Abstract
Inverted formin 2 (INF2) encodes a member of the diaphanous subfamily of formin proteins. Mutations in INF2 cause human kidney disease characterized by focal and segmental glomerulosclerosis. Disease-causing mutations occur only in the diaphanous inhibitory domain (DID), suggesting specific roles for this domain in the pathogenesis of disease. In a yeast two-hybrid screen, we identified the diaphanous autoregulatory domains (DADs) of the mammalian diaphanous-related formins (mDias) mDia1, mDia2, and mDia 3 as INF2_DID-interacting partners. The mDias are Rho family effectors that regulate actin dynamics. We confirmed in vitro INF2_DID/mDia_DAD binding by biochemical assays, confirmed the in vivo interaction of these protein domains by coimmunoprecipitation, and observed colocalization of INF2 and mDias in glomerular podocytes. We investigated the influence of this INF2_DID/mDia_DAD interaction on mDia mediated actin polymerization and on serum response factor (SRF) activation. We find that the interaction of INF2_DID with mDia_DAD inhibited mDia-mediated, Rho-activated actin polymerization, as well as SRF-responsive gene transcriptional changes. Similar assays using the disease-causing E184K and R218Q mutations in INF2_DID showed a decreased effect on SRF activation and gene transcription. The binding of INF2_DID to mDia_DAD may serve as a negative regulatory mechanism for mDias’ function in actin-dependent cell processes. The effects of disease-causing INF2 mutations suggest an important role for this protein and its interaction with other formins in modulating glomerular podocyte phenotype and function.
Formins are a group of heterogeneous actin nucleating proteins that regulate a variety of cytoskeleton-dependent cellular processes (1–5). Inverted formin 2 (INF2) is a unique member of the formin family. Although it has a domain structure similar to the diaphanous formins [a diaphanous-inhibitory domain (DID), formin homology 1 and 2 domains (FH1 and FH2), and a diaphanous autoregulatory domain (DAD)], it is capable of not only accelerating actin polymerization, but also accelerating actin depolymerization (6). INF2’s depolymerizing activity relies on the combination of its FH2 domain and C terminus, including a DAD that also serves as an actin monomer-binding WH2 domain (Wasp homology domain 2). INF2’s depolymerization activity is regulated via an autoinhibitory interaction of its DID and DAD (6). A similar intramolecular interaction between the DID and DAD also regulates the diaphanous-related formin mDia1, although, in this case, the result is to inhibit its actin polymerizing function (7). In the case of the mammalian diaphanous-related formins (mDias), the autoinhibition of the DID/DAD interaction can be relieved by the competitive binding of the small GTPase RhoA to the DID (8). Recently, GTP-bound Cdc42 has recently been shown to bind to the DID of INF2 and modulate INF2’s function in transcytosis (9).
We recently demonstrated that mutations in INF2 cause a form of autosomal-dominant focal and segmental glomerulosclerosis (FSGS) (10). INF2 is highly expressed in glomerular podocytes, terminally differentiated cells whose characteristics are highly cytoskeleton-dependent. These characteristics include a branched structure that terminates in filopodia-like foot processes, and the maintenance of a specialized cell–cell junction known as the glomerular slit diaphragm (11–13). The importance of the actin cytoskeleton in maintaining the glomerular filtration barrier is supported by the fact that mutations in α-actinin-4, an actin cross-linking protein, cause a similar form of autosomal-dominant FSGS (14). Numerous lines of evidence support the notion that podocytes are highly sensitive to perturbations in their actin cytoskeleton (15). Consistent with this, FSGS-associated mutant forms of INF2 induce distinct patterns of actin polymerization in cultured podocytes compared with WT INF2 (10). The mechanism underlying these differences remains unknown.
To date, all FSGS-associated mutations in INF2 alter highly conserved residues within the DID and result in genetically dominant disease (10). We conjectured that these mutations may result in the disruption of an interaction of the DID with a regulatory protein, and/or perturb the role of the DID of INF2 to regulate its function through its binding to the DAD. Here we report that the mDia family are INF2-interacting proteins. We show that FSGS-associated mutations in INF2 impair its ability to bind mDia_DAD. Furthermore, we find that the interaction of the INF2_DID with the mDia_DAD acts to limit mDia-mediated actin polymerization and serum response factor (SRF)-dependent transcription in a Rho- and CDC42-dependent manner.
Results
Diaphanous Formins Bind the INF2_DID in Yeast Two-Hybrid Assay.
