Abstract
The Wingless/Integrated (Wnt) signaling pathway controls multiple events during development and homeostasis. It comprises multiple branches, mainly classified according to their dependence on β-catenin activation. The Wnt/β-catenin branch is essential for the establishment of the embryonic anteroposterior (AP) body axis throughout the phylogenetic tree. It is also required for AP axis establishment during planarian regeneration. Wnt/β-catenin–independent signaling encompasses several different pathways, of which the most extensively studied is the planar cell polarity (PCP) pathway, which is responsible for planar polarization of cell structures within an epithelial sheet. Dishevelled (Dvl) is the hub of Wnt signaling because it regulates and channels the Wnt signal into every branch. Here, we analyze the role of Schmidtea mediterranea Dvl homologs (Smed-dvl-1 and Smed-dvl-2) using gene silencing. We demonstrate that in addition to a role in AP axis specification, planarian Dvls are involved in at least two different β-catenin–independent processes. First, they are essential for neural connectivity through Smed-wnt5 signaling. Second, Smed-dvl-2, together with the S. mediterranea homologs of Van-Gogh (Vang) and Diversin (Div), is required for apical positioning of the basal bodies of epithelial cells. These data represent evidence not only of the function of the PCP network in lophotrocozoans but of the involvement of the PCP core elements Vang and Div in apical positioning of the cilia.
Keywords: neural patterning, axial polarity, cilia basal body
Wingless/Integrated (Wnt) signaling controls multiple events, including cell migration, planar cell polarity (PCP), and stem cell self-renewal (1, 2). It acts through two main pathways: the canonical or β-catenin–dependent pathway and the noncanonical or β-catenin–independent pathways, which include the PCP and Wnt/calcium pathways. Dishevelled (Dvl) is common to all three pathways (3). In the β-catenin–dependent pathway, Dvl is recruited by the receptor Frizzled (Fz), promoting disassembly of the β-catenin destruction complex through the subsequent recruitment of Axin. As a consequence, β-catenin translocates to the nucleus, where it acts as a master regulator. A common role of the β-catenin/Wnt pathway is in establishing the anteroposterior (AP) axis during development (4). PCP signals are transmitted from cell to cell and are responsible for the polarization of cell structures. During PCP signaling, Dvl is also recruited by a Fz receptor and promotes the asymmetrical localization of the PCP core proteins within the cell, such that the Fz–Dvl–Diversin (Div)/Diego (Dgo) complex is oppositely localized to the Strabismus (Stbm)/Van-Gogh (Vang)–Prickle (Pk) complex. The asymmetrical subcellular localization of these elements in an epithelial sheet directs cytoskeletal reorganization that results, for instance, in a hair emerging solely from the distal edge of the cell (5–7). The same mechanism is used in mesenchymal cells to direct cell movement and migration during gastrulation (convergent and extension movements). Another context in which the PCP network has recently been implicated is in the apical docking and planar polarization of basal bodies in multiciliated epithelial cells (8). In addition, Dvl transduces different noncanonical Wnt/Fz signals, for example, to activate intracellular calcium-sensitive enzymes, such as PKC, or to mediate a wnt5-derailed/Related to tyrosine kinase (RYK)–dependent signal (1, 3).
Planarians are a classical model for regeneration studies because of their striking morphological plasticity. These platyhelminths are able to regenerate a whole organism from a piece of almost any part of their body and also to grow and degrow according to culture conditions (9). These properties rely on the neoblasts, multipotent stem cells present in adult organisms that are able to differentiate into any planarian cell type (10). The conserved role of the Wnt/β-catenin pathway in establishing the AP axis during planarian regeneration and homeostasis has been broadly demonstrated (11–15). A role for the Wnt/β-catenin–independent pathway in nervous system regeneration has also been reported in planarians silenced for Smed-wnt5 and Smed-evi/wntless (14). Nothing is known about the role of the PCP pathway in planarians or, indeed, in any lophotrocozoans, however.
Here, we analyze the phenotype of animals silenced for the two Schmidtea mediterranea Dvls (Smed-dvl-1 and Smed-dvl-2) (12). Our results demonstrate that in addition to their role in specifying the AP axis, Smed-dvl-1 and Smed-dvl-2 are involved in β-catenin–independent processes. First, they are essential for neural connectivity and mediolateral patterning of the CNS, mainly through a Smed-wnt5–mediated signal. Furthermore, Smed-dvl-2 acts together with the S. mediterranea homologs of Vangl2 and Div/Dgo (Smed-vang-1, Smed-vang-2, and Smed-div) to regulate apical positioning of the cilia basal bodies of epidermal cells, demonstrating a specific function for the PCP network in planarians.
Results and Discussion
Smed-dvl-1 and Smed-dvl-2 Are Essential for AP Patterning and Brain Morphogenesis.
