Abstract
Microbial colonization of mammals is an evolution-driven process that modulate host physiology, many of which are associated with immunity and nutrient intake. Here, we report that colonization by gut microbiota impacts mammalian brain development and subsequent adult behavior. Using measures of motor activity and anxiety-like behavior, we demonstrate that germ free (GF) mice display increased motor activity and reduced anxiety, compared with specific pathogen free (SPF) mice with a normal gut microbiota. This behavioral phenotype is associated with altered expression of genes known to be involved in second messenger pathways and synaptic long-term potentiation in brain regions implicated in motor control and anxiety-like behavior. GF mice exposed to gut microbiota early in life display similar characteristics as SPF mice, including reduced expression of PSD-95 and synaptophysin in the striatum. Hence, our results suggest that the microbial colonization process initiates signaling mechanisms that affect neuronal circuits involved in motor control and anxiety behavior.
Keywords: developmental programming, microbiome, basal ganglia, cognitive behavior, synapse
Early life environmental influences have a profound impact on the organism's later development, structure, and function. This phenomenon is called “developmental programming,” a process whereby an environmental factor acting during a sensitive or vulnerable developmental period exerts effects that impact on structure and function of organs that, in some cases, will persist throughout life (1). One such environmental factor is the gut microbiota that, because of an evolutionary process, has adapted to coexist in commensal or symbiotic relationship with mammals (2). Immediately after birth, the newborn organism is rapidly and densely populated with complex forms of indigenous microbes. This process has been shown to contribute to developmental programming of epithelial barrier function, gut homeostasis, and angiogenesis, as well as the innate and host adaptive immune function (3, 4). Recent data indicate that gut microbiota have systemic effects on liver function (5–7), thus raising the possibility that gut microbiota can have developmental effects in other organs elsewhere in the body.
The functional development of the mammalian brain is of particular interest because it has been shown to be susceptible to both internal and external environmental cues during perinatal life. Epidemiological studies have indicated an association between common neurodevelopmental disorders, such as autism and schizophrenia, and microbial pathogen infections during the perinatal period (8, 9). These findings are supported by experimental studies in rodents, demonstrating that exposure to microbial pathogens during similar developmental periods result in behavioral abnormalities, including anxiety-like behavior and impaired cognitive function (10–12). In a recent study, it was shown that the commensal bacteria, Bifidobacteria infantis, could modulate tryptophan metabolism, suggesting that the normal gut microbiota can influence the precursor pool for serotonin (5-HT) (13).
Here, we tested the hypothesis that the “normal” gut microbiota is an integral part of the external environmental signals that modulate brain development and function.
Results
Germ Free (GF) Mice Display Increased Motor Activity and Reduced Anxiety-Like Behavior.
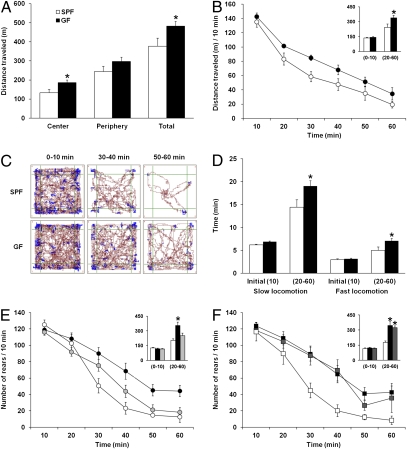
In the first set of experiments, we subjected adult GF and specific pathogen free (SPF) mice with a normal gut microbiota to a battery of tests for exploratory activity and anxiety. GF and SPF mice were placed in a novel, open-field activity box. Their spontaneous motor activity, including locomotor and rearing activities, were measured for 60 min. GF mice showed greater total distance traveled and more exploration of the center of the open field (P < 0.05; Fig. 1A). There was also a trend for GF to display higher levels of rearing activity compared with SPF (GF vs. SPF, 489 ± 43 vs. 369 ± 50, P = 0.088). Both GF and SPF mice displayed similar locomotor activity (Fig. 1B) during the initial open field exposure, indicating that the increased locomotor activity in GF mice was not triggered by novelty. Instead, significant differences between groups were detected in habituation over time (repeated measures ANOVA, main effect F(1, 70) = 6.28, P < 0.05). Thus, GF mice traveled a significantly longer distance (Fig. 1 B and C) and spent significantly (P < 0.05) more time in both slow and fast locomotion (Fig. 1D) during the 20- to 60-min interval of testing.
Fig. 1.
