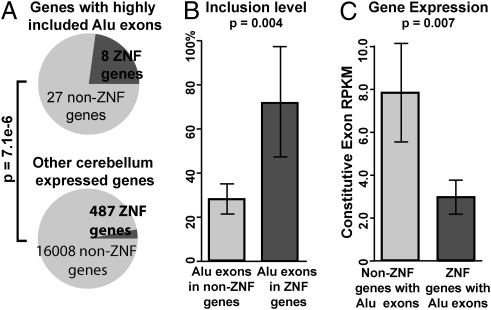
We performed additional analyses to investigate the enrichment of Alu exons in ZNF genes. For independent confirmation of the result of the DAVID analysis based on GO annotations, we collected a list of 551 ZNF genes in the human genome from Huntley and colleagues (26) (Materials and Methods). We found that 23% (8/35) of genes with a highly included Alu exon were ZNF genes, compared with only 3% among other cerebellum-expressed genes, a more than sevenfold enrichment (P = 7.1e-6; one-sided Fisher exact test; Fig. 2A). Of all nine Alu exons in ZNF genes with at least five reads mapped to at least one exon–exon junction, the average estimated transcript inclusion level was 72%, compared with 28% for Alu-derived exons in non-ZNF genes (P = 0.004; one-sided Wilcoxon test; Fig. 2B). This observation was not an artifact caused by higher expression level or increased RNA-Seq coverage of ZNF genes in the cerebellum. In fact, according to RNA-Seq gene expression estimates based on the reads per kilobase of exon model per million mapped reads (RPKM) metric (27) (Materials and Methods), among Alu-exon–containing genes, ZNF genes had lower expression levels on average than non-ZNF genes in the cerebellum (P = 0.007; one-sided Wilcoxon test) (Fig. 2C).
Fig. 2.
Enrichment of Alu exons in ZNF genes. (A) The percentage of ZNF genes is significantly higher in genes containing highly included Alu exons than in other cerebellum-expressed genes without highly included Alu exons. (B) Alu exons in ZNF genes have higher overall transcript inclusion levels than Alu exons in non-ZNF genes. (C) ZNF genes with Alu exons do not have higher overall expression levels in the cerebellum compared with non-ZNF genes with Alu exons. All exons in panels A–C have at least five reads mapped to one of the three exon–exon junctions. Error bars indicate the 95% confidence interval.
Frequent Alu Exonization Is Characteristic of Primate-Specific Genes.
Because many ZNF genes are primate-specific, we asked whether the preferential creation and establishment of Alu exons is a general characteristic of phylogenetically young genes. Based on the PhyloPat (28) phylogenetic classification of human genes (Materials and Methods), we grouped all human genes into four mutually exclusive groups with increasing phylogenetic ages: primate, mammalian, euteleostomi, and metazoan. Among genes expressed in the cerebellum, 9.3% (65) of primate genes contained internal Alu exons, compared with 4.4% (121) of mammalian genes, 4.4% (267) of euteleostomi genes, and 4.9% (346) of metazoan genes (P < 1e-5 for all one-sided Fisher exact tests between primate genes and any other age group) (Fig. S2A). We also observed a higher percentage of genes containing highly included Alu exons in the cerebellum among the primate genes (0.93%, 0.10%, 0.08%, and 0.17% in the four age groups, respectively; P < 1e-3 for all one-sided Fisher exact tests) (Fig. S2A). Moreover, among all Alu exons with at least five reads mapped to at least one of the three exon–exon junctions in the cerebellum, 75% (six of eight) in the primate gene group had at least a 50% inclusion level, a percentage higher than that of any other age group (Fig. S2B). Overall, we observed a significant anti-correlation between the phylogenetic ages of Alu-exon–containing genes and the inclusion levels of Alu exons in the cerebellum (P = 0.003, linear regression of gene age and Alu exon inclusion level). We observed the same trend in the liver RNA-Seq data set (Fig. S2 C and D).