We used a yeast two-hybrid assay to screen a human kidney cDNA library with the DID of INF2 to identify interacting proteins. Captured candidate INF2_DID binding partners included eight independent clones encoding mDia family members (Table 1): diaphanous homologue 1 (DIAPH1, mDia1), diaphanous homologue 2 (DIAPH2, mDia3) and diaphanous homologue 3 (DIAPH3, mDia2). All the captured mDia clones were DAD-containing sequences ranging from DAD alone to those containing formin homology (FH) domains, i.e., FH1-FH2-DAD. For simplicity, we refer to any single member of this family as mDia and the whole group as the mDias.
Table 1.
INF2_DAD captured mDiafamily members by using a yeast two-hybrid system
| Clone | Hits (N = 20) | Protein | Fragment, aa | Domains included |
| 1* | 3 | mDia1 | 1,181–1,262(C terminus) | DAD |
| 2 | 1 | — | 1,146–1,262 | DAD |
| 3 | 9 | — | 1,088–1,262 | FH2-DAD |
| 4 | 1 | — | 1,026–1,262 | FH2-DAD |
| 5 | 2 | — | 1,024–1,262 | FH2-DAD |
| 6* | 1 | — | 596–1,262 | FH1-FH2-DAD |
| 7 | 2 | mDia2 | 749–1,149 (C terminus) | FH1-FH2-DAD |
| 8 | 1 | mDia3 | 637–1,103 (C terminus) | FH2-DAD |
*Shortest and the longest fragments captured for each gene transcript.
INF2_DID Interacts with mDia_DAD.
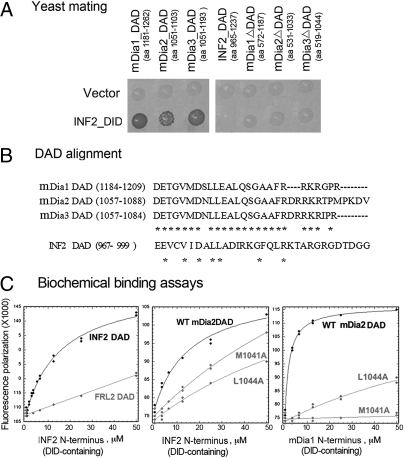
Among the 20 sequence fragments from mDia1, mDia2, or mDia3 captured by using INF2_DID as bait, all clones contained at least the full DAD sequence, with the shortest (clone 1, aa 1,181–1,262; Table 1) lacking any other defined domains. We conjectured that mDia_DAD might serve as a site for binding to INF2_DID. To confirm that this intermolecular DID/DAD binding is the basis for an association between INF2 and the mDias, we compared the binding of INF2_DID to mDia_DAD (including mDia1_DAD, mDia2_DAD, and mDia3_DAD) and FH1-FH2-C terminus constructs of the mDias lacking the DAD (including mDia1_FH1-FH2-CΔDAD, mDia2_FH1-FH2-CΔDAD, and mDia3_FH1-FH2-CΔDAD) in a yeast mating system. As shown in Fig. 1A, the mDia_DAD regions alone were sufficient for binding to INF2_DID, whereas with the deletion of DAD from the longest captured FH1-FH2-C fragments, the interactions were abolished (Fig. 1).
Fig. 1.
INF2 interacts with mDias. (A) The interactions between INF2_DID and mDia_DAD were detected in a yeast mating system. INF2_DID in the pGBKT7 vector was transformed into the AH109 yeast strain, with the empty vector as a negative control. Formin fragments mDia1_DAD, mDia2_DAD, mDia3_DAD, mDia1 FH1-FH2-CΔDAD, mDia2 FH1-FH2-CΔDAD, and mDia3 FH1-FH2-CΔDAD were cloned into pACT2 and transformed into the Y187 yeast strain. The transformants were mated and dotted on QDO agar plates. The interactions between INF2_DID and mDia truncations are displayed in the bottom row. (B) Sequence alignment of DAD in mDias and INF2. (C) DID/DAD binding measured by fluorescence polarization binding experiments by using fluorescein-labeled mDia2_DAD [WT and mutants, M1041A and L1044A], INF2-DAD or FRL2-DAD, and DID-containing N-terminal (NT) constructs of mDia1 or INF2.
Given the previously demonstrated INF2_DID/DAD intramolecular interaction (6), we were surprised that we did not find any INF2_DAD sequences among the interactors identified in our screen. To explore this further, we compared the DAD sequences of the diaphanous formins with INF2_DAD by using ClustalW software. As shown in Fig. 1B, mDia1, mDia2, and mDia3 share highly similar DAD sequences. By contrast, the INF2_DAD shows considerably less homology, suggesting that our inability to detect INF2_DAD in our screen might reflect a difference between the strength of the interactions between INF2_DID and mDia_DAD compared with INF2_DAD. To exclude the possibility that the low abundance of INF2 in the kidney cDNA library used might have led to its absence from our list of captured proteins, we expressed INF2_DAD (aa 965–1,237) in yeast, and specifically examined its interaction with INF2 DID in a yeast mating system (Fig. 1A). A specific interaction between INF2 DID and DAD was still undetectable in this system.