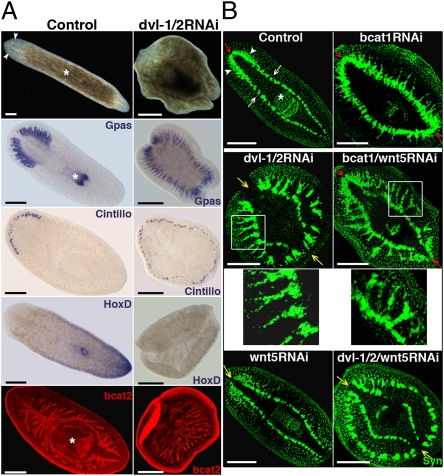
Two S. mediterranea Dvl paralogs have been identified: Smed-dvl-1 and Smed-dvl-2 (12) (Fig. S1A, phylogenetic analysis and mRNA expression pattern). It has been reported that simultaneous silencing of Smed-dvl-1 and Smed-dvl-2 generates “two-headed” planarians that resemble Smed–β-catenin1 RNAi animals (12). Because Dvl is a shared signal transducer of the three branches of the Wnt pathway, however, we expected that β-catenin–independent defects would be also observed after silencing Smed-dvl-1/2. Silencing of Smed-dvl-1/2 generates planarians with a radial morphology similar to that observed in Smed–β-catenin1 RNAi planarians (n = 164 of 166) (11). Although the animals do not differentiate ectopic eyes around the periphery, expression analysis of markers like Smed-Gpas, which labels the cephalic brain branches and the pharynx, Smed-Cintillo, which labels the chemoreceptors, and Smed-HoxD, a marker of central-posterior identity, demonstrated that Smed-dvl-1/2 RNAi animals had lost trunk and tail identities and were completely anteriorized (Fig. 1A). Staining with anti-Smed–β-catenin2, which labels the gut epithelium (16), also showed that the gut lost its AP polarity in Smed-dvl-1/2 RNAi planarians, resulting in a network of gut branches without a pharynx (n = 7 of 7) (Fig. 1A).
Fig. 1.
Silencing of dvl-1/2 generates “radial-like hypercephalized” animals with β-catenin–independent morphogenetic brain defects. (A) Stereomicroscopic view of live animals showing the radial shape and absence of eyes after dvl-1/2 RNAi; the white asterisk and arrowheads indicate the pharynx and eyes, respectively. Analysis of brain branches (Gpas) and chemoreceptors (Cintillo) revealed the differentiation of ectopic anterior structures in dvl-1/2 RNAi animals, whereas the posterior marker HoxD was absent. Anti-bcat2 staining shows the lack of polarization of the gut branches after dvl-1/2 RNAi. (B) Immunostaining with anti-Syn showing the organization of the ectopic neural structures generated by silencing dvl-1/2, wnt5, and bcat1, alone or in combination. In a control planarian, the CNS comprises bilobed CG (white arrowheads) connected by the ATC, which lies dorsal to the two VNCs (white arrows). Red and yellow arrows point to the position of the ATC (red when present and yellow when absent). White boxes indicate the area corresponding to the magnifications located just below. All images correspond to trunk pieces after 18–25 d of regeneration. The bcat2 and anti-Syn images correspond to confocal z-projections. (Left) Anterior. bcat1, Smed–β-catenin1; bcat2, Smed–β-catenin2; Cintillo, Smed-Cintillo; dvl-1/2, Smed-dvl-1/2; Gpas, Smed-Gpas; wnt5, Smed-wnt5; Syn, synapsin. (Scale bar: 300 μm.)
Immunochemistry with the panneural marker antisynapsin revealed that the cephalic ganglia (CG), which differentiate in the head region on the top of the ventral nerve cords (VNCs) in control animals, appeared as a set of isolated brain clusters around the entire periphery and were laterally displaced from the VNCs in Smed-dvl-1/2 planarians (n = 41 of 41) (Fig. 1B). Note that the anterior transverse commissure (ATC), which connects the CG in controls (Fig. 1B), was absent and that the posterior ectopic brain was not only poorly differentiated and displaced but appeared unconnected to the original VNCs. Those displacements and disconnections of the brain are never observed after silencing Smed–β-catenin1 (11) (Fig. 1B), demonstrating that in addition to its essential role in transducing canonical Wnt signaling, Smed-dvl-1/2 exerts β-catenin–independent functions that are essential for proper brain morphogenesis.
Interestingly, despite the presence of brain morphology defects in Smed-dvl-1/2 RNAi animals, differential expression of SmedOtp and SmedOtxB in the mediolateral axis of the brain (17) was observed (Fig. S2). Thus, functional domains of the brain appeared to remain conserved.
Silencing of Smed-dvl-1/2 during homeostasis also resulted in fully anteriorized animals (n = 31 of 31) with ectopic disconnected brains (n = 23 of 23) (Fig. S3). The morphogenesis of the preexisting brain was unaffected, however, and ectopic eyes were present (n = 16 of 23). This is not surprising, because antiarrestin immunostaining also revealed the differentiation of photoreceptors after lower doses of Smed-dvl-1/2 interference (Fig. S3).
To date, of the nine Wnts identified in S. mediterranea, Smed-wnt5 is the only one that has been demonstrated to signal independent of β-catenin. It is expressed from the most external region of the CG and VNCs to the lateral ends of the planarian (14, 18, 19). Interestingly, silencing of Smed-wnt5 also causes lateral expansion and displacement of the brain as well as the disappearance of the ATC (n = 16 of 18) (14, 18) (Fig. 1B). Consistent with the possibility that the displacements and disconnections observed in the brain of Smed-dvl-1/2 RNAi animals are attributable to inhibition of signaling from Smed-wnt5, we found that silencing of Smed-wnt5 together with Smed-dvl-1/2 clearly enhanced the disconnection of the brain branches from the original VNCs observed in Smed-dvl-1/2 RNAi planarians (n = 9 of 9) (Fig. 1B). Moreover, silencing of Smed-wnt5 together with Smed–β-catenin1 transformed the continuous brain of Smed–β-catenin1 RNAi animals into a discontinuous and displaced brain that was strikingly similar to the one observed in Smed-dvl-1/2 RNAi animals (Fig. 1B, higher magnification views). The ATC was present in those animals, however.
