Abstract
The ADP ribosyl transferase [poly(ADP-ribose) polymerase] ARTD3(PARP3) is a newly characterized member of the ARTD(PARP) family that catalyzes the reaction of ADP ribosylation, a key posttranslational modification of proteins involved in different signaling pathways from DNA damage to energy metabolism and organismal memory. This enzyme shares high structural similarities with the DNA repair enzymes PARP1 and PARP2 and accordingly has been found to catalyse poly(ADP ribose) synthesis. However, relatively little is known about its in vivo cellular properties. By combining biochemical studies with the generation and characterization of loss-of-function human and mouse models, we describe PARP3 as a newcomer in genome integrity and mitotic progression. We report a particular role of PARP3 in cellular response to double-strand breaks, most likely in concert with PARP1. We identify PARP3 as a critical player in the stabilization of the mitotic spindle and in telomere integrity notably by associating and regulating the mitotic components NuMA and tankyrase 1. Both functions open stimulating prospects for specifically targeting PARP3 in cancer therapy.
Keywords: mitotic division, poly(ADP ribosyl)ation, double-strand break repair
Poly(ADP ribosyl)ation is a posttranslational modification of proteins mediated by poly(ADP ribose) polymerases (PARPs). PARPs catalyze the transfer and polymerization of ADP ribose units from NAD+ to form branched polymers of ADP ribose covalently linked to heterologous acceptor proteins or PARPs themselves. PARP1, the founding and best-studied member of the PARP family, was for a long time considered to be the only enzyme that could generate poly(ADP ribose) polymers. However, in recent years, 16 additional putative PARP proteins sharing a conserved catalytic domain but with different composition of additional domains were identified (1). PARP1 and PARP2 are so far the only enzymes whose catalytic activity has been shown to be induced by DNA strand breaks, playing key shared functions in the maintenance of genome integrity. Whereas some PARP family members catalyze poly(ADP ribosyl)ation, others function as mono(ADP ribosyl)transferase or are reported as catalytically inactive. Accordingly, a new nomenclature termed the ADP ribosyltransferase diphtheria toxin-like ARTD(PARP) family has been proposed based on structural insights (2). Together, these findings highlight the importance of ADP ribosylation as a key posttranslational modification that impacts on various cellular processes and support the necessity to biochemically and physiologically define the properties of individual PARPs. For simplification, and because the official gene symbol is Parp3, we will use the term PARP3 in this study.
PARP3 was initially discovered in an EST library screening using the catalytic domain sequence of hPARP1 (3). Expression analysis revealed a tight regulation of PARP3 that differs from that of the ubiquitously expressed PARP1 and PARP2. Whereas the gene appears tissue specifically expressed in mouse adults with the highest expression in skeletal muscle (4), the protein is rather homogeneously expressed in cynomolgous monkey (5).
Further analysis of the 5′ end of the human gene revealed an alternative splicing giving rise to two proteins that differ by 7 aa at the N terminus (6). Whereas the long splice variant has been identified as a core component of the centrosome at all stages of the cell cycle, the short isoform accumulates within the nucleus. Whether both isoforms are functionally distinct remains an open question. Interestingly, the overexpression of PARP3 or its N-terminal domain interferes with the G1/S cell-cycle progression without inducing centrosomal amplification. Of note, only the shorter nuclear variant is found in mice.
Recently, efforts have been developed to define the biochemical and structural properties of PARP3 (7, 8). These studies describe PARP3 as a poly(ADP ribosyl)transferase that shares high degree of structural similarities of the PARP catalytic domains and a conserved catalytic glutamate residue with PARP1 and PARP2.
However, this protein still remains poorly characterized at the level of its in vivo cellular functions. In previous studies, human PARP3 has been found to associate with Polycomb group proteins involved in transcriptional silencing and with DNA repair networks, including base excision repair/single-strand break repair (BER/SSBR) and nonhomologous end-joining (NHEJ), suggesting an active role for PARP3 in the maintenance of genomic integrity (9).
In this study, we combined protein–protein interaction assays with the generation and characterization of human and mouse models deficient in PARP3, and provide in vivo evidences for a synergistic role of PARP3 and PARP1 in cellular response to DNA double-strand breaks (DSBs). In addition, we identify PARP3 as part of a protein network containing the mitotic factor NuMA and the telomeric protein tankyrase 1 that has fundamental implications in spindle microtubule dynamics and telomere integrity during mitosis.
Results
PARP3 Functions in Cellular Response to DNA Double-Strand Breaks.