We next used a fluorescence polarization assay to measure in vitro DID/DAD binding directly. As shown in Fig. 1C and Table 2, mDia1_DID bound mDia2_DAD [a representative mDia_DAD (16)] with a Kd of approximately 2 μM, whereas INF2_DID bound INF2_DAD weakly, with a Kd of approximately 16 μM. INF2_DID has a similar weak affinity for mDia2_DAD (Kd, 15.8 μM). INF2_DID displays some specificity for mDia2_DAD, as it binds much more weakly to the DAD region of another diaphanous formin, FRL2 (Formin-like 3, FMNL3). We used point mutants in mDia_DAD to help understand the difference between the various DID/DAD interactions. The M1041 and L1044 residues in mDia_DAD have been shown to be critical in mediating the homodimeric DID/DAD interaction of the mDias (16). Although mutating these residues strongly inhibits (L1044A) or abolishes (M1041A) the interaction of mDia2_DAD with mDia1_DID, it had a less pronounced effect on the interaction with INF2_DID, suggesting that the INF2_DID/mDia_DAD interaction may use additional contact sites.
Table 2.
Kd values for mDia1 N terminus and INF2 N terminus for DAD peptides
| Peptide | INF2 N terminus, μM | mDia1 N terminus, μM |
| mDia2_DAD WT | 15.8 | 2.1 |
| mDia2_DAD M1041A | 38.0 (2.4-fold vs. WT) | Unmeasurable* |
| mDia2_DAD L1044A | 69.0 (4.4-fold vs. WT) | 84.2 (40-fold vs. WT) |
| INF2_DAD | 16.2 | — |
| FRL2_DAD | Unmeasurable* | — |
*No measurable anisotropy increase at 50 mM of the N-terminal construct.
Cellular Interaction of INF2 and mDias Is Regulated by Rho Activity.
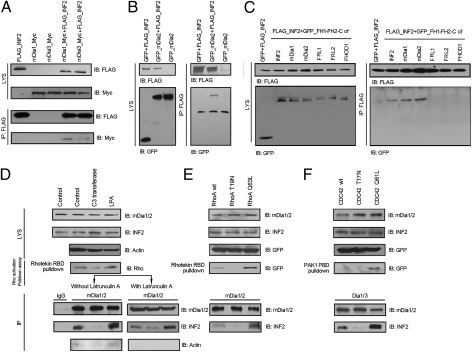
We performed coimmunoprecipitation (co-IP) experiments to determine whether INF2_DID/mDia_DAD binding reflected an in vivo interaction of mDias and INF2. In 293T cells in which FLAG-INF2 and either mDia1-Myc or mDia3-Myc were expressed, Myc-tagged mDias could be precipitated by anti-FLAG antibody (Fig. 2A). In 293T cells in which FLAG-INF2 and GFP-mDia2 were expressed, GFP-mDia2 was also precipitated by anti-FLAG antibody (Fig. 2B). To address the specificity of the intermolecular INF2/mDia interaction, we compared the binding of the INF2_DID with FH1-FH2-DAD constructs from mDia1, mDia2, INF2, and three other formins: FRL1 (Formin-like 1, FMNL1), FRL2 (FMNL3), and FHOD1. As shown in Fig. 2C, we demonstrated interactions of INF2 with the DAD-containing sequence from INF2, mDia1, and mDia2, but not with the DAD from the other three formins tested. We also demonstrated an interaction of endogenous INF2 and mDia in untransfected cells. As shown in Fig. 2D, the endogenous INF2 was precipitated by using an antibody against mDia1/2.
Fig. 2.
(A and B) In vivo interaction of INF2 and mDias detected by co-IP. In 293T cells coexpressing FLAG-INF2 and mDias (mDia1-Myc or mDia3-Myc in A, GFP-mDia2 in B), mDias were immunoprecipitated (IP) by using anti-FLAG. (C) In 293T cells coexpressing INF2-FLAG and GFP-FH1-FH2-C constructs of formins, the intermolecular interaction was detected of INF2 with mDia1, mDia2, or INF2 itself, but not with FRL1, FRL2, or FHOD1. (D–F) The endogenous interaction of INF2 and mDias in response to intracellular Rho/CDC42 activity. 293T cells pretreated with LPA (25 μM) or cell permeable C3 transferase (2 μg/ml) (D), or transfected with plasmids encoding different forms of RhoA (wt, Q63L, or T19N in E) or CDC42 (wt, Q61L, or T17N in F) . The cell lysates (LYS) were immunoprecipitated with anti-mDia1/2 and then immunoblotted with anti-INF2. To exclude the interference of actin in the binding assay, co-IP was performed in the presence/absence of 10 M of Latrunculin A, and the blots were reprobed with anti-actin (D). Rho or CDC42 activity was measured using Rhotekin RBD or PAK1PBD pull down (D–F).