GF mice display increased spontaneous motor activity. (A) Bars show cumulative distance traveled (meters) per zone and in the entire box (total) during the 60-min open field test session by SPF (open bars) and GF (filled bars) mice. (B) Average distance traveled (meters) measured in 10-min time bins across a 60-min session in an open field box. (Inset) Bars show cumulative distance traveled (meters) during the initial 10 min and the 20- to 60-min time interval of open field testing. (C) Representative tracks of movement patterns of SPF and GF mice at the 0–10, 30–40, and 50–60 min time intervals of the 60-min open field test session; distance traveled and rearing activity is shown in dark red and blue colors, respectively. (D) The time that SPF and GF mice spent in slow (>5 cm/s) or fast (>20 cm/s) locomotion during the initial 10 min of testing and the 20–60 min time interval. (E) Rearing activity of SPF (white), GF (black), and conventionalized (CON; light gray) mice. Circles show the average number of rears measured in 10-min time bins across a 60-min session in an open field box. (F) Rearing activity of SPF, GF, and adult CON mice (dark gray); lines connecting cumulative data in B, E, and F were drawn for clarity only. All data (A, B, and D–F) are presented as means (± SEM; n = 7–14 per group). *P < 0.05 compared with SPF mice.
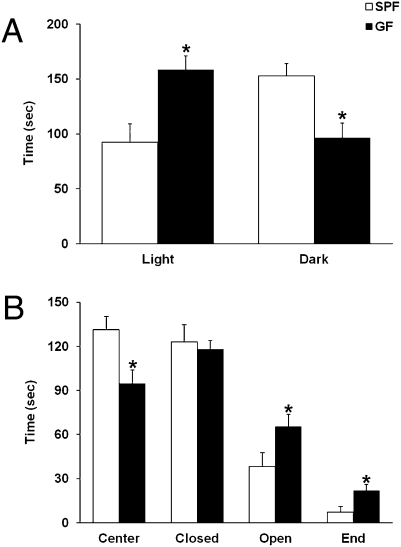
Given that certain microbial pathogens have been reported to induce anxiety-like behavior in animal models (10–12), we assessed whether the nonpathogenic gut microbiota could also affect anxiety-like behavior. For this purpose, we used two rodent tests of anxiety: the light–dark box test and the elevated plus maze (14). In the light–dark box test, GF mice spent significantly (P < 0.05) more time in the light compartment of the box than control SPF mice (Fig. 2A). In the elevated plus maze test, GF mice spent significantly (P < 0.05) more time in the open arm than SPF mice (Fig. 2B and Movies S1 and S2). The GF mice also engaged in riskier behavior than SPF as indicated by the great number of visits (GF vs. SPF: 3.9 ± 0.6 vs. 1.4 ± 0.69, P < 0.05) to the ends of the open arms. There were no significant differences in the number of entries (GF vs. SPF: 13.14 ± 0.9 vs. 14.4 ± 1.5, P > 0.1) and time spent (Fig. 2B) in the closed arm between GF and SPF mice.
Fig. 2.
GF mice display reduced anxiety-like behavior. (A) Bars show time (seconds) spent in the light and dark compartments during a 5-min light–dark box test by the SPF and GF mice. (B) Bars show time (seconds) spent in each area of the elevated plus maze by the SPF and GF mice during a 5-min test session. All data (A and B) are presented as means (±SEM; n = 7–9 per group). *P < 0.05 compared with SPF mice.
To test whether conventionalization in early life of GF mice could “normalize” the increased motor activity and alter the anxiety behavior, we conventionalized a new set of GF mice with microbiota obtained from SPF mice 30 d before mating and allowed the progeny to mature in an isolator with bacteria. Adult conventionalized offspring (CON) were behaviorally tested as described above. The rearing activity of the CON mice was significantly different from that of GF mice (P < 0.05) and did not differ from that of SPF mice (Fig. 1E).
CON mice normalized their locomotor activity to that of SPF mice during the later time interval of testing (P < 0.05; Fig. S1), whereas their initial locomotor activity was similar to that of GF mice. In the light–box test, the behavioral pattern of CON mice was similar to that of GF mice and significantly different from that of SPF mice (P < 0.05; Fig. S2). In the elevated plus maze test, the GF mice spent significantly (P < 0.05) more time exploring the open arms compared with SPF mice. Introducing microbiota early in life to GF mice showed that CON mice altered their behavior and spent less time exploring the open arms (Fig. S3).
Encouraged by the observation that early colonization of GF mice could normalize several behavioral patterns of GF mice, we explored whether there is a sensitive/critical period for the effects of the normal gut microbiota on behavior. We therefore conventionalized adult GF mice and studied their behavior in open field test as described above. Notably, conventionalization of adult mice failed to normalize the behavior of GF mice (Fig. 1F and Fig. S4).
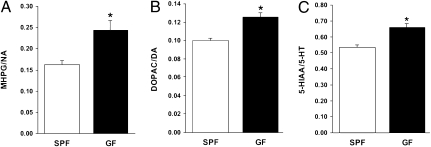
GF Mice Show Elevated Noradrenaline (NA), Dopamine (DA), and 5-HT Turnover in the Striatum.