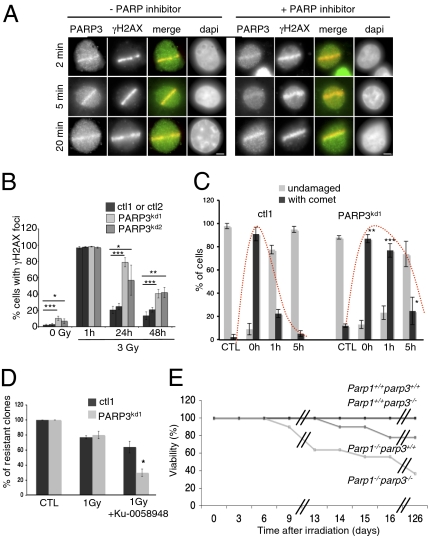
In a previous study, PARP3 was found to interact with proteins from the BER/SSBR and NHEJ pathways, suggesting a role for PARP3 in DNA repair (9). To address this question, we first examined the recruitment of PARP3 to DNA damage sites introduced by laser microirradiation. Immunofluorescence staining with a specific anti-PARP3 antibody revealed an accumulation of endogenous PARP3 at sites of DNA damage as early as 2 min and up to 20 min after microirradiation in human (HeLa), monkey (Cos1), and mouse (mouse embryonic fibroblasts, MEFs) cells (Fig. 1A and Fig. S1A). Interestingly, the treatment of Cos1 with the PARP inhibitor Ku-0058948 did not significantly affect the kinetic of PARP3 recruitment (Fig. 1A), indicating that its relocation to DNA breaks does not depend on DNA damage-induced poly(ADP ribosyl)ation.
Fig. 1.
PARP3 is involved in DNA damage response. (A) Endogenous PARP3 accumulates at laser-induced DNA damage sites detected by an anti-γH2AX antibody. Wide-field fluorescence images of Cos1 either untreated or treated with the PARP inhibitor. Fixation and immunostaining was performed at indicated time points after laser microirradiation. (Scale bar, 5 μm.) (B) Spontaneous accumulation and persistence of X-ray-induced γH2AX foci in PARP3kd cells. Quantification of the percentage of cells displaying γH2AX foci. An average of 500 cells per cell line were scored in >20 randomly selected fields. (C) Histogram showing the percentage of unrepaired (with COMET) vs. repaired (round shaped) control (ctl1) and PARP3kd1 cells as a function of time after X-irradiation. An average of 50 cells was scored for each time point. For B and C, results are averages from three independent experiments. Mean values ± SD are indicated. *P < 0.05, **P < 0.01, ***P < 0.001. (D) Clonogenic survival of untreated (CTL) or 1 Gy-X-irradiated control (ctl) and PARP3kd1 cells in the absence or in the presence of Ku-0058948 (100 nM). Experiments were performed three times, giving similar results. Mean values of triplicates ± SD are indicated. *P < 0.05. (E) Increased radiosensitivity in Parp1−/−;Parp3−/− double-knockout mice. Kaplan–Meier survival curve after 4-Gy whole-body X-irradiation. Parp1+/+;Parp3+/+ (n = 9), Parp1+/+;Parp3−/− (n = 9), Parp1−/−;Parp3+/+ (n = 9), Parp1−/−;Parp3−/− (n = 11).
To next explore the in vivo role of PARP3 in DNA damage response, we used the shRNA approach in human cells to generate stable clones depleted in PARP3 (Fig. S1B). We selected two clones showing an almost complete depletion of the previously described centrosomal and nuclear isoforms of PARP3 (PARP3kd) (6, 9) compared with the control clones expressing a nonfunctional shRNA. To directly assess the consequences of PARP3 depletion in cellular response to DNA damage, we first analyzed the level of γH2AX staining as a marker of DSBs after X-irradiation of the PARP3-depleted (PARP3kd) compared with the control (ctl) cell lines (Fig. 1B and Fig. S1C). Both PARP3kd clones displayed a significant increase in γH2AX foci in untreated cells, suggesting the presence of spontaneous strand breaks. After X-irradiation, γH2AX foci were observed in both control and PARP3-depleted cells 1 h after irradiation. Whereas the number of γH2AX decreased throughout time in control cells, a high number of foci persisted up to 48 h in PARP3kd cells, reflecting a delay in radioinduced strand break repair. To test this hypothesis further, we performed a time-course evaluation of DSB resealing using neutral COMET assays in both PARP3kd and ctl cells after X-irradiation (Fig. 1C and Fig. S1D). We detected a significant delay in the repair of radioinduced DSBs in PARP3kd compared with ctl cells. Whereas an almost complete repair was observed within 1 h postirradiation in the ctl cells, PARP3kd cells required about 5 h to repair X-ray-induced lesions. Together, these findings suggest that PARP3 is mainly required for efficient repair of DSBs.
PARP1 and PARP3 Act Synergistically in Response to X-Irradiation in Human Cells and Mouse.
To further evaluate the consequence of PARP3 depletion on long-term sensitivity to X-irradiation, we measured the colony-forming ability of the PARP3kd and ctl cells to increasing doses of X-rays (Fig. S1E). Strikingly, no significant difference between both cell lines was observed, likely suggesting a compensating repair activity over time by another DNA damage-induced PARP family member (i.e., PARP1 and/or PARP2). We verified this hypothesis by comparing the radioinduced sensitivity of PARP3kd and ctl cells to 1-Gy X-irradiation in the absence and in the presence of 100 nM of the PARP inhibitor Ku-0058948 (a concentration sufficient to inhibit PARPs 1 and 2, but not 3; Fig. 1D). The additional inhibition of the remaining PARP activity in PARP3kd cells significantly reduced their survival (by twofold) after X-irradiation compared with mock-treated PARP3kd cells, thus emphasizing a role of PARP1 and/or PARP2 as rescued factors in a PARP3-depleted context.