In parallel, we examined the response of endogenous binding of INF2 and mDia to manipulations of intracellular Rho activity. As shown in Fig. 2D, the INF2/mDia interaction was enhanced by lysophosphatidic acid (LPA), a potent Rho activator, but abolished by C3 transferase, a selective Rho inhibitor. To determine whether F-actin mediated an indirect association of INF2 and the mDias, we performed co-IP in the presence of latrunculin A (LatA), which disrupts actin polymerization. The effects of the pharmacological manipulation of Rho on the INF2/mDia interaction were similar in the presence or absence of LatA. By reprobing the blots, we found very little actin precipitating with the anti-mDia antibody (Fig. 2D). In a complementary experiment (Fig. 2 E and F), the INF2/mDia binding increased in response to RhoA or CDC42 activation induced by the overexpression of constitutively active RhoA (Q63L) or CDC42 (Q61L). By contrast, the interaction was decreased with the inhibition of RhoA or CDC42 by the expression of dominant-negative RhoA (T19N) or CDC42 (T17N).
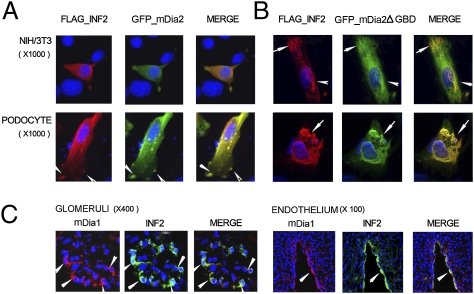
Immunofluorescence staining demonstrated colocalization of the expressed INF2 and mDia in cultured podocytes as well as in NIH 3T3 cells (Fig. 3A). Staining for both proteins in mouse kidney showed colocalization of INF2 and mDias within podocytes as well as in vessel endothelium (Fig. 3C). We also detected the prominent expression of mDia along the brush border of proximal tubular cells where the expression of INF2 was absent. We noted that in cells cotransfected with mDia2ΔGBD (GFP-FH1-FH2-C_mDia2) and INF2, INF2 showed better colocalization in a fiber-like pattern compared with the full mDia2 (Fig. 3B).
Fig. 3.
Colocalization of INF2 and mDias in vivo and in vitro. (A) NIH/3T3 cells and podocytes were cotransfected with plasmids encoding FLAG-INF2 and GFP-mDia2. These proteins were detected by using anti-FLAG and anti-GFP antibodies (red, FLAG; green, GFP; yellow, merged). The merged signal was distributed in filopodia-like structures in podocytes (A, arrows). (B) NIH/3T3 cells and podocytes were cotransfected with plasmids encoding FLAG-INF2 and GFP-FH1-FH2-C_mDia2 (GFP-mDia2ΔGBD). The merged signal localized to filament-like structures (B, arrows). (C) In vivo colocalization of endogenous INF2 and mDia1 in mouse kidney. A frozen section (4 μm) was double-stained with rabbit anti-INF2 and chicken anti-mDia1. The merged signal was distributed in podocytes and in vessel endothelium (arrows).
INF2/mDia DID/DAD Interaction Interferes with the Rho/F-Actin/SRF Pathway Mediated by mDias.
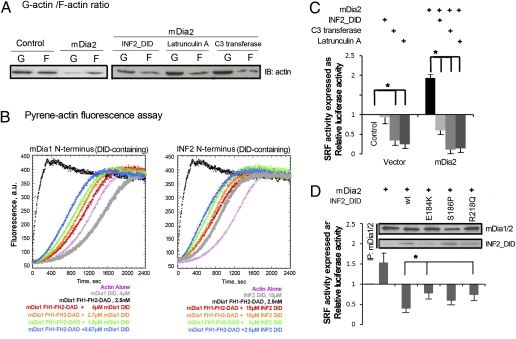
The mDias are Rho effectors that mediate Rho activated actin polymerization and the subsequent SRF signaling (1, 17). To investigate if mDia activation of F-actin formation and SRF activation are influenced by the occupation of mDia_DAD by INF2_DID, we tested the influence of INF2_DID on the polymerization activity of mDia1_FH1-FH2-DAD in a pyrene-actin fluorescence-based assay. As shown in Fig. 4B, INF2_DID inhibited the actin nucleating activity of mDia1 FH1-FH2-DAD in a dose-dependent manner. Its effect is weaker than that of the mDia1_DID construct, likely a reflection of the lower affinity of the INF2_DID/mDia_DAD interaction compared with the mDia_DID/mDia_DAD interaction. We used a luciferase reporter system to assay the effect of INF2_DID on mDia-induced SRF activity in 293T cells. Overexpression of mDia2 resulted in a decrease in the G-actin/F-actin ratio (Fig. 4A), reflecting increased incorporation of actin monomers into F-actin. As shown in Fig. 4C, mDia induced SRF activation was attenuated by the pretreatment of Rho-inhibiting exoenzyme C3 transferase or the actin depolymerizer LatA, and the coexpression with INF2_DID produced a comparable attenuation of mDia2-induced SRF activation. These inhibitory effects coincided with an increased G-actin/F-actin ratio, compared with the cells in which mDia2 is overexpressed alone (Fig. 4A). The inhibition of INF2_DID on mDia2-induced SRF activation was altered by disease-causing point mutations. As shown in Fig. 4D, mDia2-induced SRF activation was inhibited by WT INF2_DID and the mutants E184K, S186P, and R218Q. The inhibition of SRF activation in the presence of E184K or R218Q mutants was weaker than the inhibition observed with WT INF2_DID. These differences roughly corresponded to the strength of the interaction of INF2_DID mutants to mDia.