Anxiety-like behavior has been associated with alterations in monoamine neurotransmission. Therefore, we investigated potential changes in the neurochemistry of GF mice in a new set of animals. The concentration of noradrenaline (NA), dopamine (DA), and 5-HT and their major metabolites, 3-methoxy-4-hydroxyphenylglycol (MHPG), dihydroxyphenylacetic acid (DOPAC), and homovanillic acid (HVA), and 5-hydroxyindoleacetic acid (5-HIAA), were measured in frontal cortex, striatum, and hippocampus of GF and SPF mice. The turnover rate of NA, DA, and 5-HT was significantly higher in the striatum of GF mice compared with SPF mice (Fig. 3). In contrast, no significant differences were found in the frontal cortex or hippocampus (Table S1).
Fig. 3.
GF mice show elevated NA, DA, and 5-HT turnover in the striatum. The histograms depict the mean ratios (± SEM; n = 6 per group) for MHPG/NA (A), DOPAC/DA (B), and 5-HIAA/5-HT (C) in the striatum of male GF and SPF mice. Asterisks denote where GF mice differ significantly (P < 0.01) from SPF mice.
GF Mice Show Altered Expression of Synaptic Plasticity-Related Genes.
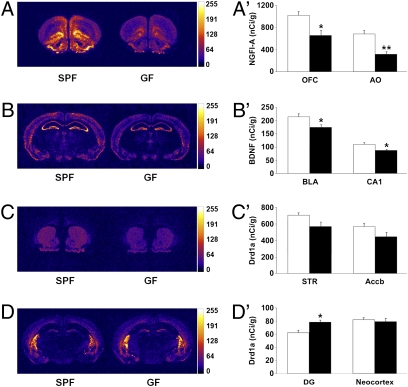
Previous molecular and behavioral studies have implicated the immediate-early gene, nerve growth factor-inducible clone A (NGFI-A), and the synaptic plasticity-related gene, brain-derived neurotrophic factor (BDNF), in the development of anxiety-like behavior (15–17). We therefore studied the expression of these genes in the frontal cortex, striatum, amygdala, and hippocampus of GF and SPF mice, by means of in situ hybridization technique. In GF mice, NGFI-A mRNA expression was significantly lower in various subregions of the prefrontal cortex, including the orbital frontal cortex (Fig. 4 A and A′); as well as in the striatum (GF vs. SPF: 329 ± 33 vs. 586 ± 18, P < 0.0001), hippocampus (CA1 region, GF vs. SPF: 258 ± 15 vs. 499 ± 22, P < 0.0001; CA3 region, GF vs. SPF: 166 ± 13 vs. 236 ± 6, P < 0.001; dentate gyrus, GF vs. SPF: 76 ± 4 vs. 113 ± 5, P < 0.0001) and amygdala (GF vs. SPF: 126 ± 17 vs. 212 ± 19, P < 0.01) compared with SPF mice. Similarly, GF mice had significantly lower BDNF mRNA expression in the hippocampus, amygdala (Fig. 4 B and B′), and cingulate cortex (GF vs. SPF: 162 ± 6 vs. 193 ± 10, P < 0.05), which are key components of the neural circuitry underlying anxiety and fear (18).
Fig. 4.
GF mice show altered expression of anxiety and synaptic plasticity-related genes. (A) Representative autoradiograms showing NGFI-A mRNA expression at the level of the frontal cortex of SPF and GF mice (OFC, orbital frontal cortex; AO, anterior olfactory region). (A') Bars show expression of NGFI-A mRNA (nCi/g) in the OFC and AO of SPF and GF mice. (B) Representative autoradiograms showing BDNF mRNA expression at the level of amygdala and dorsal hippocampus of SPF and GF mice (BLA, basolateral amygdala; CA1, CA1 region of the dorsal hippocampus). (B') Bars show expression of BDNF mRNA (nCi/g) in the BLA and CA1 region of SPF and GF mice. (C) Representative autoradiograms showing dopamine D1 receptor (Drd1a) mRNA expression at the level of the striatum and nucleus accumbens of SPF and GF mice (STR, striatum; Accb, nucleus accumbens, shell region). (C') Bars show expression of Drd1a mRNA (nCi/g) in the STR and Accb of SPF and GF mice. (D) Representative autoradiograms showing Drd1a mRNA expression at the level of the dorsal hippocampus of SPF and GF mice (DG, dentate gyrus; PtCx, parietal cortex, somatosensory area). (D') Bars show expression of Drd1a mRNA (nCi/g) in the DG and PtCx of SPF and GF mice. All data (A'–D') are expressed as means ± SEM, n = 8 per group. Filled bars represent GF mice. Open bars represent SPF mice. *P < 0.05, *P < 0.001 compared with SPF mice.
Given that DA is an important regulator of motor and cognitive functions (19), we examined the expression of dopamine receptors (D1 and D2 receptors) and intracellular signaling mechanisms (i.e., DARPP-32) in the above regions. In GF mice, DA D1 receptor mRNA was significantly (P < 0.05) higher in the hippocampus (Fig. 4 D and D'), while lower in the striatum and nucleus accumbens, albeit without significance (Fig. 4 C and C') compared with SPF mice. There were no significant differences in the expression of DARPP-32 or DA D2 receptors between GF and SPF mice.