To extend these cellular studies to an in vivo system, we generated Parp3−/− mice by breeding mice carrying a conditional Parp3 (flox) allele with mice expressing a ubiquitously active CMV-cre transgene (Fig. S2A). Considering the previously described physical and functional interaction of PARP1 and PARP3 in human cells (6) and the above results, we created and bred Parp1+/−;Parp3+/− double heterozygotes to evaluate the combined effects of the Parp1 and Parp3 null mutations (Fig. S2C). Mice of the different genotypes were obtained at approximately Mendelian frequency (Fig. S3). The mice are viable, fertile, and develop normally without any abnormal overall phenotype identified at the mean age of 15 mo. However, when we compared their sensitivity to 4-Gy whole-body X-irradiation (Fig. 1E), we observed that the combined loss of Parp1 and Parp3 significantly decreased their survival, because only 4 of 11 Parp1−/−;Parp3−/− (37%) mice were alive 126 d postirradiation compared with a mild sensitivity of the Parp1−/−;Parp3+/+ mice (7/9 alive, 78%). In contrast, the single disruption of Parp3 in mice did not increase radiosensitivity similar to PARP3 depletion in human cells (Fig. S1E) because 9/9 Parp1+/+;Parp3−/− or Parp1+/+;Parp3+/+ (100%) were still alive 1 y postirradiation. Together, these findings suggest the possibility that PARP1 and its activity might efficiently compensate for the absence of PARP3 in long-term response to DNA damage and indicate a functional synergistic cross-talk between both enzymes for maintaining genome integrity.
PARP3 Interacts with the Mitotic Components NuMA and Tankyrase 1.
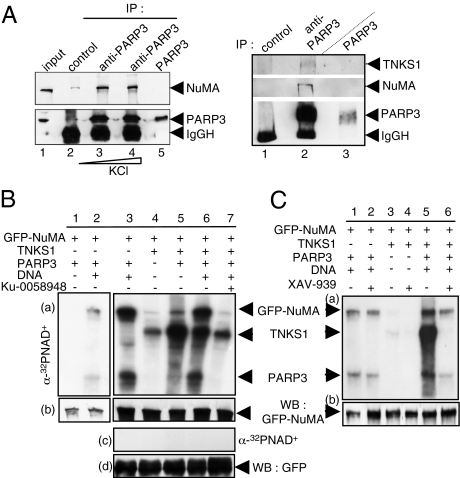
To understand the biochemical basis of PARP3 functions, we looked for PARP3-specific partners. Cos1 whole-cell extracts were immunoprecipitated with a purified anti-PARP3 antibody or an irrelevant control antibody, and coimmunoprecipitating proteins were analyzed by mass spectrometry (Fig. S4A). Among the different partners identified, we isolated 12 tryptic peptides from the large nuclear mitotic apparatus protein NuMA, a microtubule-associated protein involved in spindle dynamics (10). To further confirm the efficiency of the interaction, the anti-PARP3 immunoprecipitates were submitted to increasing stringency conditions of the washing steps and probed for the presence of NuMA by Western blot (Fig. 2A, Left). Coimmunoprecipitation of NuMA was detected after washing with buffer containing up to 500 mM KCl and 0.1% Nonidet P-40 (lanes 3 and 4). Because NuMA was previously identified as a major acceptor of tankyrase 1-mediated poly(ADP ribosyl)ation (11, 12), we tested the PARP3 immunoprecipitates for the presence of tankyrase 1 (Fig. 2A, Right). We detected significant association of both tankyrase 1 and NuMA with PARP3 in Cos1 cells (lane 2), but no association was detected using the control antibody (lane 1). Taken together, these results identify a protein complex containing PARP3, NuMA, and tankyrase 1.
Fig. 2.