Fig. 4.
INF2_DID interferes in the mDia2-mediated Rho/F-actin/SRF pathway. G-actin/F-actin distribution (A), actin polymerization assay (B), and SRF activation (C and D) were measured in 293T cells with overexpression of mDia2, with and without coexpression of INF2_DID, with cells transfected with the empty plasmid serving as a control. (A) The ratio of G-actin and F-actin representing alterations in actin transfer from G to F pool during the change of experimental conditions. Overexpression of mDia2 induced F-actin accumulation, which was reversed by INF2_DID coexpression, LatA (10 μm), or C3 transferase (2 μg/mL). (B) Pyrene-actin polymerization assays with 4 μM 5% pyrene-labeled actin monomers and 2.5 nM of mDia1_FH1-FH2-DAD in the absence or presence of mDia1_DID or INF2_DID (0–4 μM). mDia1_DID and INF2_DID inhibited mDia1_FH1-FH2-DAD–mediated actin polymerization. The inhibitory effect of INF2_DID was weaker than that of mDia1_DID. (C and D) SRF activity was measured by dual luciferase reporter gene assay and the result was expressed as relative activity normalized as fold of control. Data are expressed as mean ± SD from three independent experiments. Means of relative luciferase activity were compared in each group by one-way ANOVA. Dunnett test was performed for group INF2_DID, C3 transferase, or LatA examined in comparison with the control or mDia2 group. Background SRF activity was inhibited by LatA or C3 transferase (*P < 0.05 vs. control), but not by INF2_DID. mDia2-induced SRF activation was inhibited by INF2_DID coexpression, C3 transferase, or LatA (*P < 0.05 vs. mDia2 only). (D) SRF activation was compared in 293T cells with the coexpression of mDia2 and INF2_DID (WT or E184K, S186P, R218Q mutants). The cell lysates were precipitated with anti-mDia1/2. The interaction of mDia2 and INF2 was eliminated by E184K or R218Q mutations. mDia2-induced SRF activation was antagonized by INF2_DID WT, E184K, S186, and R218Q mutants. ΔSRF (SRFmDia2+INF2_DID-SRFmDia2) was compared by one-way ANOVA. Dunnett test was performed to compare the inhibitory effects of the mutants versus that of WT. mDia2-induced SRF by E184K or R218Q was significantly weaker than that by WT (*P < 0.05 vs. WT).
To investigate the antagonism of INF2_DID on mDia-induced SRF activation and the downstream influence on the biological phenotype of disease-relevant kidney cells, we evaluated the expression of several known SRF regulated genes by RT-PCR. By searching the Gene Expression Omnibus database, we selected 11 genes from dataset GDS1486 with transcript levels reported to be up-regulated or down-regulated in SRF-null neonatal cardiomyocytes (18). ACTN4, CTGF, CYR61, MYH9, MYL9, and TPM1 are previously reported SRF-regulated genes (19). The other five genes assayed—ANGPTL1, FAS, HIF1A, SMAD7, and Trap1, which showed increased expression in SRF-null cardiomyocytes—served here as SRF down-regulated genes. As shown in Fig. S1, consistent with the increased SRF activity measured by luciferase reporter gene assay, the transcripts of the SRF up-regulated genes (MYH9, MYL9, ACTN4, CYR61, and TPM1) were increased to various degrees by mDia2, and this increase was reversed by the coexpression of INF2_DID. Similarly, the mDia2-induced down-regulation of ANGPTL1, HIF1A, SMAD7, and TRAP1 was also diminished by INF2_DID. Thus, INF2_DID, presumably through the INF2_DID/mDia2_DAD interaction, antagonizes the effect of mDia2 on SRF-regulated genes. In addition, we found that the effects of INF2_DID on the expression of SRF-regulated genes were diminished by the disease-causing mutations, particularly E184K and R218Q, consistent with the variation in SRF activity seen in the luciferase assays (Fig. 4D).