GF Mice Show Alterations in Genes Involved in Four Canonical Pathways.
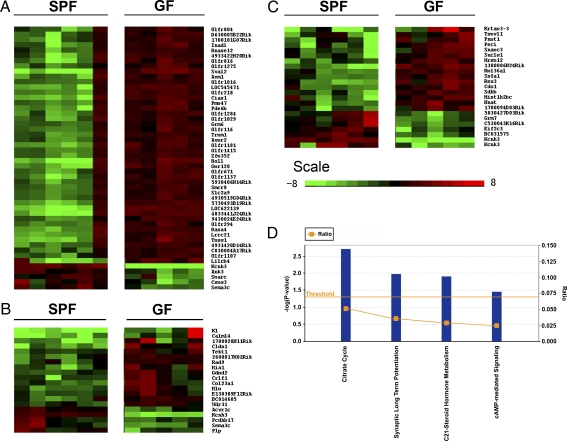
Having established that the normal gut flora could modify behavior and gene expression in key brain regions, we applied the gene expression profiling technique to assess whether other genes were subject to modulation by gut microbiota. The gene expression patterns of five brain regions of 11 mice (5 GF mice and 6 SPF mice) were profiled. Genes differentially expressed between GF and SPF mice (with significant fold changes >2) were used to generate heat maps by using Cluster and Tree View software. The gene expression profile was very consistent across independent tissue samples of the same brain region (see patterns of green or red color). In the hippocampus, we found 50 genes to be significantly differentially expressed (45 genes were higher and 5 were lower in GF mice compared with SPF mice; Fig. 5A). Moreover, 20 and 23 genes were significantly, differentially expressed in the cortex and striatum, respectively (Fig. 5B and C). In addition, 84 genes were differentially expressed in the cerebellum and only 1 gene in hypothalamus. For quantitative real-time PCR (Q-PCR) confirmation of microarray data, we selected 44 genes of the modulated genes described above and 6 candidate genes implicated in the behavioral phenotype of GF mice. We confirmed 38 of these 50 genes to be differentially expressed (35 in hippocampus, 11 in cortex, and 4 in striatum), with three genes common to all regions (Indo, Tlr1, and Gna1) (Table S2–S4). These gut microbiota-regulated genes were characterized into four statistically significant canonical pathways (Fig. 5D).
Fig. 5.
Expression profiling of GF mice and SPF mice brains. A heatmap of genes showing statistically significant (q < 5%) and fold change (>2) differences, between SPF (n = 6) and GF (n = 5) mice in the hippocampus (A), frontal cortex (B), and striatum (C). Each row represents the relative levels of expression of a single gene across all mice; each column represents the levels of expression for a single mouse. The colors red and green denote high and low expression, respectively. Differentially expressed genes were investigated for functional clustering by using Ingenuity Pathway Analysis software for canonical pathways (D), as described in Experimental Procedures.
Gut Microbiota Colonization of GF Mice Reduces Protein Expression of Synaptophysin and PSD-95 in Striatum.
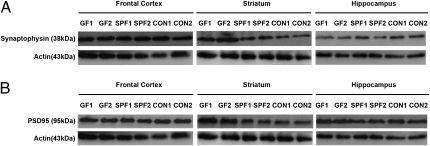
In an attempt to identify whether specific proteins connected to synaptogenesis would be subject to gut microbiota regulation and, thereby, provide a possible mechanism for the observed alteration of brain function, we investigated the protein expression of PSD-95 and synaptophysin in the frontal cortex, striatum, and hippocampus of GF, SPF, and CON mice. As can be seen in Fig. 6 and Table 1, the expression of both synaptophysin and PSD-95 in the striatum was significantly lower (P < 0.05) in SPF and CON mice compared with GF mice. However, there were no differences in the expression of synaptophysin or PSD-95 in frontal cortex and hippocampus.
Fig. 6.
GF mice show higher expression of synaptic-related proteins in the striatum compared with SPF mice. Representative Western blot films for synaptophysin (A) and PSD-95 (B) protein expression in the frontal cortex, striatum, and hippocampus of two male GF, SPF, and CON mice (for further details, see Table 1).
Table 1.