Consistent with this trend, the creation and establishment of Alu exons were enriched in primate-specific ZNF genes as compared with ancient ZNF genes (Fig. S3). Given that a large fraction of ZNF genes are primate-specific, this result suggests that the enrichments of Alu exons in ZNF genes and in primate-specific genes are coupled to a certain extent. Nonetheless, the preference in ZNF genes over non-ZNF genes holds even after controlling for phylogenetic age (Fig. S3). Indeed, we found highly included Alu exons in both evolutionarily ancient and primate-specific ZNF genes. For example, ZNF445 is conserved among most mammalian species and has a 5′-UTR Alu exon that regulates its translational efficiency (Fig. 3D). Despite the ancient origin of this gene, the strong splicing activity of this Alu exon appears to have been acquired recently, because the comparison of the human and nonhuman primate orthologous genomic regions and the consensus sequence of the corresponding AluSc subfamily revealed a human/chimpanzee-specific C-to-T substitution that created the “GT” 5′ splice site. Interestingly, the genomic region of ZNF445 appeared under positive selection during recent human evolution according to SNP-based scans of positive selection signals (29). Collectively, our results indicate that Alu exonization has played a role in both the ongoing evolution of ancient ZNF genes and the recent expansion of the gene family in the primate and human lineages.
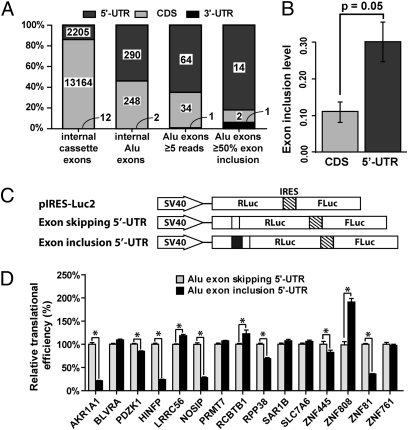
Fig. 3.
Enrichment and regulatory impact of Alu exons in the 5′-UTR. (A) The transcript location (5′-UTR, CDS, or 3′-UTR) of UCSC Known Genes alternatively spliced internal cassette exons, UCSC internal Alu exons, Alu exons with at least five reads mapped to at least one exon–exon junction, and highly included Alu exons (i.e., ≥50% exon inclusion level). (B) Alu exons in the 5′-UTR have significantly higher overall inclusion levels than Alu exons in the CDS. All Alu exons included in the plot have at least five reads mapped to one of the three exon–exon junctions in the cerebellum. (C) Schematic diagrams of the pIRES-Luc2 dual-luciferase reporter vector and the Alu exon skipping/inclusion 5′-UTR constructs. (D) Modulation of translational efficiency in HeLa cells by 5′-UTR Alu exons. The translational efficiency of each 5′-UTR construct was calculated as the ratio between the Renilla luciferase and the firefly luciferase activities. For each tested gene, the translational efficiency of the Alu exon skipping 5′-UTR construct was set as 1 in the plot. P value was calculated from at least eight independent replicate luciferase experiments. *P < 0.05. Error bars indicate the 95% confidence interval.
Preferential Establishment and Regulatory Impact of Alu Exons in the 5′-UTR.
We found a strong enrichment of Alu exons in the 5′-UTR. We compared the occurrence and transcript inclusion levels of Alu exons within different regions of protein-coding genes [5′-UTR, coding sequence (CDS), 3′-UTR]. We restricted this analysis to exons for which transcript locations could be determined unambiguously using the UCSC Known Genes annotation (14). Among all internal Alu exons collected from the UCSC Known Genes database, 290 (54%) are in the 5′-UTR, 248 (46%) are in the CDS, and two are in the 3′-UTR (Fig. 3A). Of the 99 Alu exons with at least five reads mapped to one of the three exon–exon junctions (i.e., two inclusion junctions and one skipping junction, thus already slightly biased toward cerebellum-included Alu exons), 64 (65%) are in the 5′-UTR, 34 are in the CDS, and only one is in the 3′-UTR (Fig. 3A). Moreover, of the 17 highly included Alu exons according to RNA-Seq data, an even higher percentage of exons are located in the 5′-UTR (82%) (Fig. 3A). The percentages of 5′-UTR exons in these lists of Alu exons are substantially higher than in the general population of alternatively spliced exons in human genes. For example, among the high-confidence alternatively spliced cassette exons from the Alternative Splicing Annotation Project 2 (ASAP2) database (30), 673 (19.9%) are in the 5′-UTR, 2,700 (80.0%) are in the CDS, and two (0.1%) are in the 3′-UTR. Similarly, in the Human-transcriptome DataBase for Alternative Splicing (H-DBAS) compiled from full-length cDNAs (31), 17.5% of alternatively spliced exons are in the 5′-UTR. In the UCSC Known Genes database (14), 14.3% of alternatively spliced internal cassette exons are in the 5′-UTR (Fig. 3A). In summary, from alternatively spliced cassette exons to Alu exons to highly included Alu exons, we observed a consistent increase in the percentage of 5′-UTR exons and decrease in the percentage of CDS exons. Moreover, Alu exons in the 5′-UTR had higher average transcript inclusion levels than exons in the CDS (30% vs. 11%, P = 0.05, one-sided Wilcoxon test; Fig. 3B). Together, these results suggest that the 5′-UTR not only is a hotspot for the initial creation of new Alu exons but also favors the subsequent establishment of strong splicing activities. It should be noted that this analysis was restricted to internal spliced Alu exons in the UTRs, although Alu elements also could contribute to sequences within terminal exons, including polyadenylation sites (32).