In summary, as in Smed–β-catenin1 RNAi animals, Smed-dvl-1/2 silencing abolishes the AP axis and leads to fully anteriorized animals. The CNS of those animals is poorly differentiated, however, and consists of a set of isolated and unconnected brain clusters from which eyes are unable to differentiate. Smed-wnt5 appears to be responsible for most of the β-catenin–independent defects observed in the CNS of Smed-dvl-1/2 RNAi planarians.
Smed-dvl-1/2 Directs Neural Connectivity Through Smed-wnt5 Signaling.
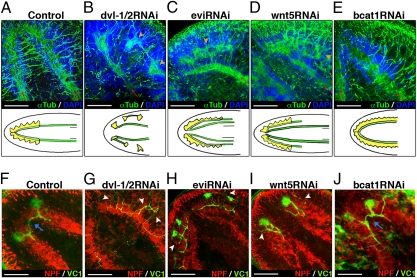
To gain further insight into the morphogenetic defects of Smed-dvl-1/2 RNAi animals, we performed a comparative analysis of the regeneration of the CNS and visual axons of animals following Smed–β-catenin1, Smed-wnt5, and Smed-evi silencing, using antitubulin and anti-arrestin antibodies, respectively. Smed-evi is the planarian homolog of evi/wls, a transmembrane protein required for secretion of Wnt ligands, and silencing of the gene results in differentiation of a displaced CNS (14). In control animals, preexisting VNCs grow beneath the regenerating CG and completely overlap (20) (Figs. 1 and 2A and Movie S1). In contrast, in Smed-dvl-1/2 RNAi planarians, the regenerating brain forms disconnected clusters that appear laterally displaced relative to the old VNCs (n = 26 of 41) (Fig. 2B and Movie S2). Likewise, in Smed-evi RNAi animals, the preexisting VNC failed to connect to the ectopic brain (n = 14 of 16) (Fig. 2C and Movie S3). The brain of Smed-wnt5 RNAi animals showed essentially the same lateral displacement and disconnection between the old VNCs and the newly developed brain (n = 11 of 11). Thus, although the disaggregation of the brain is much more extreme following Smed-dvl-1/2 silencing, it shares the main Wnt-5–dependent defects with Smed-evi RNAi animals (14) (Fig. 2D and Movie S4). In contrast, Smed–β-catenin1 RNAi animals differentiate the brain just above the VNCs, as in control planarians (Fig. 2E and Movie S5).
Fig. 2.
Dvl-1/2 and wnt5 RNAi planarians share the main neural connection defects. (A–E) Anti-αTub immunostaining reveals lateral displacement of the CNS in dvl-1/2, evi, and wnt5 RNAi animals. Red arrows indicate the growth direction of preexisting VNCs, and orange arrowheads indicate the regenerating CNS when displaced. Schematic drawings under each image represent the different phenotypes: yellow indicates CG, and green indicates VNCs. (F–J) Double immunostaining with anti-VC1, which specifically labels the planarian visual system, and anti-NPF to visualize the CNS. White arrowheads indicate aberrant projections of visual axons, and blue arrows indicate the presence of contralateral projecting axons crossing the midline. All images correspond to trunk pieces after 18–25 d of regeneration. A, D, and F–I show anterior brain, whereas B, C, and E correspond to ectopic posterior brain. Anti-αTub, anti-NPF, and anti-VC1 images correspond to confocal z-projections. Blue shows DAPI nuclear staining, which allows us to visualize the cell bodies of the CNS. (A, D, and F–I) Anterior (Left); (B, C, and E) anterior (Right). dvl-1/2, Smed-dvl-1/2; evi, Smed-evi; NPF, Neuropeptide F; wnt5, Smed-wnt5; αTub, α-tubulin; VC1, arrestin. (Scale bar: 150 μm.)
Planarian eyes consist of two cell types: pigment cup cells and photosensitive cells. In control animals, visual axons from photosensitive cells project either ipsilaterally to the most medial domain of the brain or contralaterally to form the optic chiasm (21) (Fig. 2F). Smed-dvl-1/2 interference leads to impaired photoreceptor regeneration in which there is no visible accumulation of photosensitive cells, and the visual axons, which appear dispersed among them, project in aberrant directions and never cross the midline (n = 12 of 12) (Fig. 2G). Although they are able to differentiate eyes, some of these defects are shared by Smed-evi (n = 4 of 4) and Smed-wnt5 (n = 8 of 9) RNAi planarians (Fig. 2 H and I). As expected, Smed–β-catenin1 RNAi does not severely affect either eye regeneration or targeting of the visual axons (n = 4 of 4) (Fig. 2J). These data support a β-catenin–independent role for Smed-dvl-1/2 in eye differentiation and in visual axon targeting, probably through Smed-wnt5 signaling.