Expression of synaptophysin and PSD-95 in various brain regions of GF, SPF, and CON mice
| Frontal cortex |
Striatum |
Hippocampus |
||||
| Mice | Synaptophysin | PSD-95 | Synaptophysin | PSD-95 | Synaptophysin | PSD-95 |
| GF | 0.989 ± 0.058 | 1.039 ± 0.085 | 1.803 ± 0.037* | 2.171 ± 0.042* | 0.982 ± 0.046 | 1.025 ± 0.136 |
| SPF | 1.000 ± 0.062 | 1.000 ± 0.145 | 1.000 ± 0.015* | 1.000 ± 0.036* | 1.000 ± 0.051 | 1.000 ± 0.075 |
| CON | 0.992 ± 0.022 | 1.011 ± 0.090 | 1.013 ± 0.028* | 1.001 ± 0.063* | 0.973 ± 0.039 | 1.045 ± 0.076 |
Expression of synaptophysin and PSD-95 in the frontal cortex, striatum, and hippocampus of GF (n = 4), SPF (n = 4), and CON (n = 6) mice were quantified and calculated against their respective actin expression (Fig. 6). The values were then compared against the SPF mice values and expressed as average fold increase. Values are expressed as means ± SEM.
*P < 0.05 compared with SPF mice.
Discussion
This study supports the hypothesis that normal gut microbiota can affect normal brain development and behavioral functions. Our data extend previous observations from Sudo and coworkers who demonstrated that gut microbiota could modulate the levels of adreno-corticotrophic hormone (ACTH) in young mice (20), as well as findings from the Gordon laboratory reporting elevated home-cage activity counts in GF mice (21). Here, we propose that altered expression profiles of canonical signaling pathways, neurotransmitter turnover, and synaptic-related proteins may combine and contribute to behavioral differences observed between SPF and CON mice compared with the GF mice.
Synaptophysin is a synaptic vesicle glycoprotein, which is expressed in neuroendocrine cells and in most neurons in the central nervous system. It is a hallmark of synaptic vesicle maturation (22), and it is also considered an indirect marker of synaptogenesis in the developing brain (23). Likewise, PSD-95 is involved in the maturation of excitatory synapses (24). In line with a window of opportunity early in life, it is plausible that the gut microbiota was able to modulate both synaptophysin and PSD-95 in the striatum during a sensitive period of synaptogenesis. Therefore, the modulation of these proteins by the gut microbiota could lead to long-term modulation of synaptic transmission affecting motor control and anxiety-like behavior in adult life. The perinatal period appears to be critical for this type of developmental programming because the ability of the gut microbiota to modulate, e.g., the hypothalamic-pituitary-adrenal axis, can only occur if GF mice are exposed to the gut microbiota early during postnatal development (i.e., before 6 wk of age) (20). The present data also support the notion that there is in early life a sensitive period for gut microbiota to affect later-life brain and behavior. Interestingly, nonhuman primate studies have demonstrated that administered glucocorticoids reduce synaptophysin expression in the fetal brain (25). Hence, modulation of stress hormones (e.g., ACTH and corticosterone) by the gut microbiota is another potential mechanism that could explain the present findings.
Another possible mechanism mediating the gut-brain communication, proposed here, may be via established neuronal circuits. Gut microbiota can elicit signals via the vagal nerve to the brain and vice versa (26, 27). Modulation of transmitters (e.g., serotonin, melatonin, gamma-aminobutyric acid, histamines, and acetylcholine) within the gut is yet another possible mechanism of action that could mediate the effects of the gut microbiota. For example, metabolic profiling of GF and conventionalized mice have revealed that conventionalization of GF mice by gut microbiota results in a 2.8-fold increase in plasma serotonin levels (28). This increase may arise from peripheral serotonin pools stored in the colon that, upon exposure to the incoming gut microbiota during early postnatal life, are released (29). Thus, this postnatal developmental change induced by the gut microbiota creates system conditions by which periphery pools of serotonin are carefully monitored and tightly regulated in early postnatal development. It is intriguing that the same neurotransmitter pathway is involved in the regulation of both food intake (30) bone remodeling (31) and behavioral brain functions (32). Recent data have demonstrated similar cross-talk between food intake, innate immunity, and G-coupled receptors expressed in the nervous system of Caenorhabditis elegans (33).
Although the mechanisms outlined above may help to explain how gut microbiota could modulate brain development and functions, they do not, however, explain the region-specific changes in brain observed in the present study, i.e., regulation of synaptic-associated proteins and neurotransmitter turnover specifically in the striatum. Within this context, it is worth mentioning that some bacteria (e.g., Clostridium botulinum and Clostridium tetani) are known to produce neurotoxins that specifically target a group of proteins that enable synaptic vesicles to dock and fuse with presynaptic plasma membranes. In the present study, we also found that the normal gut microbiota target two key synaptic proteins, namely synaptophysin and PSD-95 in the striatum. Recent data indicate that pattern recognition receptors expressed on the microglia surface characterize one of the primary, common pathways by which neurotoxin signals affect neuronal tissues (34). Interestingly, there seems to be a great diversity in microglia phenotype and function (35). Therefore, microglia cells could be involved in the signaling pathways induced by normal gut microbiota-derived metabolites.
Our results suggest that during evolution, the colonization of gut microbiota has become integrated into the programming of brain development, affecting motor control and anxiety-like behavior. It is tempting to speculate that the differences that we observe between GF and SPF mice are mediated by signaling initiated soon after birth at a time when the newborn mice become exposed to gut microbiota. This suggestion does not exclude the possibility that exposure to gut microbiota metabolites, generated by the flora of the pregnant mother, could also influence brain development during embryogenesis.