Physical and functional association of PARP3 with NuMA and tankyrase 1. (A Left) Coimmunoprecipitation of NuMA with PARP3 after increasing stringency conditions of washing buffers. Cos1 cell extracts were immunoprecipitated with a control antibody (lane 2) or an anti-PARP3 antibody (lanes 3 and 4) and analyzed by Western blotting using successively anti-NuMA and anti-PARP3 antibodies. Input corresponds to 1/13 of the total amount of cell extract used for immunoprecipitation. Lane 5, purified recombinant PARP3 (10 ng). (A Right) Coimmunoprecipitation of tankyrase 1 and NuMA with PARP3. Cos1 cell extracts were immunoprecipitated with a control antibody (lane 1) or an anti-PARP3 antibody (lane 2) and analyzed by Western blotting using successively anti-NuMA, anti-tankyrase 1 and anti-PARP3 antibodies. Lane 3, purified recombinant PARP3 (10 ng). (B) PARP3 induces the ADP ribosylation of NuMA both directly and through tankyrase 1. (a and b) Immunopurified GFP-NuMA was incubated with purified PARP3 and/or tankyrase 1 (TNKS1) as indicated in PARP activity buffer. The addition of Ku-0058948 (250 nM) significantly inhibits PARP3 but not tankyrase 1 (lane 7 vs. lanes 2 and 3). (c and d) In similar experimental conditions as above, no ADP ribosylation of GFP alone was detected. (a and c) Autoradiography. (b and d) Immunopurified GFP or GFP-NuMA were analyzed by Western blotting using an anti-GFP antibody. (C) PARP3 stimulates the auto-ADP ribosylation of tankyrase 1. Immunopurified GFP-NuMA were incubated with purified PARP3 or tankyrase 1 and assayed for PARP activity as above. The addition of XAV-939 (500 nM) inhibits efficiently tankyrase 1 but not PARP3 (compare lanes 4 with 3, and 2 with 1). (a) Autoradiography. (b) Immunopurified GFP-NuMA was analyzed by Western blotting using an anti-GFP antibody.
PARP3 Stimulates the ADP Ribosylation of NuMA both Directly and Through Tankyrase 1.
To gain further insights into the functional interactions governing this protein network, we next compared the ability of either PARP3 or tankyrase 1 to ADP ribosylate NuMA. Immunopurified GFP-NuMA (Fig. 2B, a and b) or GFP alone as a control (Fig. 2B, c and d) were incubated with purified PARP3, tankyrase 1, or both in the presence of α-32PNAD+ and DNase-I–treated calf thymus DNA when indicated.
No visible PARP3 catalyzed automodification and ADP ribosylation of NuMA was detected in the absence of DNA (Fig. 2B, a lane 1). However, in agreement with its previously described ability to bind DNA (6), both the automodification of PARP3 and the heteromodification of NuMA were highly stimulated by the addition of fragmented DNA in the reaction buffer, thus identifying NuMA as a major acceptor for covalent modification by PARP3 in the presence of DNA (lanes 2 and 3). Using GFP-tagged deletion domains of NuMA, we next found that NuMA is ADP ribosylated onto its C-terminal domain encompassing the nuclear localization signal and the microtubule binding region (Fig. S4B). Furthermore, in agreement with previous reports (12), we detected an ADP ribosylation of NuMA by tankyrase 1 (lane 4). Importantly, ADP ribosylation of tankyrase 1 and, in turn, of NuMA were greatly enhanced in the presence of functional PARP3 even under condition of a poor activation of PARP3 (without fragmented DNA; compare lanes 5 and 4). The addition of DNA further amplified this reaction by stimulating the direct PARP3 catalyzed heteromodification of NuMA (lane 6). In contrast, when PARP3 is inactivated by the addition of the PARP inhibitor Ku-0058948, both stimulations are lost and only the tankyrase 1-catalyzed ADP ribosylation is detected (lane 7 vs. 4). In similar experimental conditions, the ADP ribosylation of GFP alone was never observed (Fig. 2B, c and d). Together, these results indicate that PARP3 stimulates the ADP ribosylation of tankyrase 1 and increases its ability to modify NuMA.
To next verify whether PARP3 targets tankyrase 1 or stimulates its automodification, we used the specific tankyrase 1 inhibitor XAV-939 (Fig. 2C and Fig. S4C). We first verified that XAV-939 significantly inhibited tankyrase 1 activity but not PARP3 (Fig. 2C, a lane 2 vs. 1 and lane 4 vs. 3; Fig. S4C, lane 2 vs. 1 and lane 4 vs. 3). Again, a significant increase in the ADP ribosylation of tankyrase 1 and NuMA is observed in the presence of PARP3 (Fig. 2C, a lane 5). However, the specific inhibition of tankyrase 1 completely abrogated its ADP ribosylation activity and only the PARP3 catalyzed modification of NuMA was detected (Fig. 2C, a, lane 6; Fig S4C, lanes 6 and 7).
Together, these in vitro data provide a convincing demonstration that (i) active PARP3 stimulates the auto-ADP ribosylation of tankyrase 1 and in turn its ability to modify NuMA in a DNA-independent manner, and (ii) PARP3 is able to directly ADP ribosylate NuMA in a DNA-dependent manner.
PARP3 Is Required for Mitotic Spindle Integrity During Mitosis.