INF2 Mutants Interfere with Interaction Between INF2 and mDias.
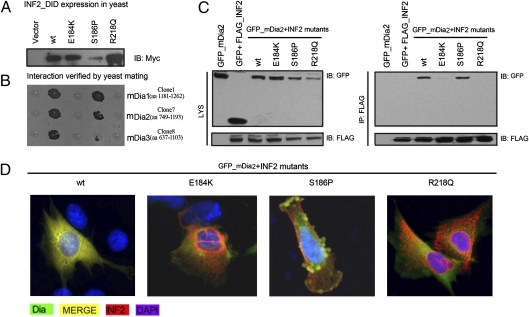
We investigated whether the presence of these mutations in INF2 would alter its ability to interact to the mDias. We compared the heterogeneous interaction of INF2_DID mutants and mDia_DAD containing fragments in a yeast mating system. As shown in Fig. 5B, the S186P mutation did not abolish the interaction, whereas INF2_DID carrying the E184K or R218Q mutation was unable to bind to mDia fragments. To further confirm the possible structure/function relationship of these disease-causing mutations, we subcloned the full-length INF2, including WT, E184K, S186P, and R218Q mutants, into a FLAG tagging vector. As shown in Fig. 5C, in 293T cells cotransfected with FLAG-INF2 constructs and GFP-mDia2, mDia2 coprecipitated with WT and S186P mutant INF2, but not with E184K or R218Q mutant INF2. Overexpressed mDia2 showed better colocalization with WT and S186P mutant INF2 compared with E184K and R218Q INF2 (Fig. 5D; Fig. S2 shows additional cell images).
Fig. 5.
Effect of disease-causing INF2 mutations on mDia interaction. (A) The expression of WT and mutants of INF2_DID in AH109 was detected by immunoblotting with an antibody against Myc-Tag. The plasmids (pACT2) carrying candidate proteins were transformed Y187 yeast strain. (B) The transformants were mated and dotted on QDO agar plates. The interactions between INF2_DID (WT) and the candidate protein fragments of mDia1, mDia2, and mDia3 were tested (column labeled WT), as were the interactions of INF_DID mutants (E184K, S186P, and R218Q) with the captured fragments. (C) 293T cells were cotransfected with plasmids encoding GFP-mDia2 and FLAG-INF2 mutants. The cell lysates were subjected to immunoprecipitation with anti-FLAG antibody. Immunoprecipitation of INF2 with the E184K and R218Q mutants did not bring down mDia2, in contrast to WT and S186P mutants. (D) Podocytes were cotransfected with plasmids encoding GFP-mDia2 and FLAG-INF2 mutants and subjected to double staining of FLAG and GFP-tagged proteins (red, FLAG; green, GFP; yellow, merged).
Discussion
By using a yeast two-hybrid screen for INF2_DID interacting proteins, we captured sequences containing the DAD region of several diaphanous-related formins. The shortest common interacting fragment mapped to the DAD region. This suggested that the mDia DAD region is a site for recognition, binding, and regulation by INF2_DID. We made truncations of mDia_DAD and mDiaΔDAD and compared their ability to interact with INF2_DID in a yeast mating system. mDia_DAD alone was sufficient for interaction with INF2_DID. With the deletion of DAD, the interactions between INF2_DID and the DAD-containing sequences of mDia were disrupted, indicating that mDia_DAD is required for the mDia/INF2 interaction. We did not observe INF2 DID/DAD binding in our yeast two-hybrid system, despite the fact that INF2_DID shows similar binding affinity for INF2_DAD and mDia_DAD in vitro. Although we do not have a definite explanation for this difference, various aspects of cellular context (e.g., the binding of actin to the INF2_DAD/WH2) are likely responsible. Interestingly, mutations that disrupt the mDia_DID/mDia_DAD interaction showed less pronounced effects on the INF2_DID/mDia_DAD interaction, suggesting that the binding sites may be different, with differing biological roles and regulatory mechanisms.
The ability of INF2 to interact with other formins appears specific to the mDia subfamily, as DAD-containing constructs of other formins FRL1, FRL2, and FHOD1 did not interact with INF2 in co-IP experiments. Copeland et al. demonstrated heterodimerization between close members of mDias based on DID/DAD interactions and demonstrated that this interaction enabled them to function as negative regulators of each other (20). Our investigation suggests a similar intermolecular interaction between more distantly related subgroups of formins. Studies using kidney disease-causing INF2_DID mutations suggest that disruption of this interaction is a possible contributor to this form of human disease.