Gut microbiota may also be able to modify expression of risk genes (16, 36) or be part of mechanisms that alter cognitive functions observed in patients with gastrointestinal diseases (37, 38). Finally, the observed behavioral changes imposed by the presence of the gut flora in rodents, reported in this paper, may have wider implications when considering psychiatric disorders in humans.
Experimental Procedures
Animals.
Newborn litters of GF and SPF NMRI mice were placed and raised in special plastic isolators until they reached 8 to 10 wk of age (Core Facility for Germ Free Research, Karolinska Institutet). Only male animals were used in these experiments. Animals were maintained on autoclaved R36 Lactamin Chow (Lactamin) and kept in 12-h light cycles. All procedures were approved by the Local Research Ethics Committee.
Behavioral Studies.
Testing took place between 0900 and 1500 hours under low illumination to reduce stress. On the day of testing, animals were brought in sterile filtered cages to an adjacent testing room and allowed to rest for 1 h before testing. Test chambers were cleaned first with disinfectant and then with 70% ethanol and water after each animal.
Open Field Test.
Animals were placed individually in the center of an open field box (48 cm × 48 cm; Acti-Mot detection system; TSE), and their spontaneous motor activity was recorded as described (39). The computer program automatically recorded the following parameters: distance traveled in the center, periphery and total (entire box), a count of rearing activity (vertical infrared photo beam breaks), and time spent in slow (>5 cm/s, with an upper range of 20 cm/s) or fast (>20 cm/s) locomotion.
Light–Dark Box Test.
In this test, the mouse was placed into the dark compartment and allowed to freely explore the apparatus (48 × 48 cm; with two zones of equal areas) for 5 min. Time spent in the dark and light compartments were measured by using photocells (TSE).
Elevated Plus Maze Test.
The elevated plus maze, made of dark gray glacial polyvinyl chloride, consisted of four arms (each 30 × 5 cm) and a central area (5 × 5 cm) elevated 50 cm above the floor. Two arms were open and two were closed with 10-cm-high walls made of the same material. Mice were individually placed in the center facing an open arm and allowed to explore for 5 min. The behavior of the animal was recorded with a video camera and later scored by two independent observers that were blind to the identity of the animals. The following behaviors were scored: open and closed arm entries, time spent in the closed and open arms and in the center, and exploration of open arm ends.
Behavioral Analysis.
The behavioral data from the open field test were analyzed by using either repeated measures analysis of variance (ANOVA; phenotype and time as main factors) or factorial ANOVA when appropriate. The behavioral data from the light–dark box and elevated-plus maze tests were analyzed by using one-way ANOVA. All post hoc comparisons were made with Bonferroni/Dunn test in the presence of significant ANOVA effects. The threshold for statistical significance was set as P ≤ 0.05.
Neurochemical Analysis.
Brain samples were analyzed at Pronexus Analytical AB (Karolinska Institutet Science Park, Stockholm). Briefly, tissue concentration of NA, MHPG, DA, DOPAC, HVA, 5-HT, and 5-HIAA were determined by reversed-phase high-performance liquid chromatography with electrochemical detection as described (40). Data were analyzed by using one-way ANOVA. Post hoc comparisons were made with Bonferroni/Dunn test.
In Situ Hybridization.
Brains were rapidly dissected, frozen on dry ice, and stored at −80 °C. Coronal sections (14 μm) of various brain regions (frontal cortex, striatum, hippocampus, and amygdala) were prepared on a cryostat and stored at −80 °C until used. Fixation, prehybridization, and hybridization were performed as described (ref. 39; see SI Experimental Procedures for information about synthesis of riboprobes and quantification procedure).
Illumima Expression Array.
Brain tissues (frontal cortex, striatum, hypothalamus, hippocampus, and cerebellum) were rapidly dissected and stored at −80 °C until used. RNA was extracted by using Qiagen RNA extraction kit. The RNA (0.5 μg) was amplified and labeled by using Illumina total prep RNA amplification kit (Ambion). cRNA (0.75 μg) were applied on Illumina chips (Mouse chips Ref.V1.1) according to the manufacturer's instruction. After hybridization and washing, each array was scanned by Illumina's scanning software (BeadScan) to produce an image in the Tagged Image File Format, along with files in a proprietary file format containing intensity and location information. Original data were extracted and normalized by using BeadStudio software. After normalization, probe signals were checked for detection against negative controls with a BeadStudio internal algorithm and missing values were introduced to replace signals under the detection limit. Probes that failed to detect signal in at least three mice were filtered out.