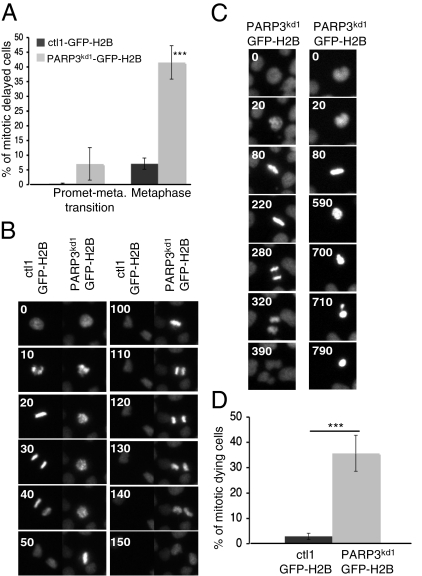
Tankyrase 1 and NuMA are now recognized as key regulators of mitotic progression (12– 14). To investigate the role of PARP3 in this process, we generated ctl and PARP3kd cell lines that constitutively and stably express GFP-H2B (ctl-GFP-H2B and PARP3kd-GFP-H2B) and followed mitosis by video time-lapse microscopy. Depletion of PARP3 induced a significant increase in mitotic duration compared with the control cell line (Fig. S5B). More precisely, PARP3kd-arrested cells fell into two distinct classes (Fig. 3A). Whereas 7% of PARP3kd mitotic cells remained delayed in the prometaphase-to-metaphase transition (Fig. 3B, Fig. S5C, and Movie S1), a major proportion (>40%) remained arrested in metaphase (Fig. 3C, Fig. S5D, and Movies S2 and S3), often resulting in mitotic cell death (Fig. 3D). A similar metaphase arrest was previously observed in tankyrase 1-depleted cells and was associated with persistent telomere associations and inappropriate NHEJ-directed fusions, but also impaired NuMA-mediated bipolar spindle assembly (11, 13, 15).
Fig. 3.
PARP3 is required for efficient mitotic progression. (A) Histogram showing an increase in the percentage of PARP3kd1-GFP-H2B mitotic cells displaying a delay in prometaphase-to-metaphase transition or a delay in metaphase compared with normal mitotic progression in ctl1-GFP-H2B. For each step, >100 mitoses were scored by life cell microscopy. ***P < 0.0001. (B) Representative time-lapse video live-cell imaging of ctl1-GFP-H2B and PARP3kd-GFP-H2B cells. Whereas a normal mitotic progression is observed in the control cell, PARP3kd-GFP-H2B remain delayed for up to 80 min in the prometaphase-to-metaphase transition. (C) Representative time-lapse video live-cell imaging of the PARP3kd-GFP-H2B cell line showing either a delay of ∼220 min in metaphase (Left) or a metaphase arrest resulting in mitotic cell death (Right). (D) Frequencies of mitotic cell death in ctl1-GFP-H2B and PARP3kd-GFP-H2B cells. More than 100 mitoses were scored by life cell microscopy. ***P < 0.0001.
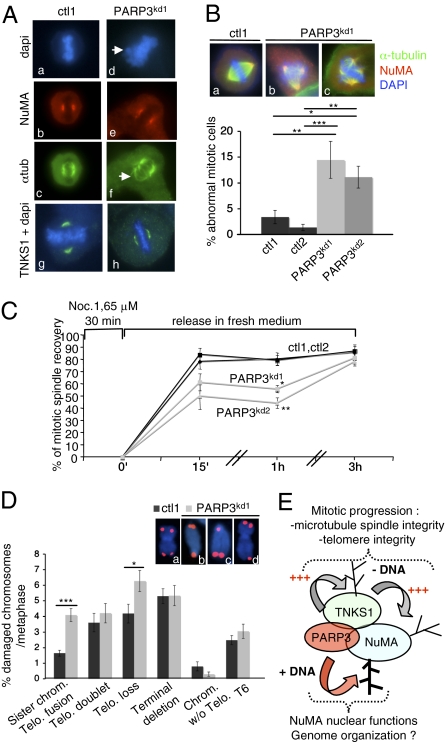
The physical and functional interaction of PARP3 with NuMA and its binding partner tankyrase 1 reported here raises the question of a role of PARP3 in the microtubule assembly activity of NuMA. We therefore probed ctl and PARP3kd cells for mitotic spindle integrity by immunofluorescence studies. The absence of PARP3 did not prevent the accumulation of NuMA or tankyrase 1 at spindle poles (Fig. 4A) but induced a significant accumulation of abnormal mitotic figures displaying an aberrant metaphase configuration with extra α-tubulin and NuMA-containing microtubule organizing centers (Fig. 4B), as detected in tankyrase 1-depleted cells (11). In addition, bipolar PARP3kd mitosis often displayed splayed microtubules associated with chromosome misalignment compared with compacted focused spindles observed in control cells, indicating a defect in the integrity of the microtubule spindle (Fig. 4A; compare lanes d and f with lanes a and c). To confirm this observation and further probe the role of PARP3 in spindle dynamics, we performed spindle regrowth assays after release from nocodazole depolymerization (Fig. 4C). Whereas microtubule assembly was restored within 15 min in control cells, up to 3 h were required for PARP3kd cells. Together, these results describe PARP3 as an additional regulator of the mammalian spindle pole function.
Fig. 4.