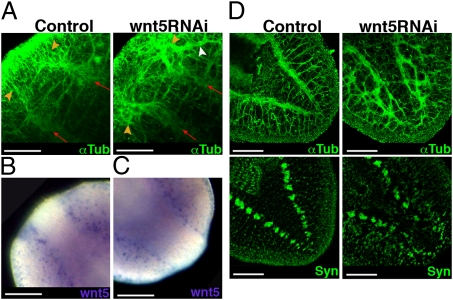
In an attempt to understand the function of Smed-wnt5 better, we analyzed CNS regeneration over time after silencing. As early as 3 d after amputation, the brain and the newly developed VNCs regenerate in a laterally displaced position. Moreover, some axons escape and project toward the lateral edge of the animal (Fig. 3A and Fig. S4A). Interestingly, at this time point of regeneration, Smed-wnt5 is expressed externally to the growing VNCs (Fig. 3B), suggesting a role in preventing lateral expansion of the regenerating CNS. Smed-wnt5 mRNA is not only expressed in the anterior blastema but is detected along the length of the VNCs up to the posterior blastema (Fig. 3C). Accordingly, in addition to the defects in the positioning of the brain, Smed-wnt5 RNAi animals regenerated a defective tail; the two preexisting VNCs grow laterally displaced into the blastema and their axons, instead of forming two distinguishable structures, appeared to separate and project in aberrant directions (Fig. 3D, Fig. S4B, and Movies S6 and S7). Moreover, the VNCs became more tangled and displaced as regeneration proceeded, and even the lateral nerves, which project from the VNCs to the edge of the animal in control animals, appeared extremely disorganized (Fig. S4 C and D). Antisynapsin immunostaining of these regenerating tails further demonstrates the lateral expansion and disorganization of the neuronal synapses (Fig. 3D).
Fig. 3.
Wnt5 expression supports its role in restricting CNS positioning along the mediolateral axis. (A) Anti-αTub staining reveals lateral displacement of the anterior CNS after 3 d of regeneration in wnt5 RNAi animals. Red arrows indicate the growth direction of preexisting VNCs, and orange arrowheads indicate the regenerating CNS. The white arrowhead indicates lateral aberrant projection of some axons. Expression of wnt5 at 3 d after amputation in anterior (B) and posterior (C) blastemas. (D) Anti-αTub and anti-Syn staining shows the disorganization of the axon projections and connections in the 18-d-old regenerated tail of a wnt5 RNAi planarian. All images correspond to regenerating trunk pieces. Anti-αTub and anti-Syn images correspond to confocal z-projections. (Left) Anterior. wnt5, Smed-wnt5; Syn, synapsin; αTub, α-tubulin. (Scale bar: 150 μm.)
Our results are consistent with the suggestion that Smed-wnt5 defines the planarian mediolateral axis (18), at least with regard to the positioning of the neural structures. Although the molecular mechanisms through which Smed-wnt5 regulates the positioning of neural structures remain to be elucidated, it is likely to act together with slit, because Smed-slit silencing leads to midline collapse of the CNS (22). One possibility is that Smed-wnt5 controls the position of the regenerating CNS along the mediolateral axis by restricting the migration of neural precursor cells to more medial regions. Alternatively, given the evolutionarily conserved role of Wnt5 in providing repulsive guidance information to axons (23, 24), Smed-wnt5 could restrict the direction of the axon growth and establish the position of the newly developed neural tissues.
In summary, the lateral displacement and disconnections of the CNS as well as the aberrant projections of the visual axons observed in Smed-dvl-1/2 RNAi animals are shared with Smed-evi and Smed-wnt5 RNAi animals and are not observed after Smed–β-catenin1 interference. These results suggest that the mediolateral neural connection defects observed in Smed-dvl-1/2 RNAi animals could reflect the signal input of Smed-wnt5, which is essential for the mediolateral patterning of neural structures.
Functional Specialization of Smed-dvl-1 and Smed-dvl-2: Smed-dvl-2 Is Involved in the Stabilization of Intracellular β-Catenin.
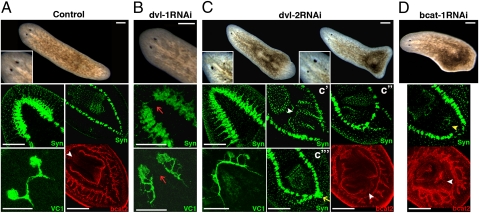
When analyzing the function of the two Smed-dvl paralogs separately, Smed-dvl-1 RNAi planarians appeared normal. Molecular markers revealed the absence of the ATC, however, along with impaired regeneration of visual axons similar to that previously described for Smed-wnt5 RNAi planarians (Figs. 1B and 4B). In contrast, regenerating trunks of Smed-dvl-2 RNAi animals had multiple defects. During anterior regeneration, the CG as well as the visual axons expanded posteriorly, together with the development of ectopic eyes (Fig. 4C), which were not well differentiated. The two anterior eyes exhibited clear asymmetry in terms of the degree of differentiation and AP position (n = 258 of 272) (Fig. 4C, live animals).
Fig. 4.
Smed-dvl paralogs are functionally specialized. (A) Stereomicroscopic image of a control animal (magnified view of the eye region shown at Bottom Left), with anti-Syn staining of the anterior and posterior regions, anti-VC1 staining of the visual system, and anti-bcat2 staining showing the two posterior branches and the pharynx. The white arrowhead indicates the anchoring of the pharynx. (B) dvl-1 RNAi animal lacking the ATC and with aberrant visual system regeneration (red arrows). (C) Analysis of a dvl-2 RNAi animal. Note the poor and asymmetrical differentiation of the eyes. An ectopic posterior brain is indicated with a yellow arrow, and the anchoring of the ectopic pharynges is marked with white arrowheads. (D) Low-dose bcat-1 RNAi animal showing the differentiation of an ectopic mouth (yellow arrowhead) and an ectopic pharynx primordium (white arrowhead). All images correspond to regenerating trunk pieces. (Left) Anterior. Anti-Syn, bcat2, and anti-VC1 staining images correspond to confocal z-projections. bcat1, Smed–β-catenin1; bcat2, Smed–β-catenin2; dvl-1, Smed-dvl-1; dvl-2, Smed-dvl-2; Syn, synapsin; VC1, arrestin. (Scale bar: 250 μm.)