We demonstrated an in vivo interaction between the full-length proteins, with colocalization of INF2 and mDias in podocytes. Our in vitro fluorescence polarization assays suggest that the interaction of mDia_DAD with mDia_DID is greater than the interaction of INF2_DID with mDia_DAD. This is consistent with the speculation that the INF2/mDia transinteraction is not the predominant state in vivo, and that the mDia intramolecular DID/DAD interaction is favored.
Immunofluorescence staining showed greater colocalization of INF2 with a GTPase binding domain (GBD) absent form of mDia2 (mDia2ΔGBD), suggesting that the presence of this GBD might interfere with the accessibility of mDia2 for INF2_DID. In the model depicted in Fig. S3, the binding of active Rho (RhoA-GTP) to the N-terminal GBD of mDia family members inhibits the DID/DAD interaction, leading to disautoinhibition (7, 16). INF2 may have more access to the mDias in the disinhibited conformation, favoring the INF2_DID/mDia_DAD interaction. To test this, we perturbed intracellular Rho activity by using chemical compounds and overexpression of plasmids encoding Rho (constitutively active or dominant-negative) and found that the INF2/mDia association is regulated by Rho activation. We also found that the interaction of INF2 and mDias was regulated by CDC42 activation.
The interaction of INF2 with mDias may allow these molecules to function as regulators of each other. As mDias are effectors of Rho in regulating cytoskeleton dynamics, we investigated the INF2/mDia interaction on actin polymerization and on SRF-regulated gene transcription. mDias are essential effectors mediating Rho-induced activation of the megakaryoblastic leukemia 1/SRF pathway (Fig. S3). In this model, the autoinhibited state of mDia is relieved by binding to active RhoA, and then mediates actin polymerization with the exposure of the functional FH2 domain. mDia activation promotes G-actin incorporating into F-actin, releasing megakaryoblastic leukemia 1, which then binds SRF and activates gene transcription.
In our model, the occupation of mDia functional sites by the heterogeneous INF2_DID/mDia_DAD interaction interferes with actin polymerization and consequent SRF activation. Thus, this INF2_DID/mDia_DAD interaction serves as a potential negative-regulatory mechanism for Rho activation of mDia. This negative regulation affects SRF activation. Several of these SRF-regulated genes have known roles in the development of FSGS and/or podocyte biology (21, 22). Here we found that the enhanced SRF activity and SRF-targeted gene transcription induced by mDia were largely reversed by the coexpression of INF2_DID, confirming that the heterogeneous DID/DAD interaction contributes to the negative modulation of Rho/mDia/SRF pathway activation and the consequent phenotype of target cells.
Two disease-causing mutations in INF2_DID, E184K and R218Q (10), disrupted the interaction with mDia_DAD binding as detected by yeast mating and co-IP. We also found that the inhibitory effects of INF2_DID on mDia-induced SRF activity and SRF-regulated gene transcription were disrupted by these mutations. E184 and R218 thus appear to be critical for the interaction between the two domains, and the pathogenic significance of these mutants may lie in their impaired ability to interact with and regulate mDia-mediated pathways.
In conclusion, we have shown that binding of INF2_DID to mDia_DAD binding mediates an association between INF2 and mDias. The in vivo interaction of these proteins depends on Rho GTPase signaling. In podocytes, Rho signaling may be triggered by various extracellular stimuli. When the autoinhibitory effect of of mDia_DID on mDia_DAD is relieved by active Rho, INF2 is able to interact with mDia_DAD. This regulatory activity inhibits Rho/mDia-mediated actin polymerization, and thus modulates the cell's responses downstream of Rho pathway activation. Disease-causing mutations in INF2_DID can disrupt this INF2_DID/mDia_DAD interaction, alter the regulation of the mDias, and lead to a disturbance in actin dynamics. This leads also to altered SRF activity and SRF responsive gene transcription and, perhaps, dysregulated podocyte behavior. The full physiological and the pathophysiological significance of the INF2/mDia interaction in vivo is not yet clear. This interaction between INF2 and mDias may provide a mechanism for the subtle regulation of formin activity in cytoskeletal regulation. In the glomerular podocyte, where INF2 mutations lead to an overt human phenotype, this INF2-mediated cytoskeletal regulation may be particularly important.
Materials and Methods
Please refer to the SI Methods for detailed materials and methods.
Plasmids.
Plasmid constructs were generated as described in SI Methods.
Protein Preparation and Fluorescence Polarization.