PARP3 is required for microtubule spindle dynamics and telomere integrity. (A) Spindle defects in PARP3-depleted cells. (a–f) Representative images of NuMA (red)/α tubulin (green) coimmunostaining of control (ctl1) and PARP3kd metaphases, counterstained with DAPI (blue). Note the splayed microtubules in PARP3kd cells (arrow) compared with a focused spindle in ctl1 cells, despite a wild-type–like accumulation of NuMA to the spindle poles (e and f vs. b and c). (g and h) Representative images of tankyrase 1 (TNKS1) (green) immunostaining of control (ctl1) and PARP3kd metaphases showing a normal targeting of TNKS1 to the spindle poles. DNA is counterstained with DAPI (blue). (B) Accumulation of abnormal mitotic cells in PARP3kd cells. Percentage of abnormal mitotic figures with supernumerary spindle poles in PARP3kd cells (b and c) vs. ctl cells (a) determined by coimmunodetection of α-tubulin and NuMA and DNA staining with DAPI. An average of 65 mitotic cells were scored per cell line in >20 randomly selected immunofluorescence fields. Results are averages of five independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Spindle microtubule regrowth is delayed in PARP3kd cells. Microtubules were depolymerized by nocodazole treatment as indicated, and repolymerized at 37 °C. At the indicated time points, cells were fixed, coimmunostained for NuMA and α-tubulin, and scored for the formation of regular compact spindle. Over 45 cells were scored for each independent cell line. Results are averages from three independent experiments. *P < 0.05, **P < 0.01. (D) Spontaneous increase in sister telomere fusions and sister telomere loss in PARP3kd1 compared with ctl1 cells. Telomere aberrations were detected by FISH on metaphase spreads and expressed as percentages of damaged chromosomes per metaphase. (Insets) Sister telomere fusions (b and c) and telomere loss (d) identified in PARP3kd metaphases compared with normal telomeres (a) observed in ctl1 cells. (E) Working model posing the dual functions of PARP3 in association with NuMA. Within the mitotic protein network containing tankyrase 1 and NuMA, PARP3 stimulates (indicated by +++) the tankyrase 1 catalyzed auto-ADP ribosylation and hetero-ADP ribosylation of NuMA to favor telomere integrity and spindle dynamics in a DNA-independent manner. In addition, PARP3 is able to directly ADP ribosylate NuMA in a DNA-dependent manner, a possible means of regulating its functions in the interphase nucleus. Gray arrow, tankyrase 1-catalyzed poly(ADP ribosyl)ation; red arrow, PARP3-catalyzed ADP ribosylation.
PARP3 Is Required for Telomere Stability.
As described above (Fig. 1B), PARP3kd cells display spontaneous genome instability. In addition to their localization and function at spindle poles, NuMA and tankyrase 1 localize to the nuclear matrix and telomeres, respectively, where they play a role in genome integrity (16, 17). Furthermore, PARP3 associates with proteins from the NHEJ (9), a process causing sister telomere fusions in the absence of tankyrase 1 (15). To explore a potential role for PARP3 in genome stability specifically at telomeres in mitosis, we monitored spontaneous telomere aberrations using FISH analysis on metaphase spreads. As shown in Fig. 4D, PARP3kd cells displayed a significant and specific increase in sister telomere fusions, as observed previously in tankyrase 1-depleted cells (15). In addition, we observed sister telomere loss evidencing spontaneous genomic instability, as reported previously (Fig. 1B). Other chromosome abnormalities, such as dicentric chromosomes, telomere doublets, and terminal deletions, were not affected. Together, these results are consistent with a role of PARP3 in telomere function and stability.
Discussion
Although recent studies have provided insights into the biochemical and structural properties of PARP3 (7, 8), its physiological functions are unknown. In this study, we provide in vivo evidence for two distinct roles of PARP3 in genome maintenance and mitotic progression.
A potential role of PARP3 in cellular response to DNA damage has been suggested by its interaction with components of the BER/SSBR and NHEJ repair machineries (9). Consistent with this hypothesis, we show here that endogenous PARP3 accumulates at laser-induced DNA damage sites in human, monkey, and mouse cells independently of PARP activity. In addition, PARP3 automodification appears to be efficiently stimulated by fragmented DNA in agreement with its ability to bind DNA in a Southwestern assay (6). Together, these data suggest a role of PARP3 in cellular response to DNA damage. Accordingly, we found that the knockdown of human PARP3 confers specific susceptibility to DSBs as revealed by prolonged persistence of unrepaired X-rays that induced γH2AX foci, whereas SSBR remained unaffected (18). Therefore, although the basic features of DNA damage recognition by PARP3 still needs to be addressed, our data emphasize a particular role of PARP3 in cellular response to DSBs.
What could be the function of PARP3 in this process? FACS analysis shows that PARP3 is dispensable for G2/M cell-cycle checkpoint after ionizing radiation (Fig. S5A). Based on recent biochemical studies, it has been proposed that PARP3 interacts with and activates PARP1 (6, 18). One interesting scenario would be that PARP3 serves to accelerate PARP1-dependent DSB repair. In line with this idea, the loss of PARP3 in either human cells or mice did not significantly impact long-term survival after X-irradiation. In contrast, the additional inhibition of PARP1 in PARP3-depleted cells and the genetic ablation of both Parp1 and Parp3 in mice clearly increased hypersensitivity to ionizing radiation. Together, these findings suggest a functional synergy of PARP1 and PARP3 in cellular response to DNA damage.