Posterior regeneration was also affected in Smed-dvl-2 RNAi animals. Antisynapsin and anti-Smed–β-catenin2 staining revealed the development of ectopic pharynges. Although their orientation varied, it was always located posterior to the original one. To gain further insights into this phenotype, we compared planarians that had regenerated for less than 30 d with those that had regenerated for longer periods (Fig. S5A). In the first group, most ectopic pharynges were orientated in the same direction as the original one (69%) (Fig. 4C′). Longer periods of regeneration led to a significant increase in the proportion of ectopic pharynges with the opposite orientation (from 3% to 56%) (Fig. 4C′′). Interestingly, the orientation of the ectopic pharynges was closely linked to the gradual anteriorization of Smed-dvl-2 RNAi animals. Thus, when the pharynx had the same orientation as the original one, the VNCs remained unconnected to the posterior end (Fig. 4C′). When the ectopic pharynx had an opposite orientation, however, Smed-dvl-2 RNAi planarians had either a “no-tail” phenotype with closure and thickening of the VNCs (Fig. 4C′′), similar to the phenotype described after low doses of Smed–β-catenin1 silencing (63%) (11), or the appearance of a posterior brain (37%) (Fig. 4C′′′).
In situ hybridization with the posterior identity marker Smed-AbdBA and the central marker Smed-TCEN revealed a significant reduction in tissue with a posterior identity after Smed-dvl-2 knockdown (Fig. S5B). Note that, as with anterior structures, Smed-AbdBA expression was asymmetrical (n = 5 of 12). These data, together with the observation that nearly 40% (n = 73 of 188) of the head fragments become biheaded after Smed-dvl-2 RNAi treatment (Fig. S5C), demonstrate that Smed-dvl-2 transduces a β-catenin–dependent Wnt signaling.
To explain the overall phenotype of Smed-dvl-2 RNAi animals, we hypothesized that a low-level inhibition of Smed–β-catenin1 activity leads to displacement of anterior and central identity toward the posterior, with posterior identity considerably reduced or switched to anterior after a long period of silencing (Fig. S5D). Accordingly, very low-dose RNAi for Smed–β-catenin1 partially reproduced the phenotype caused by Smed-dvl-2 knockdown. In most Smed–β-catenin1 RNAi planarians that did not regenerate an ectopic posterior head, the VNCs were disconnected posteriorly (74%, n = 30), and half of them developed posterior ectopic mouths accompanied by pharyngeal primordia (Fig. 4D).
Interestingly, silencing of Smed-dvl-2 in intact animals resulted in the differentiation of ectopic brain branches toward the posterior, which were unconnected to the original brain. The pharynx was also displaced posteriorly, and the VNCs were disconnected at the posterior end (Fig. S5E). The overall phenotype of intact planarians is consistent with the hypothesis that Smed-dvl-2 causes a slight reduction in Smed–β-catenin1 activity that leads to posterior displacement of more anterior identities.
Whereas most animals develop a complete ectopic pharynx after Smed-dvl-2 silencing, very low doses of Smed–β-catenin1 RNAi only resulted in a complete second pharynx in 1 of 30 animals. The capacity to differentiate ectopic pharynges could thus involve a β-catenin–independent role of Smed-dvl-2. Conversely, Smed-dvl-2 silencing led to eye differentiation defects different from those observed after Smed-evi/wls or Smed-wnt5 silencing (14) (Fig. S6), suggesting that this Smed-dvl-2 activity may be independent of Wnt. This would be consistent with the failure of eye differentiation observed in Smed-dvl-1/2 RNAi but not in Smed-evi or Smed–β-catenin1/wnt5 RNAi animals (Fig. S6).
Our results demonstrate a functional specialization of the Smed-dvl paralogs in which only Smed-dvl-2 appears to be involved in β-catenin–dependent signal transduction. Accordingly, the phenotypes described after silencing canonical Wnts, such as Smed-wnt11-2, Smed-wnt-1, and Smed-Wnt11-6, are clearly reproduced after Smed-dvl-2 silencing. Conversely, Smed-dvl-1 seems to transduce some of the noncanonical branches in which Smed-wnt5 is involved. Nevertheless, comparison of the strong phenotype observed after Smed-dvl-1/2 silencing with the phenotypes after their individual silencing reveals functional redundancy, as described in vertebrates (3). Further studies will elucidate whether, in a WT situation, both paralogs are involved in signal transduction for a single Wnt ligand.
Smed-dvl-2 Functions Alongside Smed-vang-1/2 and Smed-div as a PCP Network Component and Is Crucial for the Apical Positioning of the Basal Body in Epithelial Cells.
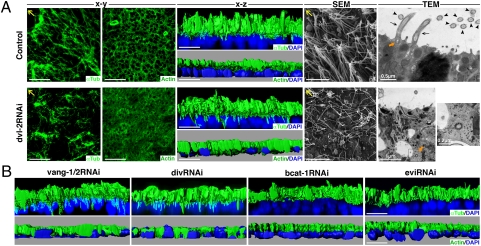
The epithelium of planarians is composed of mucociliary cells from which multiple motile cilia emerge (25, 26). To address whether Smed-dvl-1/2 functions as a component of the PCP network, we analyzed the polarization of the ventral epithelium after Smed-dvl-1/2 interference. Anti–α-tubulin staining revealed a reduction in the density of cilia and a loss of polarization after Smed-dvl-2 (Fig. 5A) and Smed-dvl-1/2 RNAi but never after Smed-dvl-1 silencing.
Fig. 5.