N-terminal mDia1 [mouse, aa 1 -548(DID)], FH1-FH2-DAD, and IND2_DID were expressed in bacteria and purified (5, 7). The mDia2-DAD peptide (acetyl-CDETGVMDSLLEALQSGAAFRRKRG-amide) was synthesized and labeled with fluorescein-maleimide as described previously (7). Proteins were dialyzed into binding buffer (10 mM imidazole-HCl, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 0.5 mM DTT,and 0.02% polyethylene glycoldodecyl ether) before measurements. DID/DAD binding was measured by fluorescence polarization using fluorescein-labeled mDia2_DAD [WT and mutants, M1041A and L1044A], INF2-DAD or FRL2-DAD, and DID-containing N-terminal (NT) constructs of mDia1 or INF2.
Actin Polymerization by Fluorescence Spectroscopy.
Pyrene-actin fluorescence assay was performed as described previously (7).
Dual-Luciferase Reporter Gene Assays.
SRF activity was detected by a dual-luciferase reporter gene assay (Promega) as detailed in SI Methods.
Cellular G-Actin/F-Actin Assay.
Changes in the G-actin/F-actin ratio in response to expression of proteins were studied by using a G-actin/F-actin in vivo assay kit (Cytoskeleton).
Rho/CDC42 Activation.
The Rho/CDC42 activation was detected by pull-down assay by using Rho and CDC42activation kits (Upstate Biotechnology) as detailed in SI Methods.
Acknowledgments
This work was supported by Nature Science Foundation of China Grant 81000282/H0502, Research Fund for Young Teachers at Shanghai University Grant JDY08061 (to H.S.), and National Institutes of Health Grants DK080947 (to J.S.S.), K12HD052896 (to E.J.B.), GM069818 (to H.N.H.), and DK088826 (to M.R.P. and H.N.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017010108/-/DCSupplemental.
References
- 1.Krebs A, Rothkegel M, Klar M, Jockusch BM. Characterization of functional domains of mDia1, a link between the small GTPase Rho and the actin cytoskeleton. J Cell Sci. 2001;114:3663–3672. doi: 10.1242/jcs.114.20.3663. [DOI] [PubMed] [Google Scholar]
- 2.Gupton SL, Eisenmann K, Alberts AS, Waterman-Storer CM. mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J Cell Sci. 2007;120:3475–3487. doi: 10.1242/jcs.006049. [DOI] [PubMed] [Google Scholar]
- 3.Bindschadler M, McGrath JL. Formin’ new ideas about actin filament generation. Proc Natl Acad Sci USA. 2004;101:14685–14686. doi: 10.1073/pnas.0406317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang C, et al. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN. INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci. 2009;122:1430–1440. doi: 10.1242/jcs.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chhabra ES, Higgs HN. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281:26754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- 7.Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280:6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- 8.Rose R, et al. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- 9.Madrid R, et al. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell. 2010;18:814–827. doi: 10.1016/j.devcel.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Brown EJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42:72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang PY, He JC. Signaling in regulation of podocyte phenotypes. Nephron, Physiol. 2009;111:9–15. doi: 10.1159/000191075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Agati VD. The spectrum of focal segmental glomerulosclerosis: New insights. Curr Opin Nephrol Hypertens. 2008;17:271–281. doi: 10.1097/MNH.0b013e3282f94a96. [DOI] [PubMed] [Google Scholar]
- 13.Barisoni L, Mundel P. Podocyte biology and the emerging understanding of podocyte diseases. Am J Nephrol. 2003;23:353–360. doi: 10.1159/000072917. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan JM, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 15.Faul C, Asanuma K, Yanagida-Asanuma E, Kim K, Mundel P. Actin up: Regulation of podocyte structure and function by components of the actin cytoskeleton. Trends Cell Biol. 2007;17:428–437. doi: 10.1016/j.tcb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Wallar BJ, et al. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J Biol Chem. 2006;281:4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- 17.Copeland JW, Treisman R. The diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization. Mol Biol Cell. 2002;13:4088–4099. doi: 10.1091/mbc.02-06-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balza RO, Jr., Misra RP. Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J Biol Chem. 2006;281:6498–6510. doi: 10.1074/jbc.M509487200. [DOI] [PubMed] [Google Scholar]
- 19.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Copeland SJ, et al. The diaphanous inhibitory domain/diaphanous autoregulatory domain interaction is able to mediate heterodimerization between mDia1 and mDia2. J Biol Chem. 2007;282:30120–30130. doi: 10.1074/jbc.M703834200. [DOI] [PubMed] [Google Scholar]
- 21.Kunishima S, Saito H. Advances in the understanding of MYH9 disorders. Curr Opin Hematol. 2010;17:405–410. doi: 10.1097/MOH.0b013e32833c069c. [DOI] [PubMed] [Google Scholar]
- 22.Sawai K, et al. Expression of CCN1 (CYR61) in developing, normal, and diseased human kidney. Am J Physiol Renal Physiol. 2007;293:F1363–F1372. doi: 10.1152/ajprenal.00205.2007. [DOI] [PubMed] [Google Scholar]