The high level of Alu exon establishment in the 5′-UTR raised an interesting question about the regulatory impact of these exons. It is well known that the 5′-UTRs contain regulatory signals of mRNA stability and protein translation (33). In several genes the alternative splicing of 5′-UTR exons regulates protein translation (34–36). To determine if Alu exonization in the 5′-UTR could affect the translation of host mRNAs broadly, we selected 15 Alu exons and compared the translational efficiency of the exon inclusion 5′-UTR versus the exon skipping 5′-UTR using a dual luciferase reporter construct pIRES-Luc2 (Materials and Methods). For each 5′-UTR isoform, the resulting reporter construct expressed both the firefly luciferase and the Renilla luciferase bicistronicly. The Renilla luciferase was fused downstream of the cloned 5′-UTR isoform, whereas the firefly luciferase translation was driven independently by the internal ribosome entry site (IRES) and was not regulated by the cloned 5′-UTR (Fig. 3C). The translational efficiency of each 5′-UTR construct was calculated as the ratio between the Renilla luciferase and the firefly luciferase activities. In four genes encoding ZNF transcription factors [ZNF445, zinc finger protein 808 (ZNF808), ZNF81, HINFP/ZNF743], the Alu exon significantly altered the translational efficiency in HeLa cells when included in the 5′-UTR (Fig. 3D). We also observed a significant effect of the Alu exon on the translational efficiency of the non-ZNF genes aldo-keto reductase family 1, member A1 (AKR1A1), PDZ domain containing 1 (PDZK1), leucine-rich repeat containing 56 (LRRC56), nitric oxide synthase interacting protein (NOSIP), regulator of chromosome condensation and BTB domain containing protein 1 (RCBTB1), and ribonuclease P protein subunit p38 (RPP38) (Fig. 3D). In total, 10 of the 15 tested Alu exons altered translational efficiency. We tested nine exons in HEK293 cells and obtained similar results (Fig. S4).
Molecular Mechanisms of Translational Regulation by Alu Exons.
Newly created 5′-UTR Alu exons potentially could regulate translational efficiency through a variety of molecular mechanisms, such as upstream ORF (uORF), IRES, secondary structure, and 5′-UTR length (37). We decided to test hypotheses reflecting two prevalent mechanisms of translational regulation by 5′-UTR elements; one would decrease and the other would increase translational efficiency. In the first hypothesis, Alu exons may introduce or elongate uORFs before the primary start codons, thus repressing the translation of the primary ORFs (37, 38). In the second hypothesis, Alu exons could introduce cellular IRES elements to the 5′-UTRs and thus increase translational efficiency (37).