Defective positioning of the cilial actin network in dvl-2, vang-1/2, and div RNAi planarians. (A) Anti-αTub and anti-Actin immunostaining of control and dvl-2 RNAi planarians. The x-y and x-z confocal projections are shown. SEM of the planarian epidermis showing the differences in cilial density and orientation. Ultrastructural analysis by TEM of epithelial cells of the planarian epidermis. Several cilia anchored to the apical membrane can be seen in longitudinal (black arrows) and transverse (black arrowheads) sections in a control planarian. A basal body is indicated with an orange arrow. A basal body found isolated in the cytoplasm, outside a vesicle, is shown at two different magnifications in the image corresponding to a dvl-2 RNAi planarian (orange arrow). Yellow arrows indicate anterior. (B) Anti-αTub and anti-Actin immunostaining of vang-1/2, div, bcat1, and evi RNAi planarians. The x-z confocal projections are shown. DAPI nuclear staining is shown in blue. bcat1, Smed–β-catenin1; div, Smed-div; dvl-2, Smed-dvl-2; evi, Smed-evi; αTub, α-tubulin; vang-1/2, Smed-vang-1/2. (Scale bar: 10 μm unless otherwise indicated.)
It has long been established that planarian movement is governed by ventral motile cilia and by a muscle network (27). In Smed-dvl-2 RNAi animals, movement appeared to be mediated by muscle contraction and the gliding movement mediated by cilia was absent (Movies S8 and S9). This “inching” behavior has also been described following silencing of SmedIguana, which is involved in ciliogenesis (28), and silencing of the structural proteins of the cilia, Smed-ift88 and Smed-kif3a (26). Thus, Smed-dvl-2 RNAi animals show fewer, shorter, and less well-organized cilia, which are not fully functional.
It has been reported that Dvl controls the apical positioning of the basal bodies and the planar polarization of the cilia in the epithelium of Xenopus embryos, which is a mucous and multiciliated epithelium similar to that of planarians (8). Because the positioning of the basal bodies is governed by actin assembly, we analyzed the role of Smed-dvl-1/2 in the assembly of the actin network and in the docking of the basal bodies in the planarian epithelium. The actin filaments are arranged as a network that defines each cell border and contains regular openings to allow mucus secretion by the rhabdomeres (25). Silencing of Smed-dvl-2 dramatically affected the arrangement of this network, resulting in an irregular net in which the cell borders and rhabdomere openings were not visible (Fig. 5A). Furthermore, 3D reconstructions of α-tubulin and actin staining in cross-section showed that cilia emerge from the apical-most side of the cells in WT animals and that the actin filaments are also located apically on top of the nucleus (Fig. 5A). In contrast, after Smed-dvl-2 silencing, microtubules fail to localize to the apical surface and actin filaments appear disorganized and embedded in the nuclear layer (Fig. 5A).
Next, we used electron microscopy to analyze the ultrastructure of the cilia and their docking to the epithelial cells in Smed-dvl-2 RNAi animals. SEM revealed a decrease in cilial density and a loss of cilial polarization in Smed-dvl-2 RNAi animals, exactly as seen with α-tubulin staining (Fig. 5A). Transmission electron microscopy (TEM) in control planarians demonstrated that the basal bodies of the cilia are docked at the apical side of the epithelial cells, from which the ciliary axonemes project. In contrast, Smed-dvl-2 RNAi animals had fewer and shorter cilia and also had isolated basal bodies, with the typical 9 + 0 structure, inside the cytoplasm, a feature that we never observed in control animals. Dvl has been implicated in secretory vesicle trafficking through its association with the exocyst component Sec8 (8). Accordingly, in control animals, basal bodies are visible inside vesicles, whereas after Smed-dvl-2 silencing, a high concentration of empty vesicles was detected in the apical cytoplasm, suggesting that the failure of the basal bodies to dock could be a consequence of defective vesicle trafficking. Taken together, these results demonstrate that Smed-dvl-2 is essential for actin network assembly and apical positioning of the cilia. This may be mediated by an effect of Dvl on vesicle trafficking, which has been reported previously in the context of the PCP pathway (8). A mechanism directing Dvl localization to the plasma membrane, such as the one exerted by Syndecan-4 during convergent and extension movements in Xenopus embryos (29), could be also involved.
To test whether the defects observed in the apical positioning of the cilia are governed by a PCP network in our model, we characterized Smed-vang and Smed-div, other components of the core PCP network. We have identified two homologs of Vangl2 in the genome of S. mediterranea, Smed-vang-1 and Smed-vang-2, and one homolog of Div/Dgo, Smed-div (Fig. S7, sequence analysis and mRNA expression pattern). Analysis of α-tubulin and actin filaments revealed that simultaneous silencing of both Smed-vang paralogs (Smed-vang-1/2) and Smed-div led to disorganization of the cilia and actin network within the plane of the epithelium (Fig. S8A). Furthermore, the apical positioning of α-tubulin and the actin network was similar to that observed for Smed-dvl-2 (Fig. 5B). As expected, treated animals also displayed the impaired movement governed by muscle instead of cilia (Movies S10 and S11). Interestingly, when the same analysis was done in Smed–β-catenin1 and Smed-evi RNAi animals, the cilia and the actin network were correctly organized (Fig. S8B) and located at the apical-most part of the ventral epithelial cells (Fig. 5B).