Further data analysis was applied on Gene Spring, where probes with low detection values (<50) or low confidence detection (P < 0.99) were further filtered out. Genes differentially expressed more than twofold were selected and used for statistical analysis (one-way ANOVA) to select for significance (P < 0.05). Lists of statistically, differentially expressed genes were then analyzed by using Ingenuity Pathway Analysis, Cluster, and Tree View software.
Western Immunoblotting.
Brain tissues were dissected on ice frozen by liquid nitrogen and stored at −80 °C. Tissue samples were homogenized in ice-cold RIPA lysis buffer [50 mM Tris at pH 7.4, 150 mM NaCl, 1% (wt/vol) Nonidet P-40, 0.5% (wt/vol) sodium deoxycholate, 1 mM EDTA, 0.1% (wt/vol) SDS] in the presence of protease inhibitors (1 mM PMSF; 10 μg/mL chymostatin, leupeptin, antipain and pepstatin A; and 2 μg/mL aprotinin). After centrifugation at 4 °C, 25 μg of protein were mixed with reducing sample buffer, denatured, and subjected to electrophoresis (7.5–15% SDS/PAGE gels) followed by electroblotting onto presoaked PVDF membranes (BioRad). Blots were blocked in 0.1% Tween-20, 5% milk in PBS, at room temperature. Primary antibodies synaptophysin and PSD-95 (Cell Signaling) were incubated with the blot o/n at 4 °C. Incubation with horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia) was performed at room temperature. Protein bands were visualized by chemiluminescence by using Super Signal West Pico Chemiluminescent Substrate (Pierce) reagent. Protein expression of synaptophysin and PSD-95 were then quantified by using ImageJ software. Group differences were analyzed by using separate Student's t test from each protein.
Q-PCR.
Expression of genes of interest, including genes from microarray results (Table S1–S3), were quantitatively measured by using TaqMan real-time PCR system as previously described (4). The unpaired Student's t test was used for statistical analyses. The level of statistical significance was set at P ≤ 0.05.
Acknowledgments
This work was supported by the Swedish Research Council, Vinnova, and the Swedish Foundation for Strategic Research (S.P. and H.F.); the Söderberg Foundation and the 7th EU program TORNADO (S.P.); Foundation Olle Engkvist Byggmästare (H.F.); and Foundation Frimurare Barnhuset (R.D.H.). F.A. holds a postdoctoral fellowship from A*STAR Singapore. M.L.H. and S.W. were supported by A*STAR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010529108/-/DCSupplemental.
References
- 1.Seckl JR, Meaney MJ. Glucocorticoid programming. Ann N Y Acad Sci. 2004;1032:63–84. doi: 10.1196/annals.1314.006. [DOI] [PubMed] [Google Scholar]
- 2.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 4.Lundin A, et al. Gut flora, Toll-like receptors and nuclear receptors: A tripartite communication that tunes innate immunity in large intestine. Cell Microbiol. 2008;10:1093–1103. doi: 10.1111/j.1462-5822.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 5.Björkholm B, et al. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS ONE. 2009;4:e6958. doi: 10.1371/journal.pone.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claus SP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finegold SM, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002;35(Suppl 1):S6–S16. doi: 10.1086/341914. [DOI] [PubMed] [Google Scholar]
- 9.Mittal VA, Ellman LM, Cannon TD. Gene-environment interaction and covariation in schizophrenia: The role of obstetric complications. Schizophr Bull. 2008;34:1083–1094. doi: 10.1093/schbul/sbn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goehler LE, Park SM, Opitz N, Lyte M, Gaykema RP. Campylobacter jejuni infection increases anxiety-like behavior in the holeboard: Possible anatomical substrates for viscerosensory modulation of exploratory behavior. Brain Behav Immun. 2008;22:354–366. doi: 10.1016/j.bbi.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilbo SD, et al. Neonatal infection induces memory impairments following an immune challenge in adulthood. Behav Neurosci. 2005;119:293–301. doi: 10.1037/0735-7044.119.1.293. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan R, et al. The International Society for Developmental Psychobiology annual meeting symposium: Impact of early life experiences on brain and behavioral development. Dev Psychobiol. 2006;48:583–602. doi: 10.1002/dev.20170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desbonnet L, Garrett L, Clarke G, Bienenstock J, Dinan TG. The probiotic Bifidobacteria infantis: An assessment of potential antidepressant properties in the rat. J Psychiatr Res. 2008;43:164–174. doi: 10.1016/j.jpsychires.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57:809–818. doi: 10.1016/j.neuron.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Govindarajan A, et al. Transgenic brain-derived neurotrophic factor expression causes both anxiogenic and antidepressant effects. Proc Natl Acad Sci USA. 2006;103:13208–13213. doi: 10.1073/pnas.0605180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10:1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, et al. cDNA microarray analysis of gene expression in anxious PVG and SD rats after cat-freezing test. Exp Brain Res. 2003;149:413–421. doi: 10.1007/s00221-002-1369-1. [DOI] [PubMed] [Google Scholar]