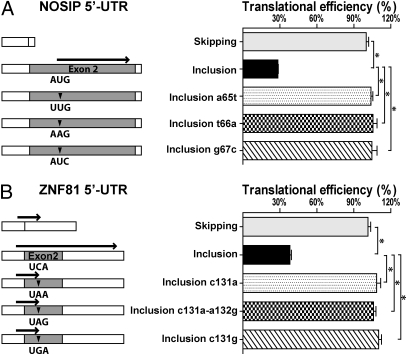
To test the first hypothesis involving uORFs, all 15 Alu exons tested by luciferase assays (Fig. 3D) were scanned for differences in uORFs between the Alu exon inclusion and exon skipping 5′-UTRs. We identified three genes in which the Alu exons either created a single new uORF (NOSIP and RPP38) or elongated an existing uORF (ZNF81). In NOSIP, the Alu exon introduced a new uORF to the 5′-UTR (Fig. 4A). We used site-directed mutagenesis to disrupt the uORF by mutating the upstream AUG (uAUG) start codon within the Alu exon to UUG (a65t), AAG (t66a), or AUC (g67c). All three mutant constructs completely reversed the translational repression by the Alu exon (Fig. 4A). Similar results were obtained for the RPP38 Alu exon (Fig. S5). In ZNF81, the Alu exon caused a frame shift of an existing uORF (51 nt), resulting in a fourfold longer uORF (267 nt) (Fig. 4B). The Alu exon inclusion 5′-UTR caused a >60% decrease in translational efficiency compared with the exon skipping 5′-UTR (Fig. 4B). We introduced three different stop codons, UAA, UAG, or UGA, within the Alu exon to shorten the uORF back to its original length (51 nt). All three mutations resulted in a full recovery of the translational efficiency (Fig. 4B). Together, these mutational studies reveal a general mechanism of translational repression by Alu exons through the creation or elongation of uORFs in the 5′-UTR.
Fig. 4.
Alu exons repress translation by creating or elongating uORFs. The wild-type and mutant pIRES-Luc2 dual-luciferase reporter constructs of (A) NOSIP and (B) ZNF81 Alu exon skipping/inclusion 5′-UTRs were tested for translational efficiency. (Left) Schematic diagrams of the wild-type and mutant constructs. ▼ indicates site of mutations. Bold arrows indicate uORFs. (Right) Estimated translational efficiency. *P < 0.0001.
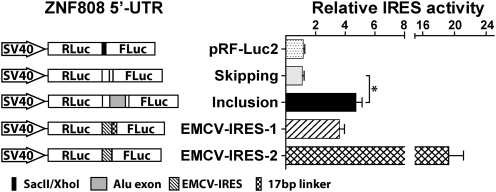
To test the second hypothesis involving potential creation or disruption of IRES by Alu exons, a bicistronic reporter system was adapted using pRF-Luc2 as the empty vector backbone to generate a series of IRES-activity reporter constructs (Materials and Methods and SI Materials and Methods). Test sequences were cloned and inserted between the upstream Renilla luciferase stop codon and the downstream firefly luciferase start codon. IRES activity of the test sequence was indicated by the ratio between firefly luciferase and Renilla luciferase activities. We selected three genes [ZNF808, HINFP, and protein arginine methyltransferase 7 (PRMT7)] to test in this system. These genes represented increased (ZNF808), decreased (HINFP), or constant (PRMT7) translational efficiency after Alu exon inclusion (Fig. 3D). Among all tested 5′-UTR isoforms, most constructs showed a very low firefly/Renilla luciferase ratio that was not above the empty vector (pRF-Luc2) control (Fig. S6). Only the Alu exon inclusion 5′-UTR construct of ZNF808 showed a significant firefly/Renilla ratio comparable to the Encephalomyocarditis virus (EMCV)-IRES-1 positive control and much higher (approximately fourfold) than its Alu exon skipping counterpart (Fig. 5 and Fig. S6). This result is consistent with the observation that the Alu exon inclusion 5′-UTR of ZNF808 showed a significant increase in translational efficiency (Fig. 3D), suggesting a potential IRES created by Alu exon inclusion. However, to confirm the identity of the IRES element definitively and to define its exact location in the Alu exon inclusion 5′-UTR requires substantial additional experimental efforts (39, 40).
Fig. 5.
IRES activity assay of the ZNF808 Alu exon 5′-UTRs. Sequences of the ZNF808 Alu exon inclusion or skipping 5′-UTRs were tested for potential IRES activity using the pRF-Luc2 dual-luciferase reporter vector. (Left) Schematic diagrams of the reporter constructs. (Right) Ratios of the firefly luciferase activity to the Renilla luciferase activity. *P < 0.0001.
Together, these experiments reveal potential molecular mechanisms (uORF, IRES) by which Alu exons modulate translational efficiency. We note that the mechanisms of translational regulation by 5′-UTRs are complex (37), and it is entirely possible that other Alu exons regulate translation through alternative mechanisms.