Summarizing, the defects in actin and α-tubulin apical organization caused by silencing Smed-dvl-2 were reproduced by silencing the two core components of the PCP network, Smed-vang-1/2 and Smed-div, indicating that Smed-dvl-2 may function as a component of the PCP network. Furthermore, the effect was independent of β-catenin signaling and Wnt ligand activation. Although a common feature of PCP core proteins is their asymmetrical localization inside the cells and their role as organizers of the actin cytoskeleton, a dependence on prior Wnt activation has only been found in vertebrates (30, 31).
Following Smed-vang-1/2 or Smed-div RNAi, treated planarians had an apparently normal appearance in vivo and immunostaining with antisynapsin revealed no apparent defects in brain morphogenesis (Fig. S8C). Thus, the PCP core network appears not to be involved in the effects of Smed-dvl-1/2 on brain morphogenesis.
Our data suggest that a PCP network acts in the multiciliated epithelium of planarians to control apical docking of the basal bodies and are consistent with a mechanism involving the activation of the actin regulator RhoA, as reported for the multiciliated epithelial cells of Xenopus embryos (8). Further analysis with markers for the ciliary rootlet or basal bodies will be necessary to confirm the involvement of the PCP network in the final polarization of the cilia in the planarian epithelium.
In conclusion, our data represent functional evidence that Dvl, as a component of a PCP network, is essential for proper positioning of the actin cytoskeleton and for the docking of basal bodies to the apical side of epithelial cells in invertebrates. Moreover, although Vangl2 and Div proteins have been shown to localize to the base of the cilia in vertebrate epithelial cells (8, 32, 33), we provide functional evidence that those core members of the PCP pathway are also required.
Conclusions
Analysis of the complex phenotype generated after silencing of Smed-dvls indicates their role in at least three different pathways. First, they are involved in β-catenin–dependent signaling, which has been broadly documented as functionally conserved in the establishment of the AP axis. Second, Smed-dvls are required for proper neural connectivity and mediolateral patterning of the planarian CNS via β-catenin–independent/Wnt-dependent signaling. Future experiments must address whether this Smed-wnt5–mediated role occurs through the restriction of cell migration or via the control of axon growth through the wnt5-drl/RYK pathway, which is already known to be involved in axon guidance. Finally, Smed-dvl-2 and the two PCP core elements Smed-vang-1/2 and Smed-div control apical positioning of the cilia basal body in a β-catenin– and Wnt-independent manner during planarian regeneration. Dvl, through its active role in actin remodeling, is required for proper vesicle trafficking of the basal bodies, and therefore for ciliogenesis and, subsequently, planar polarization of the cilia. Taking into account the fact that Vangl2 and Div are located in the base of the cilia (32, 33) and that, according to our results, they are required for basal body docking, we propose that these two PCP core elements function together with Dvl to regulate directional trafficking of the basal bodies and polarization of the cilia within the epithelial sheet.
Materials and Methods
Organisms.
Planarians of the asexual strain of S. mediterranea were collected in Montjuïch, Barcelona, Spain and were maintained as described (34).
Whole-Mount in Situ Hybridization.
Gene expression analysis was carried out essentially as described (17). Details are provided in SI Materials and Methods.
RNAi Silencing.
For regeneration experiments, animals were amputated pre- and postpharyngeally after injection of dsRNA on 3 consecutive days (one round of injection). In the case of Smed-wnt5, Smed-evi/Wntless, Smed-vang-1/2, and Smed-div, we injected a second round of dsRNA 1 wk after the first round. For low-dose RNAi experiments, we injected dsRNA just 1 d before amputation. For homeostasis experiments, we followed the same protocol of injection but without amputation. Details are provided in SI Materials and Methods.
Motility Analysis.
Details are provided in SI Materials and Methods.
Immunostaining and Confocal Imaging.
Immunostaining was carried out as described (22). Details are provided in SI Materials and Methods.
Electron Microscopy.
SEM and TEM were carried out essentially as described (35). Details are provided in SI Materials and Methods.
Acknowledgments
We thank all members of the F. Cebrià and E. Saló groups, especially M. D. Molina and M. Iglesias, J. M. Martín-Durán, and A. Repiso, for discussion and suggestions; F. Cebrià for providing Cintillo and anti-Neuropeptide F; M. Iglesias for providing Smed-Gpas clones; H. Orii and Prof. K. Watanabe for providing antiarrestin; M. Handberg-Thorsager for providing the SmedOtp riboprobe; M. Bosch for helping with confocal analysis; N. Cortadellas and A. García for helping with electron microscopy; and I. Patten for advice on English style in a previous version. The monoclonal antibodies anti-SYNORF1, anti–α-tubulin and antiactin JLA20 were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa, Iowa City, IA. This work was supported by Grant BFU2008-01544 from the Ministerio de Educación y Ciencia, Spain (to M.A-C.) and Grant 2009SGR1018 from the Agència de Gestió d'Ajuts Universitaris i de Recerca (to E.S.). M.A.-C. received a Formación de Personal Investigador fellowship from the Ministerio de Educacuón y Ciencia.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. HQ141795 (Smed-Dvl-1), EHQ141796 (Smed-Dvl-2), HQ141793 (Smed-Div), HQ141797 (Smed-Vang-1), and HQ141794 (Smed-Vang-2)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012090108/-/DCSupplemental.