- 18.Lau JY, Pine DS. Elucidating risk mechanisms of gene-environment interactions on pediatric anxiety: Integrating findings from neuroscience. Eur Arch Psychiatry Clin Neurosci. 2008;258:97–106. doi: 10.1007/s00406-007-0788-1. [DOI] [PubMed] [Google Scholar]
- 19.Bateup HS, et al. Distinct subclasses of medium spiny neurons differentially regulate striatal motor behaviors. Proc Natl Acad Sci USA. 2010;107:14845–14850. doi: 10.1073/pnas.1009874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudo N, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becher A, et al. The synaptophysin-synaptobrevin complex: A hallmark of synaptic vesicle maturation. J Neurosci. 1999;19:1922–1931. doi: 10.1523/JNEUROSCI.19-06-01922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulfig N, Setzer M, Neudörfer F, Bohl J. Distribution of SNAP-25 in transient neuronal circuitries of the developing human forebrain. Neuroreport. 2000;11:1259–1263. doi: 10.1097/00001756-200004270-00023. [DOI] [PubMed] [Google Scholar]
- 24.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 25.Antonow-Schlorke I, Schwab M, Li C, Nathanielsz PW. Glucocorticoid exposure at the dose used clinically alters cytoskeletal proteins and presynaptic terminals in the fetal baboon brain. J Physiol. 2003;547:117–123. doi: 10.1113/jphysiol.2002.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borovikova LV, et al. Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature. 2000;405:458–462. doi: 10.1038/35013070. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, et al. Evidences for vagus nerve in maintenance of immune balance and transmission of immune information from gut to brain in STM-infected rats. World J Gastroenterol. 2002;8:540–545. doi: 10.3748/wjg.v8.i3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uribe A, Alam M, Johansson O, Midtvedt T, Theodorsson E. Microflora modulates endocrine cells in the gastrointestinal mucosa of the rat. Gastroenterology. 1994;107:1259–1269. doi: 10.1016/0016-5085(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 30.Gruninger TR, LeBoeuf B, Liu Y, Garcia LR. Molecular signaling involved in regulating feeding and other motivated behaviors. Mol Neurobiol. 2007;35:1–20. doi: 10.1007/BF02700621. [DOI] [PubMed] [Google Scholar]
- 31.Kawai M, Rosen CJ. Minireview: A skeleton in serotonin's closet? Endocrinology. 2010;151:4103–4108. doi: 10.1210/en.2010-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cools R, Roberts AC, Robbins TW. Serotoninergic regulation of emotional and behavioural control processes. Trends Cogn Sci. 2008;12:31–40. doi: 10.1016/j.tics.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Aballay A. Neural regulation of immunity: Role of NPR-1 in pathogen avoidance and regulation of innate immunity. Cell Cycle. 2009;8:966–969. doi: 10.4161/cc.8.7.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Block ML, Zecca L, Hong JS. Microglia-mediated neurotoxicity: Uncovering the molecular mechanisms. Nat Rev Neurosci. 2007;8:57–69. doi: 10.1038/nrn2038. [DOI] [PubMed] [Google Scholar]
- 35.de Haas AH, Boddeke HW, Biber K. Region-specific expression of immunoregulatory proteins on microglia in the healthy CNS. Glia. 2008;56:888–894. doi: 10.1002/glia.20663. [DOI] [PubMed] [Google Scholar]
- 36.Laurin N, et al. Investigation of the G protein subunit Galphaolf gene (GNAL) in attention deficit/hyperactivity disorder. J Psychiatr Res. 2008;42:117–124. doi: 10.1016/j.jpsychires.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Addolorato G, et al. State and trait anxiety and depression in patients affected by gastrointestinal diseases: Psychometric evaluation of 1641 patients referred to an internal medicine outpatient setting. Int J Clin Pract. 2008;62:1063–1069. doi: 10.1111/j.1742-1241.2008.01763.x. [DOI] [PubMed] [Google Scholar]
- 38.Nikolov RN, et al. Gastrointestinal symptoms in a sample of children with pervasive developmental disorders. J Autism Dev Disord. 2009;39:405–413. doi: 10.1007/s10803-008-0637-8. [DOI] [PubMed] [Google Scholar]
- 39.Diaz Heijtz R, Scott L, Forssberg H. Alteration of dopamine D1 receptor-mediated motor inhibition and stimulation during development in rats is associated with distinct patterns of c-fos mRNA expression in the frontal-striatal circuitry. Eur J Neurosci. 2004;19:945–956. doi: 10.1111/j.0953-816x.2004.03154.x. [DOI] [PubMed] [Google Scholar]
- 40.Weikop P, Yoshitake T, Kehr J. Differential effects of adjunctive methylphenidate and citalopram on extracellular levels of serotonin, noradrenaline and dopamine in the rat brain. Eur Neuropsychopharmacol. 2007;17:658–671. doi: 10.1016/j.euroneuro.2007.02.014. [DOI] [PubMed] [Google Scholar]