An alternative hypothesis can be based on its previously described association and colocalization with the epigenetic chromatin modifiers, PcG proteins (9). Recently, emerging studies implicate the PcG proteins in DSBs responses (19). The work by Hong et al. (20) identified a Ku70/Ku80-dependent recruitment of the human PcG member PHF1 to DSB where it contributes in the efficiency of the NHEJ pathway. By analogy, a possible molecular mechanism underlying a PARP3/PcG network that works in DSBs repair remains an exciting issue to be investigated.
In this study, we also discovered essential functions of PARP3 in orchestrating the progression through mitosis by at least two mechanisms that are not necessarily exclusive: (i) influencing spindle microtubule organization and stabilization and (ii) promoting telomere integrity. Furthermore, we identify PARP3 as part of a protein network containing tankyrase 1 and NuMA. Tankyrase 1 was previously described as the PARP member that polymerizes spindle-associated poly(ADP ribose) and ADP ribosylates NuMA (11, 12, 21). Cellular studies proposed a role of this modification in the assembly of the bipolar spindle and/or the release of telomere from the nuclear matrix or spindle poles for exit from anaphase. From this and our in vivo and biochemical investigations, we speculate that the role of PARP3 in this protein complex is to act as a positive regulator of tankyrase 1-mediated poly(ADP ribosyl)ation of NuMA in a DNA-independent manner, that in turn controls specific mitotic functions (i.e., spindle stabilization and telomere function; Fig. 4E). According to this model, we found that the depletion of human PARP3 results in both tankyrase 1 and NuMA-like mitotic phenotypes, although expressed to different extents (11, 14, 22). The loss of PARP3 induced mild spindle defects characterized by the appearance of either supernumerary poles or bipolar pole with splayed microtubules and delayed spindle assembly. However, we did not observe the detachment of centrosomes from mitotic spindles and the resulting microtubule defocusing as identified in NuMA-disrupted MEFs (14). In addition, similar to tankyrase-depleted cells (15), we also detected sister telomere fusions in PARP3kd cells, which, consistent with its association with proteins from the NHEJ pathway, suggests a role of PARP3 in the regulation of inappropriate NHEJ-directed telomere fusions. Notably, both tankyrase 1 and NuMA localized properly to spindle poles. Thus, PARP3 might not be essential for all aspects of tankyrase and NuMA functions. As such, the striking mitotic cell death detected in PARP3kd cells might result from the combined effects of PARP3 on NuMA, tankyrase 1, and yet-to-be-identified novel mitotic acceptors.
What is then the functional role of the markedly DNA-stimulated modification of NuMA by PARP3? In addition to its location at mitotic spindle poles, NuMA has been reported as an abundant component of interphase nuclei (16, 23). Similarly, we identified both a nuclear and centrosomal human PARP3 isoform (this work and ref. 6). Although the precise nuclear functions of NuMA remain unclear, cumulative evidences point toward potential roles in nuclear organization and gene regulation as a structural constituent of the nuclear matrix (10). On these bases, an attractive scenario suggests that the DNA-activated nuclear PARP3 might contribute to genome reorganization through the ADP ribosylation of NuMA, in addition to its active role in genome integrity, reported above (Fig. 4E). Furthermore, this hypothesis would suggest that the nuclear and centrosomal PARP3 are qualitatively different. Whether only the centrosomal isoform of PARP3 associates with NuMA and tankyrase 1 to regulate mitotic functions remains to be investigated.
In summary, our findings identify PARP3 as (i) a new player in cellular response to DNA damage and (ii) a key component of a protein complex containing NuMA and tankyrase 1, in which the tankyrase 1-catalyzed ADP riboslation of NuMA is stimulated to facilitate the formation and maintenance of the mitotic spindle and genome integrity. Importantly, recent work uncovered a requirement of NuMA in centrosome clustering, an adapted mechanism developed by mature cancer cells to circumvent multipolar divisions that can lead to aneuploidy and cell death (24, 25). Therefore, our findings create fascinating prospects for PARP3 in new, targeted anticancer strategies to suppress centrosome clustering. Finally, the DNA-dependent (ADP ribosyl)ation of NuMA by PARP3 creates challenging new avenues in the search for a potential role of PARP3 activity in the nuclear functions of NuMA in interphase cells (Fig. 4E).