Discussion
The origin and evolution of new exons have attracted considerable interest in recent years (6). Although lineage-specific exons are common in mammalian genomes, identifying those exons that have genuine biological functions is a major challenge. Although a growing list of studies report new exons with strong splicing activities and functional roles, a global search for such new exons remains difficult. Most genomic technologies, such as EST sequencing and exon array, are not capable of defining exon inclusion levels in individual tissues. For example, our previous Affymetrix exon array analysis of Alu exons (9) was seriously limited by the incomplete coverage of the exon array design for Alu exons and the potential cross-hybridization of Alu exon probes to other Alu-containing transcripts. Moreover, we cannot directly quantify the exon inclusion level in individual tissues because of the confounding factor of microarray probe affinity (41).
In this work, we analyzed transcriptome profiles generated by RNA-Seq to systematically identify Alu exons with high splicing activities. Using the RNA-Seq data, we estimated the transcript inclusion levels of 287 Alu exons in the human cerebellum. The RNA-Seq estimates were strongly correlated with RT-PCR measurements of 46 Alu exons. Forty-five of the 46 RT-PCR–tested exons were spliced in the cerebellum, including 23 at the medium or higher levels. In total, our RNA-Seq analysis identified 197 internal Alu exons (≤ 250 bp and flanked by constitutive exons) spliced in the cerebellum, including all eight such exons discovered previously by the Affymetrix exon array (9). Our RT-PCR analysis of nine additional tissues indicates that a minor fraction (5/45, 11%) of these exons have notable tissue-specific shifts in splicing activities. Thus our study reveals a much expanded list of Alu exons with high splicing activities, indicating widespread establishment of Alu exons in human genes.
We found a significantly higher level of Alu exonization in genes encoding ZNF transcription factors. Such enrichment was observed for ZNF genes of different phylogenetic age groups. ZNF genes comprise a large family of transcription factors that underwent rapid evolution in primates and humans. Approximately 25% of ZNF genes in the human genome are primate-specific, and the majority of ZNF gene groups with primate-specific expansion were subject to strong positive selection according to nonsynonymous/synonymous substitution rate (dN/dS) analysis (18). Moreover, over one-third of transcription factor genes differentially expressed between human and chimpanzee brains encode ZNF transcription factors (20). It has been proposed that these genes function as master regulators of transcriptome evolution and could contribute to the transcriptional and phenotypic differences between humans and nonhuman primates (18, 20). Our results reveal Alu exonization as an important mechanism in the lineage-specific regulatory evolution of ZNF genes. In four ZNF genes (ZNF445, ZNF81, HINFP/ZNF743, and ZNF808), the first three of which are conserved between primates and other mammals, we demonstrate experimentally that the primate-specific Alu exons in their 5′-UTRs significantly alter the translational efficiency of the host mRNAs. These results suggest an interesting evolutionary scenario: that the creation and establishment of Alu exons could impact the primate and human transcriptomes globally by fine-tuning the protein production of master transcriptional regulators in a lineage-specific manner.
Our study has broad implications for understanding gene regulation and transcriptome evolution in primates and humans. Importantly, we observed a significant enrichment of Alu exons, especially those with high splicing activities, in the 5′-UTR. Moreover, of the 15 Alu exons tested by luciferase reporter assays, 10 exons altered mRNA translational efficiency when included in the 5′-UTR (Fig. 3D). These results provide genomic evidence that a major functional consequence of Alu exonization is the lineage-specific evolution of the 5′-UTR and translational regulation. Consistent with our data pointing to the importance of the 5′-UTR in human evolution, a recent study revealed 5′-UTR exons as the hotspot of human-specific acceleration of nucleotide substitutions (42). Together, these findings suggest that the evolution of the 5′-UTR is an essential aspect of human genome evolution and may contribute to the acquisition of species-specific traits. Our finding on the widespread translational impact of Alu exons also provides insight into the function of 5′-UTR alternative splicing events in general. Our data imply that alternative splicing of 5′-UTR exons could be a prevalent mechanism of gene regulation in humans that affects phenotypes or modulates disease pathogenesis.
Materials and Methods
RNA-Seq and RT-PCR Analysis of Alu Exons.
The locations of Alu elements in the human genome were downloaded from the UCSC Genome Browser database (43). The locations of internal spliced exons in human genes were taken from the UCSC Known Genes database (14). To eliminate long exonic regions possibly resulting from intron-retention events, we removed exons longer than 250 bp as in ref. 9. We defined an exon as Alu-derived if the Alu element covered at least 25 bp of the exon and more than 50% of the total exon length. To avoid complications in RNA-Seq analysis arising from complex alternative splicing patterns of flanking exons, we focused on “simple” Alu exons with constitutive flanking exons.