References
- 1.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 2.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao C, Chen YG. Dishevelled: The hub of Wnt signaling. Cell Signal. 2010;22:717–727. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 4.Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139:1056–1068. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 5.Vladar EK, Antic D, Axelrod JD. Planar cell polarity signaling: The developing cell's compass. Cold Spring Harb Perspect Biol. 2009;1:a002964. doi: 10.1101/cshperspect.a002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montcouquiol M, Crenshaw EB, III, Kelley MW. Noncanonical Wnt signaling and neural polarity. Annu Rev Neurosci. 2006;29:363–386. doi: 10.1146/annurev.neuro.29.051605.112933. [DOI] [PubMed] [Google Scholar]
- 7.McNeill H. Planar cell polarity: Keeping hairs straight is not so simple. Cold Spring Harb Perspect Biol. 2010;2:a003376. doi: 10.1101/cshperspect.a003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan TH. Experimental studies of the regeneration of Planaria maculata. Arch Entw Mech Org. 1898;7:364–397. [Google Scholar]
- 10.Saló E. The power of regeneration and the stem-cell kingdom: Freshwater planarians (Platyhelminthes) Bioessays. 2006;28:546–559. doi: 10.1002/bies.20416. [DOI] [PubMed] [Google Scholar]
- 11.Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–1221. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- 12.Gurley KA, Rink JC, Sánchez Alvarado A. β-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen CP, Reddien PW. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- 14.Adell T, Salò E, Boutros M, Bartscherer K. Smed-Evi/Wntless is required for β-catenin-dependent and -independent processes during planarian regeneration. Development. 2009;136:905–910. doi: 10.1242/dev.033761. [DOI] [PubMed] [Google Scholar]
- 15.De Robertis EM. Wnt signaling in axial patterning and regeneration: Lessons from planaria. Sci Signal. 2010;3:pe21. doi: 10.1126/scisignal.3127pe21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chai G, et al. Complete functional segregation of planarian beta-catenin-1 and -2 in mediating Wnt signaling and cell adhesion. J Biol Chem. 2010;285:24120–24130. doi: 10.1074/jbc.M110.113662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umesono Y, Watanabe K, Agata K. Distinct structural domains in the planarian brain defined by the expression of evolutionarily conserved homeobox genes. Dev Genes Evol. 1999;209:31–39. doi: 10.1007/s004270050224. [DOI] [PubMed] [Google Scholar]
- 18.Gurley KA, et al. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol. 2010;347:24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsal M, Pineda D, Saló E. Gtwnt-5 a member of the wnt family expressed in a subpopulation of the nervous system of the planarian Girardia tigrina. Gene Expr Patterns. 2003;3:489–495. doi: 10.1016/s1567-133x(03)00058-9. [DOI] [PubMed] [Google Scholar]
- 20.Cebrià F, Newmark PA. Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development. 2007;134:833–837. doi: 10.1242/dev.02794. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto K, Takeuchi K, Agata K. Neural projections in planarian brain revealed by fluorescent dye tracing. Zoolog Sci. 2005;22:535–546. doi: 10.2108/zsj.22.535. [DOI] [PubMed] [Google Scholar]
- 22.Cebrià F, Guo T, Jopek J, Newmark PA. Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev Biol. 2007;307:394–406. doi: 10.1016/j.ydbio.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bovolenta P, Rodriguez J, Esteve P. Frizzled/RYK mediated signalling in axon guidance. Development. 2006;133:4399–4408. doi: 10.1242/dev.02592. [DOI] [PubMed] [Google Scholar]
- 24.Endo Y, Rubin JS. Wnt signaling and neurite outgrowth: Insights and questions. Cancer Sci. 2007;98:1311–1317. doi: 10.1111/j.1349-7006.2007.00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smales LR, Blankespoor HD. The epidermis and sensory organs of Dugesia tigrina (Turbellaria: Tricladida). A scanning electron microscope study. Cell Tissue Res. 1978;193:35–40. doi: 10.1007/BF00221599. [DOI] [PubMed] [Google Scholar]
- 26.Rompolas P, Patel-King RS, King SM. An outer arm Dynein conformational switch is required for metachronal synchrony of motile cilia in planaria. Mol Biol Cell. 2010;21:3669–3679. doi: 10.1091/mbc.E10-04-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glazer AM, et al. The Zn finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Dev Biol. 2010;337:148–156. doi: 10.1016/j.ydbio.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muñoz R, Moreno M, Oliva C, Orbenes C, Larraín J. Syndecan-4 regulates non-canonical Wnt signalling and is essential for convergent and extension movements in Xenopus embryos. Nat Cell Biol. 2006;8:492–500. doi: 10.1038/ncb1399. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence PA, Casal J, Struhl G. Towards a model of the organisation of planar polarity and pattern in the Drosophila abdomen. Development. 2002;129:2749–2760. doi: 10.1242/dev.129.11.2749. [DOI] [PubMed] [Google Scholar]
- 31.Chen WS, et al. Asymmetric homotypic interactions of the atypical cadherin flamingo mediate intercellular polarity signaling. Cell. 2008;133:1093–1105. doi: 10.1016/j.cell.2008.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh K, Jenny A, Mlodzik M, Sokol SY. Centrosomal localization of Diversin and its relevance to Wnt signaling. J Cell Sci. 2009;122:3791–3798. doi: 10.1242/jcs.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross AJ, et al. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet. 2005;37:1135–1140. doi: 10.1038/ng1644. [DOI] [PubMed] [Google Scholar]
- 34.Molina MD, Saló E, Cebrià F. The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev Biol. 2007;311:79–94. doi: 10.1016/j.ydbio.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 35.Martín-Durán JM, Amaya E, Romero R. Germ layer specification and axial patterning in the embryonic development of the freshwater planarian Schmidtea polychroa. Dev Biol. 2010;340:145–158. doi: 10.1016/j.ydbio.2010.01.018. [DOI] [PubMed] [Google Scholar]