Materials and Methods
Detailed information on the experimental procedures and reagents used are provided in SI Materials and Methods. Mice carrying a conditional Parp3 allele (with loxP-flanked exons 3–5) were engineered as described in Fig. S2 and crossed with CMV-cre transgenic mice producing Parp3+/− offspring. Mice were X-irradiated using a Pantak Seifert X-ray system operating at 200 kV, 4.5 mA, and equipped with a 1-mm-thick aluminum filter. ctl and PARP3kd MRC5 cells were generated as detailed in SI Materials and Methods. For GFP-H2B–expressing cells, ctl1 and PARP3kd1 MRC5 clones were transfected with GFP-H2B and selected as described in SI Materials and Methods. For immunofluorescence, cells were fixed with 4% paraformaldehyde in PBS and stained with the indicated antibodies as described in SI Materials and Methods. For local DNA damage induction, cells were sensitized using BrdU (10 μg/mL). Microirradiation was carried out as detailed in SI Materials and Methods. Cells were X-irradiated using a Pantak Seifert X-ray system operating at 100 kV, 4.5 mA. Colony-forming assays and neutral COMET assays were performed as detailed in SI Materials and Methods. Live videomicroscopy was carried out as described in SI Materials and Methods. Metaphase spreads and analysis of telomere aberrations were performed as described in SI Materials and Methods. Cos1 cells were immunoprecipitated using a purified anti-PARP3 (4698) or a rabbit anti-mouse antibody as control as detailed in SI Materials and Methods. Coprecipitated proteins were analyzed by Nano-LC-MS/MS experiments or by standard immunoblotting using the appropriate antibodies as detailed in SI Materials and Methods. In vitro poly(ADP ribosyl)ation assays were performed using immunopurified GFP-NuMA or GFP, purified PARP3, and/or purified tankyrase 1 in activity buffer containing α-32PNAD+ and DNase I-activated calf thymus DNA as detailed in SI Materials and Methods. Statistical analyses were determined by ANOVA tests as indicated by P values using StatView software.
Acknowledgments
The authors thank C. Cleveland for the NuMA expression construct, N. Martin for Ku-0058948, H. Leonhardt for access to the microirradiation platform, and N. Magroun, A. Noll, and V. Gasser for technical assistance. The mouse mutant line was established at the Mouse Clinical Institute in the Department of Targeted Mutagenesis and Transgenesis, Illkirch, France. This work was supported by Electricité de France, l'Agence Nationale pour la Recherche, Centre National de la Recherche Scientifique, and Région Alsace.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016574108/-/DCSupplemental.
References
- 1.Schreiber V, Dantzer F, Ame J-C, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Johansson M. A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologues. Genomics. 1999;57:442–445. doi: 10.1006/geno.1999.5799. [DOI] [PubMed] [Google Scholar]
- 4.Urbánek P, Paces J, Králová J, Dvorák M, Paces V. Cloning and expression of PARP-3 (Adprt3) and U3-55k, two genes closely linked on mouse chromosome 9. Folia Biol (Praha) 2002;48:182–191. [PubMed] [Google Scholar]
- 5.Rouleau M, El-Alfy M, Lévesque MH, Poirier GG. Assessment of PARP-3 distribution in tissues of cynomolgous monkeys. J Histochem Cytochem. 2009;57:675–685. doi: 10.1369/jhc.2009.953380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augustin A, et al. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci. 2003;116:1551–1562. doi: 10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- 7.Lehtiö L, et al. Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J Med Chem. 2009;52:3108–3111. doi: 10.1021/jm900052j. [DOI] [PubMed] [Google Scholar]
- 8.Rulten SL, et al. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Rouleau M, et al. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J Cell Biochem. 2007;100:385–401. doi: 10.1002/jcb.21051. [DOI] [PubMed] [Google Scholar]
- 10.Radulescu AE, Cleveland DW. NuMA after 30 years: The matrix revisited. Trends Cell Biol. 2010;20:214–222. doi: 10.1016/j.tcb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 12.Chang W, Dynek JN, Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J. 2005;391:177–184. doi: 10.1042/BJ20050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 14.Silk AD, Holland AJ, Cleveland DW. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J Cell Biol. 2009;184:677–690. doi: 10.1083/jcb.200810091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiao SJ, Smith S. Sister telomeres rendered dysfunctional by persistent cohesion are fused by NHEJ. J Cell Biol. 2009;184:515–526. doi: 10.1083/jcb.200810132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abad PC, et al. NuMA influences higher order chromatin organization in human mammary epithelium. Mol Biol Cell. 2007;18:348–361. doi: 10.1091/mbc.E06-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 18.Loseva O, et al. PARP-3 is a mono ADP ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem. 2010;285:8054–8060. doi: 10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck SA, Falconer E, Catching A, Hodgson JW, Brock HW. Cell cycle defects in polyhomeotic mutants are caused by abrogation of the DNA damage checkpoint. Dev Biol. 2010;339:320–328. doi: 10.1016/j.ydbio.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 20.Hong Z, et al. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 2008;36:2939–2947. doi: 10.1093/nar/gkn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 22.Haren L, Gnadt N, Wright M, Merdes A. NuMA is required for proper spindle assembly and chromosome alignment in prometaphase. BMC Res Notes. 2009;2:64. doi: 10.1186/1756-0500-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merdes A, Cleveland DW. The role of NuMA in the interphase nucleus. J Cell Sci. 1998;111:71–79. doi: 10.1242/jcs.111.1.71. [DOI] [PubMed] [Google Scholar]
- 24.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 25.Kwon M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]