We downloaded Illumina RNA-Seq data of human cerebellum and liver from published datasets. The cerebellum dataset consists of 123 million reads for six human cerebellum samples (13). The liver dataset consists of 90 million reads merged from two studies (13, 16). We mapped RNA-Seq reads to the human genome (hg18) and all exon–exon junctions supported by the UCSC Known Genes annotations (14), using the software ELAND allowing up to 2 bp mismatches. Each mapped exon–exon junction sequence required at least 5 bp from any side of the exon junction. We removed reads that mapped to either the human genome (hg18) or multiple junctions. For each Alu exon, its transcript inclusion level in a given sample was calculated using the number of reads that uniquely mapped to its upstream junction (UJ), downstream junction (DJ), and skipping junction (SJ) as (UJ+DJ)/(UJ+DJ+2*SJ) (as in ref. 13). The overall expression levels of human genes in the cerebellum were calculated using the RPKM metric (27) within the constitutive exons. Cerebellum-expressed genes were defined as genes with at least one unique cerebellum RNA-Seq read mapped to their constitutive exons.
Total RNA samples of 10 human tissues were purchased from Clontech. For each tested Alu exon, we designed a pair of forward and reverse PCR primers at flanking constitutive exons. The RT-PCR PAGE gel images were analyzed by densitometry using the ImageQuant TL software (GE). Final Alu exon inclusion levels were grouped into five categories: no exon inclusion (0%), minor (1–30%), medium (30–70%), major (70–99%), and constitutive (100%). To confirm further the exon inclusion events of Alu exons with weak exon inclusion PCR bands, we also designed a pair of PCR primers with one primer located within the Alu exon and the other primer located in a flanking constitutive exon. All RT-PCR primer sequences are described in Tables S3 and S4.
Collection of ZNF Genes in the Human Genome.
We collected a list of 551 UCSC Known Genes loci encoding ZNF transcription factors, using a catalog of ZNF genes compiled by Huntley et al. (26). Specifically, we intersected the ZNF genes in Huntley et al. with the UCSC Known Genes annotations, with the requirement that at least 70% of the genomic region of a ZNF gene defined by Huntley et al. be covered by a UCSC Known Genes transcript.
Phylogenetic Age Analysis.
We used the PhyloPat database (28) to determine the phylogenetic ages of human genes. PhyloPat classifies the phylogenetic lineages of human genes using Ensembl orthology annotations (28). We grouped all human genes into four mutually exclusive groups based on the PhyloPat classification: primate, mammalian, euteleostomi, and metazoan. For example, primate genes refer to human genes present in primate species but absent from other nonprimate mammalian species.
Dual Luciferase Reporter Vector Construction and Dual Luciferase Reporter Assay.
The psiCHECK-2 vector (Promega) was modified to construct the dual luciferase reporter pIRES-Luc2 and pRF-Luc2 vector backbones. The pIRES-Luc2 reporter was used to assess the translational efficiency of Alu exon inclusion or skipping 5′-UTRs. The pRF-Luc2 reporter was used to test the potential IRES activity of the 5′-UTR isoforms. Details of the reporter vector construction and dual-luciferase reporter assay are supplied in SI Materials and Methods.
Acknowledgments
We thank Dr. Russ Carstens for the pIRES-Blast-delta-Int vector and Jennifer Dozier for technical assistance. This work was supported by National Institutes of Health Grants R01HG004634 and R01GM088342 (to Y.X.) and P01NS050210 (to B.L.D.), and by the Roy J. Carver Trust (to B.L.D.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012834108/-/DCSupplemental.
References
- 1.Häsler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006;34:5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev. 2010;20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 5.Makałowski W, Mitchell GA, Labuda D. Alu sequences in the coding regions of mRNA: A source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 6.Sorek R. The birth of new exons: Mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mersch B, Sela N, Ast G, Suhai S, Hotz-Wagenblatt A. SERpredict: Detection of tissue- or tumor-specific isoforms generated through exonization of transposable elements. BMC Genet. 2007;8:78. doi: 10.1186/1471-2156-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin L, et al. Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet. 2008;4:e1000225. doi: 10.1371/journal.pgen.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singer SS, Männel DN, Hehlgans T, Brosius J, Schmitz J. From “junk” to gene: Curriculum vitae of a primate receptor isoform gene. J Mol Biol. 2004;341:883–886. doi: 10.1016/j.jmb.2004.06.070. [DOI] [PubMed] [Google Scholar]
- 11.Gerber A, O'Connell MA, Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 12.Krull M, Brosius J, Schmitz J. Alu-SINE exonization: En route to protein-coding function. Mol Biol Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 13.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu F, et al. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 15.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 16.Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowick K, Hamilton AT, Zhang H, Stubbs L. Rapid sequence and expression divergence suggest selection for novel function in primate-specific KRAB-ZNF genes. Mol Biol Evol. 2010;27:2606–2617. doi: 10.1093/molbev/msq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci USA. 2009;106:22358–22363. doi: 10.1073/pnas.0911376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo K, et al. Activation of transcriptional activities of AP1 and SRE by a novel zinc finger protein ZNF445. Gene. 2006;367:89–100. doi: 10.1016/j.gene.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Bleibel WK, Roe CA, Cox NJ, Eileen Dolan M. Gender-specific differences in expression in human lymphoblastoid cell lines. Pharmacogenet Genomics. 2007;17:447–450. doi: 10.1097/FPC.0b013e3280121ffe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colombo J, et al. Gene expression profiling reveals molecular marker candidates of laryngeal squamous cell carcinoma. Oncol Rep. 2009;21:649–663. [PubMed] [Google Scholar]
- 24.Kleefstra T, et al. Zinc finger 81 (ZNF81) mutations associated with X-linked mental retardation. J Med Genet. 2004;41:394–399. doi: 10.1136/jmg.2003.016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekimata M, Takahashi A, Murakami-Sekimata A, Homma Y. Involvement of a novel zinc finger protein, MIZF, in transcriptional repression by interacting with a methyl-CpG-binding protein, MBD2. J Biol Chem. 2001;276:42632–42638. doi: 10.1074/jbc.M107048200. [DOI] [PubMed] [Google Scholar]
- 26.Huntley S, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: Insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 28.Hulsen T, Groenen PM, de Vlieg J, Alkema W. PhyloPat: An updated version of the phylogenetic pattern database contains gene neighborhood. Nucleic Acids Res. 2009;37(Database issue):D731–D737. doi: 10.1093/nar/gkn645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang K, Thornton KR, Stoneking M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim N, Alekseyenko AV, Roy M, Lee C. The ASAP II database: Analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007;35(Database issue):D93–D98. doi: 10.1093/nar/gkl884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeda J, et al. Large-scale identification and characterization of alternative splicing variants of human gene transcripts using 56,419 completely sequenced and manually annotated full-length cDNAs. Nucleic Acids Res. 2006;34:3917–3928. doi: 10.1093/nar/gkl507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JY, Ji Z, Tian B. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res. 2008;36:5581–5590. doi: 10.1093/nar/gkn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheper GC, van der Knaap MS, Proud CG. Translation matters: Protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- 34.Wang G, Guo X, Floros J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol. 2005;289:L497–L508. doi: 10.1152/ajplung.00100.2005. [DOI] [PubMed] [Google Scholar]
- 35.Shalev A, et al. A proinsulin gene splice variant with increased translation efficiency is expressed in human pancreatic islets. Endocrinology. 2002;143:2541–2547. doi: 10.1210/endo.143.7.8920. [DOI] [PubMed] [Google Scholar]
- 36.Lin L, et al. Evolution of alternative splicing in primate brain transcriptomes. Hum Mol Genet. 2010;19:2958–2973. doi: 10.1093/hmg/ddq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 38.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang H, Coleman J, Miskimins R, Srinivasan R, Miskimins WK. Cap-independent translation through the p27 5′-UTR. Nucleic Acids Res. 2007;35:4767–4778. doi: 10.1093/nar/gkm512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baranick BT, et al. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc Natl Acad Sci USA. 2008;105:4733–4738. doi: 10.1073/pnas.0710650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irizarry RA, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 42.Kostka D, Hahn MW, Pollard KS. Noncoding sequences near duplicated genes evolve rapidly. Genome Biol Evol. 2010;2:518–533. doi: 10.1093/gbe/evq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhead B, et al. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res. 2010;38(Database issue):D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]